Abstract
Male germ cell genome integrity is critical for spermatogenesis, fertility and normal development of the offspring. Several DNA repair pathways exist in male germ cells. One such important pathway is the Fanconi anemia (FANC) pathway. Unlike in somatic cells, expression profiles and the role of the FANC pathway in germ cells remain largely unknown. In this study, we undertook an extensive expression analyses at both mRNA and protein levels of key components of the FANC pathway during spermatogenesis in the mouse. Herein we show that Fanc mRNAs and proteins displayed developmental enrichment within particular male germ cell types. Spermatogonia and pre-leptotene spermatocytes contained the majority of the FANC components examined i.e. complex I members FANCB, FANCG and FANCM, complex II members FANCD2 and FANCI, and complex III member FANCJ. Leptotene, zygotene and early pachytene spermatocytes contained FANCB, FANCG, FANCM and FANCD2. With the exception of FANCL, all FANC proteins examined were not detected in round spermatids. Elongating and elongated spermatids contained FANCB, FANCG, FANCL and FANCJ. qPCR analysis on isolated spermatocytes and round spermatids showed that Fancg, Fancl, Fancm, Fancd2, Fanci and Fancj mRNAs were expressed in both of these germ cell types, indicating that some degree of translational repression of these FANC proteins occurs during the transition from meiosis to spermiogenesis. Taken together, our findings raise the possibility that the assembly of FANC protein complexes in each of the male germ cell type is unique and may be distinct from the proposed model in mitotic cells.
Abbreviations
| ATR | = | Ataxia telangiectasia and Rad3-related |
| DDR | = | DNA damage response and repair |
| DSBs | = | DNA double strand breaks |
| eST | = | elongating spermatid |
| FANC | = | Fanconi anemia |
| HPG | = | Hypothalamic-Pituitary-Gonadal |
| qPCR | = | quantitative PCR |
| Pl | = | Pre-leptotene spermatocytes |
| PTM | = | Peritubular myoid cells |
| PS | = | Pachytene spermatocyte |
| SC | = | Sertoli cells |
| Sg | = | spermatogonia |
| RNPs | = | Ribonucleoprotein complexes |
| rST | = | round spermatids |
Introduction
In mammals, spermatogenesis begins at puberty and continues throughout life. This process involves mitotic divisions and self-renewal of a stem cell population within the spermatogonia pool, meiotic division of spermatocytes and the terminal differentiation of haploid spermatids into highly polarized spermatozoa in a process known as spermiogenesis. In the mouse, each spermatogenic cycle takes ∼35 d. Sperm subsequently undergo a post-testicular maturation process within the epididymis.Citation1 In addition to germ cells, the testis contains specialized somatic cells including Sertoli and Leydig cells. Sertoli cells provide the isolated environment and paracrine regulation necessary for development of germ cells.Citation2 Leydig cells are located within the interstitial tissue of the testis. Through a complex regulatory network of the hypothalamic-pituitary-gonadal (HPG) axis, Leydig cells produce testosterone to the seminiferous tubules to drive spermatogenesis.Citation1
In order to assure the health of offspring, the integrity of the sperm genome must be maintained throughout spermatogenesis. The male germ cell genome can be damaged by various exogenous insults including exposure to ionizing radiation, endocrine disrupters or oxidative stress.Citation3,4 Moreover, physiological processes such as DNA replication errors that occur during mitotic divisions of spermatogonia, DNA double strand breaks (DSBs) that occur during meiotic recombination Citation5-7 and chromatin remodelling in spermatids Citation8-12 can also trigger male germ cell DNA damage. As such, several DNA repair pathways exist in male germ cells to simultaneously facilitate an efficient DNA damage response and repair (DDR) thus preserving genome integrity.Citation13 DDR comprises an elaborate network of signaling pathways and the assembly of macromolecular DDR proteins at the vicinity of the DNA lesions.Citation14 Defects in DDR pathways have been shown to be associated with infertility, gamete aneuploidy and disease risk in offspring.Citation4 Moreover, germ line mutations in DDR genes have been shown to be associated with cancers and genome instability syndromes e.g. ataxia telangiectas,Citation15 Nijmegen breakage syndrome,Citation16 Bloom's syndrome Citation17 and Fanconi anemia.Citation18 Together, these findings highlight the importance of DNA repair pathways in maintaining genome stability in both somatic and germ cells.
