Abstract
The nuclear envelope consists of 2 membranes separated by 30–50 nm, but how the 2 membranes are evenly spaced has been an open question in the field. Nuclear envelope bridges composed of inner nuclear membrane SUN proteins and outer nuclear membrane KASH proteins have been proposed to set and regulate nuclear envelope spacing. We tested this hypothesis directly by examining nuclear envelope spacing in Caenorhabditis elegans animals lacking UNC-84, the sole somatic SUN protein. SUN/KASH bridges are not required to maintain even nuclear envelope spacing in most tissues. However, UNC-84 is required for even spacing in body wall muscle nuclei. Shortening UNC-84 by 300 amino acids did not narrow the nuclear envelope space. While SUN proteins may play a role in maintaining nuclear envelope spacing in cells experiencing forces, our data suggest they are dispensable in most cells.
Introduction
The nucleus is enclosed by a double membrane structure known as the nuclear envelope. The two membranes of the nuclear envelope, the inner nuclear membrane (INM) and outer nuclear membrane (ONM), are separated by a uniform distance of 30–50 nm.Citation1 To facilitate the import and export of cargos, the INM and ONM meet at junctions containing nuclear pore complexes (NPCs). Furthermore, the ONM is contiguous with the ER. In addition to its role in containing the genome, the nuclear envelope has been implicated in cell signaling and regulation of many important cellular functions, such as localization of nuclear proteins, organization of heterochromatin, and DNA repair.Citation2 The role of nuclear envelope defects in a wide-ranging collection of human diseasesCitation2 is drawing increased attention to this complex cellular structure.
In addition to NPCs, connections between the INM and ONM are also formed by linkers of nucleoskeleton and cytoskeleton (LINC) complexes, consisting of Sad1p, UNC-84 (SUN) proteins in the INM interacting with Klarsicht, ANC-1, and Syne homology (KASH) proteins (also known as nesprins) in the perinuclear space (PNS).Citation3,4 This protein complex is evolutionarily conserved from yeasts to plants and humans. SUN and KASH proteins interact via conserved domains at their C-termini in the PNS.Citation5 SUN proteins interact with lamins in the nucleoplasmCitation6,7 and KASH proteins recruit cytoskeletal components to the ONM.Citation8 In this way, LINC complexes stabilize the nuclear envelope against cytoplasmic forces and facilitate nuclear positioning in a variety of cellular processes, including cell division, establishment of cellular polarity, fertilization, cellular migration, and differentiation. Furthermore, LINC complexes have been hypothesized to maintain the even spacing between the 2 membranes of the nuclear envelope.Citation9-11
In Caenorhabditis elegans somatic cells, the SUN protein UNC-84 interacts with the KASH protein UNC-83, which recruits microtubule motors kinesin and dynein to facilitate nuclear migration.Citation12,13 In the early embryo, hypodermal precursors line up in 2 rows across the dorsal midline. As cells intercalate and elongate to form one row of 16 cells, the nuclei migrate contra-laterally to the opposite side of the dorsal midline.Citation14 The hypodermal cells eventually fuse, and in the resulting syncytia, hypodermal nuclei are anchored evenly throughout. In unc-84 or unc-83 mutant embryos, the elongation and intercalation of cells proceeds normally, but the nuclei fail to move from their initial positions.Citation13,15 As the embryo continues to develop, mutant nuclei that fail to migrate are passively pushed toward the dorsal midline by underlying muscle cell migrations and can be observed by DIC microscopy in the dorsal cord of the L1 larva.Citation15 In the adult hypodermal syncytia, UNC-84 interacts with ANC-1, which interacts with actin to anchor nuclei .Citation16 After cell fusion, in unc-84 or anc-1 mutants, the nuclei are unanchored and are free to drift throughout the syncytia, often clustering in groups.Citation16,17
SUN Proteins and Nuclear Envelope Spacing
Most SUN proteins are large, with more than half of the protein residing in the PNS. The C-terminal ∼200 amino acids contain the conserved SUN domain, which trimerizes into a cloverleaf structure with the N-terminal stalk formed by a right-handed trimer coiled-coil.Citation5,18 While the structure of the linker domain between the transmembrane domain and the trimeric coil is unknown, it has been proposed that the trimeric coiled-coil continuously extends to the transmembrane domain, forming a rod approximately 45 nm long. Such an extended coil structure would be suitable to span the space between the ONM and the INM and serve as a "molecular ruler" to regulate even spacing of the nuclear envelopeCitation10,11 (). Interestingly, several divergent SUN proteins, including human SUN3-5,Citation19-21 which are restricted to the testis, and C. elegans SUN-1,Citation22,23 which is restricted to the germ line, are predicted to have much smaller luminal domains than those of their somatic counterparts. The prediction is that tissues expressing these SUN proteins would display narrower nuclear envelope spacing (), but nuclear envelope spacing in those tissues has not been directly examined.
Figure 1. SUN proteins are predicted to regulate nuclear envelope spacing. In this figure, SUN proteins are depicted in the inner nuclear membrane (INM) with their nucleoplasmic domain in yellow and their conserved SUN domain in red. SUN domains bind KASH proteins (blue) in the outer nuclear membrane (ONM). NPC are nuclear pore complexes where the inner and outer nuclear membranes are connected. (A) The linker domains of human SUN1/2 and C. elegans UNC-84, between the trans-membrane span and the SUN domain, are predicted to form trimeric rods that span the 40–50 nm distance between the inner and outer nuclear membranes. (B) Shorter SUN proteins (human SUN3–5 and C. elegans SUN-1) are predicted to have shorter luminal domains and, as a result, narrower nuclear envelope spaces. (C) In the absence of LINC complexes, lack of connection of the cytoskeleton to the nucleoskeleton is expected to cause the ONM to separate from the INM.Citation9
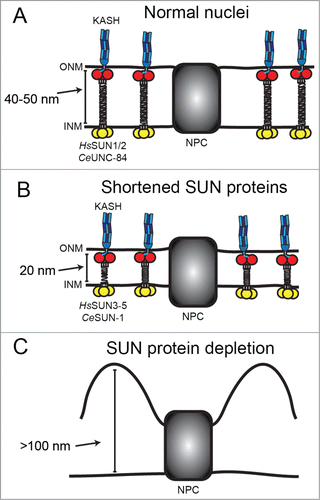
The hypothesis that LINC complexes may serve as molecular rulers for nuclear envelope spacing was primarily based on electron micrographs of HeLa cells either depleted for SUN1 and SUN2 or expressing a soluble dominant-negative SUN domain fragment, showing large distortions of the PNS and ∼100 nm separation of the ONM away from the INMCitation9 (). However, HeLa cells in culture are in a very different environment than most nuclei in vivo. Because they make extracellular contacts to a hard surface, the cytoskeleton is under increased strain. Stresses at the cell surface can cause long-range force propagation, extending to the nucleus and beyond.Citation24 As a result, nuclei in cell culture are flattened, like a pancake.Citation25 In live tissue, nuclei are usually more spherical and are presumably not subject to the same intracellular forces.