Fanconi anemia is characterized by progressive bone marrow failure (aplastic anaemia), head and neck cancers, leukemia susceptibility, multiple and variable congenital malformations and infertility.Citation18 The majority of Fanconi anemia patients die in early childhood as a result of bone marrow failure, leukemia and/or other cancers. Cells derived from Fanconi anemia patients show elevated levels of chromosomal breakage and an increased sensitivity to agents that produce interstrand DNA crosslinks, such as mitomycin C (MMC).Citation18 To date, 14 FANC genes have been identified in the human genome: FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ, FANCL, FANCM, FANCN and FANCP.Citation19-21 Based on their function, the 14 FANC proteins have been classified into 3 groups. The first group is composed of 8 proteins (FANCA, B, C, E, F, G, M, and L) that assemble into a nuclear complex known as the FANC core complex.Citation22,23 This complex, through the E3 ubiquitin ligase activity of FANCL, mediates monoubiquitination of FANCD2 and FANCI (the second group of FANC proteins) in response to DNA damage signals. A critical step in this pathway is the monoubiquitination and deubiquitination of the FANCD2/FANCI complex.Citation22,23 In addition to activation by the FANC core complex, the recruitment of the FANCD2/FANCI complex to the sites of DNA damage requires a signaling cascade induced by the activation of ATR (ataxia telangiectasia and Rad3-related) kinase and the subsequent phosphorylation of histone H2AX (γH2AX).Citation24 In the presence of γH2AX, the monoubiquitinated FANCD2/FANCI complex localizes to the damage sites. At this point, the complex interacts either directly or indirectly with the third group of FANC proteins: FANCD1 (alias BRCA2), FANCJ (alias BRIP1), FANCN (alias PALB2), FANCP (alias SLX4) and many BRCA1-interacting proteins to facilitate the repair by which the precise mechanism remains unclear.Citation19
In addition to genome instability and cancer susceptibility, ablations of Fanc genes in mice have been shown to compromise male and/or female fertility. FANCA, FANCC, FANCL and FANCP have been implicated in primordial germ cell proliferation and their ablation reduces the spermatogonial pool in the early postnatal testis.Citation25-27 Ablation of Fanca or Fancd2 leads to meiotic chromosome mispairing.Citation27,28 The mechanism by which these FANC proteins regulate spermatogonial proliferation and meiotic chromosome pairing remains undefined. Recently, however, Crossan and colleagues demonstrated a role for FANCP in male meiotic DSB repair wherein its ablation resulted in delayed DSB repair and increased apoptosis of prophase I spermatocytes.Citation21 Taken together, these findings highlight the importance of the FANC pathway in male germ cell development and function.
Unlike in somatic cells, the components and regulation of the FANC pathway is largely unknown in male germ cells. Moreover, the sites of each FANC protein expression during spermatogenesis are largely undefined. In this study, we performed an extensive survey of FANC component expression and localization during spermatogenesis in the mouse. Our data suggest that the assembly of FANC complex proteins in each male germ cell types is unique and distinct from the proposed model in mitotic cells.
Results
Fanc genes are differentially expressed during spermatogenesis
Based on Ensembl databases, all 14 Fanc genes were identified in the mouse genome (). Sequence comparison showed that these proteins are conserved between the mouse and human i.e., 47–80% sequence identity (), suggesting that these complexes play an evolutionally conserved role in the mouse and human.