The C. elegans system is particularly suited to address this question directly. UNC-84 is the only SUN protein expressed in somatic tissue, yet null mutants are viable and fertile. In unc-84(null) tissues, both UNC-83 and ANC-1 fail to localize, which allowed us to examine nuclei completely lacking LINC complexes.Citation12,16 High pressure freezing and electron microscopy techniques are well established in C. elegans,Citation26 which allowed us to expand our observations to a variety of tissues and developmental stages. The defects of the unc-84(null) mutant can be fully rescued by an UNC-84 transgene,Citation12 which allowed us to express truncated forms of UNC-84 as the sole somatic SUN protein in an otherwise null background and to examine their effects on nuclear migration. We previously used this technique to locate the transmembrane domain in UNC-84, as well as multiple sorting motifs in the N-terminus required for INM localization.Citation27
UNC-84 Maintains the NE Spacing in Force-bearing Cells but is Dispensable in other Tissues
The absence of LINC complexes caused significant deformations of the nuclear envelope in HeLa cell culture, but whether a similar phenotype would be observed in an animal lacking LINC complexes remained an open question. The hypothesis that LINC complexes serve as molecular rulers suggests that a SUN-less animal would display gross nuclear envelope defects in most or all tissues throughout development. We therefore carried out a series of experiments to characterize the role of the SUN protein UNC-84 in the architecture of the nuclear envelope.Citation28 We began by examining pre-morphogenesis embryos, near the stage where UNC-84 and UNC-83 function together to move nuclei contra-laterally across the dorsal midline of the developing hypodermis. In most wild type nuclei, the PNS is consistently 30–50 nm in width, in agreement with previously published results.Citation29 Surprisingly, nuclei from the null mutant unc-84(n369) strain are not significantly different than the N2 wild type laboratory strain. The PNS of most unc-84 mutant nuclei are evenly spaced throughout, and are not significantly wider than wild type. Similarly, in the first larval stage, most nuclei, including those of the pharynx and hypodermis, have normal nuclear envelope spacing in both wild type and unc-84(n369) animals (). Therefore, in contrast to HeLa cells, where an absence of LINC complexes results in extreme separation of the ONM away from the INM, in most C. elegans tissues, removal of LINC complexes has little to no effect on nuclear envelope spacing. Cells in the interior of the animal likely have less rigid cell-cell contacts than tissue culture cells, resulting in less overall strain to the cytoskeleton.
Figure 2. UNC-84 is required for even nuclear envelope spacing only in body wall muscle cells. (A) Hypodermal larval nuclei in the unc-84(n369) animal do not display blebbing of the nuclear envelope. (B-C) However, large distortions of the nuclear envelope were observed at the ends of body wall muscle nuclei (arrows in B) and occasionally, along the sides (arrow in C). Scale bars are 1μm.
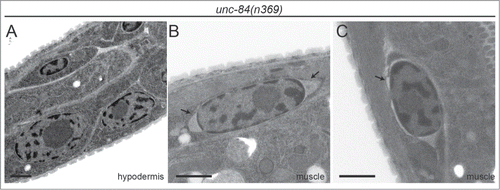
In the C. elegans larva, the best candidates for a cell type with increased intracellular tension resulting from cellular shape changes are striated body wall muscles. In contrast to most other tissues where nuclei are nearly spherical, muscle nuclei are oblong, with the long axis oriented parallel to the muscle fibers.Citation30 The PNS of muscle nuclei are an even 40–50 nm width all around the nuclei, although in some nuclei, the PNS narrowed along the long axis and/or widened at the ends. We then examined unc-84(n369) muscle nuclei to determine if LINC complexes play a role in stabilizing the nuclear envelope space in regions where forces appear to be highest (). Indeed, unc-84 mutant muscle nuclei have PNS widths of 100–500 nm. As with wild type nuclei, the largest distortions are at the ends of nuclei, although on occasion, smaller blebs are observed on the long axes (arrow in ). Each nucleus was observed over multiple serial sections, which allowed for observation of the change in shape of the blebs section by section. In the z-plane, the blebs start out small, grow larger near the centers of nuclei, and are imperceptible at the bottom surfaces of nuclei. Therefore, body wall muscle is the only C. elegans tissue of the wide variety of embryonic and larval tissues we observed that produces large nuclear envelope distortions similar to those observed in HeLa cells.Citation9
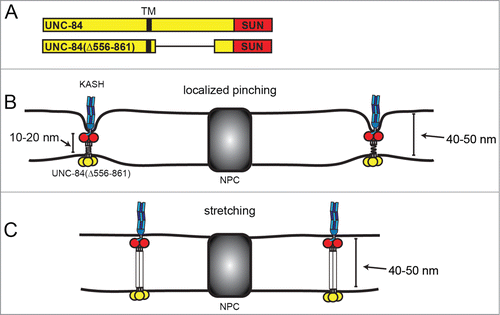