Table 1. The location and sequence analysis of the mouse and human FANC genes
To define expression profiles of Fanc genes in the mouse testis, we performed quantitative PCR (qPCR) analysis using total testis RNA extracted from postnatal day 0, 7, 14, 18, 20, 30 and 70 mice. These ages were chosen as they represent major events that occur during male germ cell development (). At postnatal day 0 (the day of birth), the majority of cells in the seminiferous epithelium are immature Sertoli cells (somatic cells) and gonocytes. We used postnatal day 0 as a reference point for gene expression during the initiation of spermatogenesis i.e. the establishment of a spermatogonial pool. At postnatal day 7, the majority of cells are undifferentiated and differentiating spermatogonia, and immature Sertoli cells. Postnatal day 7 is used as a reference point for gene expression in mitotic spermatogonia and immature Sertoli cells. Following several rounds of mitotic divisions, a subpopulation of differentiating spermatogonia enter meiosis Citation29 to form primary spermatocytes at around postnatal day 10, and thus we used postnatal days 14 and 18 as an indication of early and late gene expression in meiosis, respectively. At postnatal day 20, the majority of secondary spermatocytes have completed meiosis and converted into haploid round spermatids, which will terminally differentiate into highly polarized spermatozoa i.e., the development of characteristic head shape and motile sperm tail (spermiogenesis). We used postnatal day 20 as a time point marking late expression of meiotic genes and early expression of haploid genes. At postnatal day 30 elongated spermatids have formed and we used postnatal day 30 as an indication of gene expression in elongating spermatids. At postnatal day 70 (adult), there are mixtures of all stages of germ cells and mature Sertoli cells.
Figure 1. qPCR analyses of Fanc mRNA expression during spermatogenesis in the mouse. (A) Schematic of time points for major events during the first wave of spermatogenesis in the mouse. Sg: spermatogonia; Sp: spermatocyte, MI: meiosis I; MII: meiosis II, rST: round spermatid; eST: elongating and elongated spermatids. mRNA expression of complex I members Fanca, Fancb, Fancc, Fance, Fancf, Fancg, Fancl and Fancm (B); complex II members Fancd2 and Fanci (C); and complex III member Fancj (D) across different aged testes and in isolated spermatocytes and round spermatids as determined by qPCR analysis.
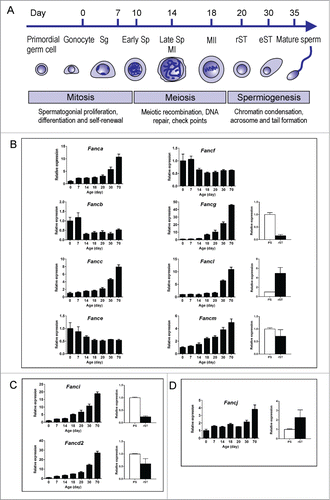
Expression of each of the Fanc mRNA was normalized against Hprt (hypoxanthine guanine phosphoribosyl transferase) and data were presented into 3 functional groups i.e. complex I (core complex), complex II (ID complex) and complex III. Our results indicated that all 11 Fanc mRNAs examined were expressed throughout spermatogenesis ().
The complex I members Fanca, Fancc, Fancl and Fancm were expressed at similar levels from postnatal day 0 to day 20 and their expression levels began to peak at postnatal day 30 then continued to increase through to adult (). These data suggest these Fanc mRNAs were expressed in multiple cell types within the testis but were enriched in haploid germ cells. Expression of Fancg mRNA followed a similar pattern, however, its expression level began to peak at postnatal day 18 then continued to increase through to adult (). This result suggests that Fancg mRNA was expressed in multiple cell types but highly enriched in late spermatocytes and spermatids. By contrast, expression of Fancb, Fance and Fancf mRNAs were enriched at early postnatal development i.e., days 0 and 7, and were down-regulated as spermatogenesis proceeded (). These results suggest these Fanc mRNAs were enriched in spermatogonia and/or Sertoli cells.
Expression of the complex II members Fancd2 and Fanci were gradually increased from day 0 through to adult whereby their expression levels began to peak at postnatal day 30 and day 18, respectively (). These results suggest that Fancd2 mRNA was expressed in multiple cell types but enriched in haploid germ cells, and Fanci mRNA was enriched in late spermatocytes and spermatids.
The complex III member Fancj showed a similar level of expression from postnatal day 0 to day 30 and was up-regulated in adulthood (day 70) (), suggesting that Fancj is expressed in multiple cell types but enriched in spermatids.
Uncoupling of transcription and translation of FANC complexes proteins during spermatogenesis
To gain further insights into the role of the FANC pathway in spermatogenesis, we next examined the localization of each of the FANC proteins using immunohistochemistry on wild-type adult mouse testis sections. We tested 18 FANC antibodies obtained from commercial sources. The majority of these antibodies either showed no staining or showed extremely high backgrounds i.e. all cells types were found strongly stained. Seven FANC antibodies showed consistent staining patterns across multiple seminiferous tubules and across different testis sections from 3 wild-type mice and were chosen for this study. These included FANCB, FANCG, FANCL, FANCM, FANCD2, FANCI and FANCJ (see materials and methods). The localization of each protein across various stages of the seminiferous epithelium was analyzed, and the data is summarised in . Representative images of each protein are shown in and Supplementary Figure 1.
Table 2. Summary of FANC protein localization in the adult mouse testis
Figure 2. FANC protein localization in the adult mouse testis. (A) FANCB was localized in the nucleus and cytoplasm of Sertoli cells (SC), in the cytoplasm of type and Intermediate spermatogonia (Sg) and of pachytene spermatocytes (PS) in the early stages of spermatogenesis. Round spermatids (rST) showed no discernible staining, however elongated spermatids (eST) showed cytoplasmic staining. (B) FANCG was detected in the cytoplasm of type A (A) and type B spermatogonia (B), and in the cytoplasm of pachytene spermatocytes (PS). Nuclear staining of elongating spermatids (eST) was also evident. Peritubular myoid cells (PTM) were also immunostained. (C) FANCL was only detected in the nuclei of spermatids just prior to, and during elongation, with the immunostaining becoming more intense in step 10 (see arrows in stage X tubule) compared to step 9 (see arrows in stage IX tubule). The Roman numerals indicate the stage of the tubule whereas the arrows indicate an elongating spermatid within each tubule (D) FANCM was localized in the cytoplasm of Sertoli cells (SC) and in the cytoplasm and Golgi of leptotene-zygotene spermatocytes (L-Z). Late pachytene spermatocytes (PS) were not obviously immunostained above background, nor were round spermatids (rST). Peritubular myoid cells (PTM) showed some staining. (E) FANCD2 was present in the Sertoli cell (SC) nuclei at stage VII and in spermatogonia (Sg), preleptotene spermatocytes (Pl) and pachytene spermatocytes (PS). Round spermatids (rST) were not obviously immune-labeled. (F) FANCI was present in the nucleus (red arrows) and cytoplasm (black arrow) of Sertoli cells (SC). It was also apparent in the nucleus of spermatogonia (Sg) but was absent from pre-leptotene – leptotene (P-L) spermatocytes. Other cells including pachytene spermatocytes (PS), round spermatids (rST) and elongated spermatids (eST) were not obviously immunostained. G. FANCJ was evident in the nucleus of some spermatogonia (Sg), including type B spermatogonia (B) but showed little immunostaining in Sertoli cells (SC). (H, I) Negative controls for the immunohistochemistry, whereby the primary antibody was omitted. The bar in each micrograph represents 50 μM.
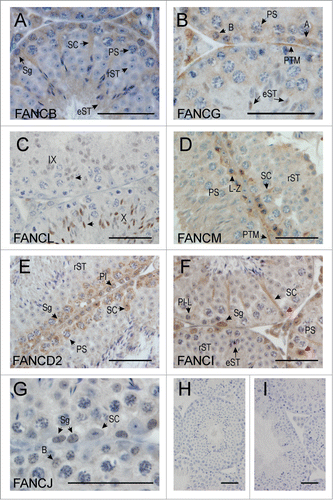
FANCB localized in the nucleus and cytoplasm of Sertoli cells, and in the cytoplasm and Golgi (compare Supplementary Fig. 1 panel A vs. J) of spermatogonia through to early pachytene spermatocytes (, Supplementary Figs. 1A and ), consistent with mRNA expression data whereby highest levels of Fancb mRNA were observed in postnatal days 0 and 7 testes (). No FANCB protein was detected in late pachytene spermatocytes or round spermatids ( and Supplementary Fig. 1A). FANCB, however, re-appeared in the cytoplasm of elongating spermatids from steps 10–16 ().