The observation that most nuclei in the unc-84(n369) display normal nuclear envelope spacing does not in and of itself contradict the hypothesis that LINC complexes set the dimensions of the PNS. Because UNC-84 is required to recruit UNC-83 to the ONM, unc-84(n369) nuclei do not have UNC-83 at the ONM and therefore lack a direct connection to motors that pull on the nuclear envelope in wild type nuclei. A direct test of this hypothesis required the introduction of an abnormally sized SUN protein that retains the ability to recruit and interact with KASH proteins. The hypothesis predicts that the PNS should then change to accommodate this mutant SUN protein in all tissues where it is expressed. The majority of the linker domain of UNC-84 in the PNS between the trans-membrane span and the conserved SUN domain bears little sequence similarity to other SUN proteins. Hydropathy analysis revealed multiple regions of increased hydrophobicity in the linker domain of UNC-84. Additionally, a hybrid protein with the C. elegans UNC-84 nucleoplasmic domain fused to the human SUN1 luminal domain, which contains 2 predicted coiled-coil domains,Citation31 functioned normally in hyp7 nuclear migration, suggesting the linker domain is functionally conserved from worms to mammals. These two findings are consistent with a model where the linker domain of UNC-84 assumes a helical structure. Also consistent with this model is our finding that deletion of 15 amino acids immediately preceding the SUN domain of UNC-84, which correspond to the 15 amino acids of human SUN2 that form the coiled stalk of the trimer, completely disrupts recruitment of UNC-83. Indirectly, this also suggests that UNC-84 functions as an oligomer in vivo. Intriguingly, deletion of approximately 300 amino acids of the UNC-84 linker domain (residues 556–861 between the transmembrane span and the conserved SUN domain; ) nonetheless produced a functional UNC-84 protein that was able to recruit UNC-83 to the ONM and move nuclei. This UNC-84 truncation therefore provided the perfect tool to directly test the hypothesis that SUN proteins set nuclear envelope spacing. The nuclear envelope spacing in embryos expressing unc-84(Δ556–861) is not noticeably narrower than wild type. Nor is there significant narrowing of PNS width in the muscle nuclei of larvae.
Figure 3. Two hypotheses for how UNC-84(Δ556–861) interacts with KASH proteins without narrowing the perinuclear space. SUN and KASH proteins are depicted as in . (A) Schematics of full-length UNC-84 and the functional truncation mutant UNC-84(Δ556–861). The transmembrane domain (TM) is depicted in black. (B) LINC complexes may be spaced far apart and localized pinching together of the 2 membranes may not be resolvable by TEM. (C) The luminal domain of UNC-84(Δ556–861) may be fully extended to accommodate the normal 40–50 nm perinuclear space.
Our results suggest that rather than setting nuclear envelope spacing, SUN proteins have evolved to span the distance between the INM and ONM. The question remains: if SUN proteins do not determine nuclear envelope spacing, what does? NPCs certainly play a role, as the perinuclear space narrows around NPCs in nuclei without LINC complexes. However, our results,Citation28 as well as SUN protein knockdown in tissue culture,Citation9 indicate that NPCs are not sufficient to maintain 40–50 nm spacing away from the pores. The answer may lie in the nuclear envelope's closest relative – the ER. The membranes of the nuclear envelope share many properties with the ER and the shape of the nuclear envelope echoes the shape of ER. During interphase, ER membranes are mostly in sheets, and polyribosomes are thought to provide the force to keep them flat.Citation32 ER sheets also have a characteristic spacing, although it is observed to be approximately twice as large as that of the perinuclear space in mammalian cells.Citation33 The morphology of ER sheets is maintained by the luminal spacer Climp63; overexpression of Climp63 results in an increase of ER sheets when overexpressed. Intriguingly, and in agreement with our results, depletion of Climp63 does not, in fact, result in blebbing of ER membranes. Rather, the intermembrane distance in the ER is reduced by half and more closely approximates the nuclear envelope distance, which is unaffected by Climp63 levels.Citation33 This suggests that Climp63 functions to expand the width of ER sheets to make them bigger than the default state found in the nuclear envelope. As the ONM is contiguous with the ER, polyribosomes could similarly be maintaining its flatness in the absence of LINC complexes. Chromatin could provide a similar force on the nucleoplasmic side of the nuclear envelope. In most tissues, these forces may be stronger than forces applied on the nucleus by the rest of the cell, such that even in the absence of SUN proteins, the nature of the membranes is to lay flat and evenly spaced. Therefore, the even spacing of the nuclear envelope in the absence of LINC complexes may be similar to the default spacing for ER sheets lacking Climp63. Alternatively, there may be other inner nuclear membrane proteins with large luminal domains that mediate perinuclear spacing. By contrast, body wall muscle cells, or cells adhered to a dish, experience more mechanical strain than most in vivo cells. These forces are presumably strong enough to overcome the forces working to keep the nuclear envelope flat in the absence of LINC complexes.
Another question is if the distance between the ONM and INM is an inherent property of the membranes, how can UNC-84(Δ556–861) mutant interact with KASH proteins without narrowing the PNS? Membranes could be pinched inward in areas too small to be resolved by EM (). Little is known about the distribution of LINC complexes along the nuclear envelope. Very few SUN/KASH interactions might be required to stabilize the nuclear envelope. If only a few, sparse LINC complexes are required to move a nucleus (), the pinching that results might not be resolvable under the sectioning and electron microscopy conditions we used. Alternatively, the remaining 60–80 residues could be fully stretched out. While the full-length UNC-84 protein may contain a trimeric helical rod that spans the distance between the ONM and INM, the UNC-84(Δ556–861) mutant might not assume the same conformation. It is possible that instead of a rod, each linker region is an unordered polypeptide chain, with a greater capacity to extend than the full-length version (). If each residue could fully extend to 3.8 Å in an open peptide backbone,Citation34 the remaining residues in the linker domain could reach 20–25 nm. Adding on an additional 14–15 nm (5 each for the ONM and INM and 4–5 for the SUN domain), gives a total of ∼40 nm, similar to the distances observed in nuclei expressing UNC-84(Δ556–861).
Nuclear Envelope Morphology and Disease
The observation that the absence of LINC complexes causes nuclear envelope architecture defects in cells under strain could have implications for understanding some human diseases. In C. elegans, nuclear envelope defects were only observed in striated body wall muscle cells. Interestingly, the largest group of laminopathies consists of those affecting striated muscle tissues.Citation2 One such disease, Emery-Dreifuss Muscular Dystrophy (EDMD), is associated with mutations in several nuclear envelope proteins, including Nesprin-1 and -2 and SUN1.Citation35,36 EDMD patients display early tightening of the elbows, Achilles tendons, and neck, followed by progressive muscle wasting in the limbs and associated dilated cardiomyopathy.Citation37 Mutations in nesprin-1 have also been linked to dilated cardiomyopathy without skeletal muscle defects.Citation38 The mechanism by which these mutations cause disease in a tissue-specific manner with variations from patient to patient is poorly understood. Our results suggest a possible involvement of defects in nuclear envelope structure. We compared the swimming motility of unc-84(n369) larvae, near the same stage where nuclear envelope defects were observed by TEM, to wild type control animals. Using both manual and computational scoring techniques the unc-84(n369) animals have an irregular swimming pattern compared to wild type. Mutant animals often coil for extended periods and have extra twitches. Notably, the motility defects we observed cannot be attributed to the loss of ventral cord neurons that has been previously described,Citation15 as the required nuclear migration event in precursor (P) cells occurs at a later stage in development. Thus, we have identified a novel locomotion defect in the unc-84(n369) mutant consistent with a muscle contraction defect. As the structure of the muscle fibers in the mutant animal were not distinctly different from wild type, the nuclear envelope architecture defects could contribute in some way to the locomotion defects in live animals. However, a neuronal defect in a different cell lineage could lead to the locomotion disorder. A potential candidate is the lumbar ganglion neuron PVQ, which is mispositioned in 15% of unc-84(n369) animals.Citation39 Nonetheless, our results open a new area for exploration in better understanding the relationship between muscle disease and nuclear envelope architecture.
Conclusion
Our work has clarified the prevailing model for the role of SUN proteins in nuclear envelope spacing. SUN proteins are necessary to maintain the even spacing between the INM and ONM in cells under high mechanical strain, but are not needed in other cells, where the inherent characteristics of ER-derived membranes are sufficient to keep them flat and evenly spaced. We also have also uncovered a potential functional consequence of nuclear envelope architecture defects in muscle tissue. Further experiments are needed to better understand the nature of this connection and the role it might play in human disease. Specifically, examination of nuclear envelope ultrastructure in tissues from mouse muscular dystrophy models, as well as patient samples, should be informative.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Erin Tapley (UC Davis), Kent McDonald (UC Berkeley), and Benjamin Cain (UC Davis), our co-authors on the original manuscript, for their wonderful scientific collaborations. We thank members of the Starr lab for comments on the manuscript.
Funding
This work was supported by NIH/NIGMS grant R01 GM073874 to DA. Starr. NE. Cain is supported by American Cancer Society Illinois Division postdoctoral fellowship PF-13-094-01-CGC.
References
- Franke WW, Scheer U, Krohne G, Jarasch ED. The nuclear envelope and the architecture of the nuclear periphery. J Cell Biol 1981; 91:39s-50s; PMID:7033243; http://dx.doi.org/10.1083/jcb.91.3.39s
- Burke B, Stewart CL. Functional architecture of the cell's nucleus in development, aging, and disease. Curr Top Dev Biol 2014; 109:1-52; PMID:24947235; http://dx.doi.org/10.1016/B978-0-12-397920-9.00006-8
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 2010; 26:421-44; PMID:20507227; http://dx.doi.org/10.1146/annurev-cellbio-100109-104037
- Gundersen GG, Worman HJ. Nuclear positioning. Cell 2013; 152:1376-89; PMID:23498944; http://dx.doi.org/10.1016/j.cell.2013.02.031
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 2012; 149:1035-47; PMID:22632968; http://dx.doi.org/10.1016/j.cell.2012.03.046
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol 2006; 26:3738-51; PMID:16648470; http://dx.doi.org/10.1128/MCB.26.10.3738-3751.2006
- Bone CR, Tapley EC, Gorjanacz M, Starr DA. The C. elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Mol Biol Cell 2014; 25(18):2853-65
- Luxton GW, Starr DA. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol 2014; 28:69-75; PMID:24704701; http://dx.doi.org/10.1016/j.ceb.2014.03.002
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 2006; 172:41-53; PMID:16380439; http://dx.doi.org/10.1083/jcb.200509124
- Sosa BA, Kutay U, Schwartz TU. Structural insights into LINC complexes. Curr Opin Struct Biol 2013; 23:285-91; PMID:23597672; http://dx.doi.org/10.1016/j.sbi.2013.03.005
- Rothballer A, Schwartz TU, Kutay U. LINCing complex functions at the nuclear envelope: what the molecular architecture of the LINC complex can reveal about its function. Nucleus 2013; 4:29-36; PMID:23324460; http://dx.doi.org/10.4161/nucl.23387
- McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell 2006; 17:1790-801; PMID:16481402; http://dx.doi.org/10.1091/mbc.E05-09-0894
- Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol 2010; 191:115-28; PMID:20921138; http://dx.doi.org/10.1083/jcb.201004118
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 1983; 100:64-119; PMID:6684600; http://dx.doi.org/10.1016/0012-1606(83)90201-4
- Sulston JE, Horvitz HR. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev Biol 1981; 82:41-55; PMID:7014288; http://dx.doi.org/10.1016/0012-1606(81)90427-9
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 2002; 298:406-9; PMID:12169658; http://dx.doi.org/10.1126/science.1075119
- Hedgecock EM, Thomson JN. A gene required for nuclear and mitochondrial attachment in the nematode Caenorhabditis elegans. Cell 1982; 30:321-30; PMID:6889924; http://dx.doi.org/10.1016/0092-8674(82)90038-1
- Zhou Z, Du X, Cai Z, Song X, Zhang H, Mizuno T, Suzuki E, Yee MR, Berezov A, Murali R, et al. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Biol Chem 2012; 287:5317-26; PMID:22170055; http://dx.doi.org/10.1074/jbc.M111.304543
- Gob E, Schmitt J, Benavente R, Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PloS one 2010; 5:e12072; PMID:20711465; http://dx.doi.org/10.1371/journal.pone.0012072
- Shao X, Tarnasky HA, Lee JP, Oko R, van der Hoorn FA. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev Biol 1999; 211:109-23; PMID:10373309; http://dx.doi.org/10.1006/dbio.1999.9297
- Xing XW, Li LY, Liu G, Fu JJ, Tan XJ, Lu GX. Identification of a novel gene SRG4 expressed at specific stages of mouse spermatogenesis. Acta Biochim Biophys Sin 2004; 36:351-9; PMID:15156277; http://dx.doi.org/10.1093/abbs/36.5.351
- Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, Feinstein N, Cohen M, Wilson KL, Gruenbaum Y. Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc Natl Acad Sci USA 2004; 101:6987-92; PMID:15100407; http://dx.doi.org/10.1073/pnas.0307880101
- Minn IL, Rolls MM, Hanna-Rose W, Malone CJ. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol Biol Cell 2009; 20:4586-95; PMID:19759181; http://dx.doi.org/10.1091/mbc.E08-10-1034
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 2009; 10:75-82; PMID:19197334; http://dx.doi.org/10.1038/nrm2594
- Magidson V, O'Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 2011; 146:555-67; PMID:21854981; http://dx.doi.org/10.1016/j.cell.2011.07.012
- McDonald K. Cryopreparation methods for electron microscopy of selected model systems. Methods Cell Biol 2007; 79:23-56; PMID:17327151; http://dx.doi.org/10.1016/S0091-679X(06)79002-1
- Tapley EC, Ly N, Starr DA. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol Biol Cell 2011; 22:1739-52; PMID:21411627; http://dx.doi.org/10.1091/mbc.E10-08-0733
- Cain NE, Tapley EC, McDonald KL, Cain BM, Starr DA. The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. J Cell Biol 2014; 206:163-72; PMID:25023515; http://dx.doi.org/10.1083/jcb.201405081
- Cohen M, Tzur YB, Neufeld E, Feinstein N, Delannoy MR, Wilson KL, Gruenbaum Y. Transmission electron microscope studies of the nuclear envelope in Caenorhabditis elegans embryos. J Struct Biol 2002; 140:232-40; PMID:12490171; http://dx.doi.org/10.1016/S1047-8477(02)00516-6; dx.doi.org/10.3908/wormatlas.1.7
- Altun ZF, Hall DH. Muscle system, somatic muscle. In WormAtlas 2009. doi:10.3908/wormatlas.1.7
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci 2005; 118:3419-30; PMID:16079285; http://dx.doi.org/10.1242/jcs.02471
- Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol 2007; 179:895-909; PMID:18056408; http://dx.doi.org/10.1083/jcb.200705112
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell 2010; 143:774-88; PMID:21111237; http://dx.doi.org/10.1016/j.cell.2010.11.007
- Chakraborty S, Venkatramani R, Rao BJ, Asgeirsson B, Dandekar AM. Protein structure quality assessment based on the distance profiles of consecutive backbone Calpha atoms. F1000Research 2013; 2:211; PMID:24555103
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet 2007; 16:2816-33; PMID:17761684; http://dx.doi.org/10.1093/hmg/ddm238
- Li P, Meinke P, Huong le TT, Wehnert M, Noegel AA. Contribution of SUN1 mutations to the pathomechanism in muscular dystrophies. Hum Mutat 2014; 35:452-61; PMID:24375709; http://dx.doi.org/10.1002/humu.22504
- Worman HJ. Nuclear lamins and laminopathies. J Pathol 2012; 226:316-25; PMID:21953297; http://dx.doi.org/10.1002/path.2999
- Puckelwartz MJ, Kessler EJ, Kim G, Dewitt MM, Zhang Y, Earley JU, Depreux FF, Holaska J, Mewborn SK, Pytel P, et al. Nesprin-1 mutations in human and murine cardiomyopathy. J Mol Cell Cardiol 2010; 48:600-8; PMID:19944109; http://dx.doi.org/10.1016/j.yjmcc.2009.11.006
- Johnson RP, Kramer JM. Neural maintenance roles for the matrix receptor dystroglycan and the nuclear anchorage complex in Caenorhabditis elegans. Genetics 2012; 190:1365-77; PMID:22298703; http://dx.doi.org/10.1534/genetics.111.136184