FANCG was detected at low levels in the cytoplasm of type A and B spermatogonia, pre-leptotene and pachytene spermatocytes (, Supplementary Fig. 1B and ). Staining was also observed in peritubular myoid (PTM) cells (, Supplementary Fig. 1B and ). Similar to FANCB, FANCG protein was not detected in round spermatids (Supplementary Fig. 1B) but was, however, seen in the nucleus of elongating spermatids in steps 9–11 (, ). As shown in , Fancg mRNA was expressed throughout spermatogenesis but its expression level began to peak at day 18 testis through to adult. The absence of FANCG protein in round spermatids suggests that transcription and translation of FANCG may be un-coupled. To further investigate this, we performed qPCR analysis using total RNA isolated from spermatocytes and round spermatids. Fancg mRNA was detected in both germ cell types (). These results suggest that transcription and translation of Fancm mRNA were uncoupled whereby translation was repressed in round spermatids and re-activated in during the elongation phase of spermiogenesis in transcriptionally inactive elongating spermatids.
FANCL protein first appeared in the nucleus of round spermatids at step 8 and persisted through elongating spermatids from step 9 onwards, however, the protein disappeared in elongated spermatids during approximately step 11 (, Supplementary Fig. 1C and ). This data was consistent with Fancl mRNA, which was expressed at low levels in postnatal days 0–20 and upregulated in postnatal day 30 testes, and in round spermatids ().
FANCM localized in the cytoplasm of Sertoli cells and the cytoplasm and Golgi of spermatogonia through to early pachytene spermatocytes, however immunostaining was faint or absent beyond pachytene spermatocytes in stage IX (, Supplementary Figs. 1D and E, and ). Fancm mRNA was highly enriched in postnatal day 30 through to adult testis () but the protein was not detected in germ cells beyond late meiosis (, Supplementary Fig. 1D and E, and ). Despite this, qPCR analysis on isolated germ cells showed that Fancm mRNA was detected in spermatocytes and round spermatids (). These results suggest that transcription and translation of Fancm mRNA were uncoupled whereby translation was shut down prior to spermiogenesis.
FANCD2 localized in the cytoplasm of spermatogonia through to late pachytene spermatocytes in stages IX-X, after which no protein was detected and haploid germ cells were also immuno-negative (, Supplementary Fig. 1F and ). Most Sertoli cell nuclei were immunonegative (Supplementary Fig. 1F) but immuno-positive nuclei were apparent in stage VII-VIII (, and ). Fancd2 mRNA was expressed throughout spermatogenesis whereby its expression level began to peak at postnatal day 30 through to adult (). Consistently, qPCR analysis of isolated spermatocytes and round spermatids showed that Fancd2 mRNA was detected in both germ cell types (). Similar to FANCM, the absence of FANCD2 protein beyond late meiosis suggests that transcription and translation of Fancd2 mRNA were uncoupled whereby translation was shut down beyond meiosis.
FANCI localized in both the cytoplasm and nucleus of Sertoli cells, and the nuclei of spermatogonia through to pre-leptotene spermatocytes, but disappeared as spermatocytes made the transition from preleptotene to leptotene (, Supplementary Fig. 1G and ). Similar to Fancd2, Fanci mRNA was expressed throughout spermatogenesis and its expression began to peak at postnatal day 30 through to adult (). Consistent with this, qPCR analysis of isolated spermatocytes and round spermatids showed expression of Fanci mRNA in both germ cell types (). Thus the absence of FANCI protein beyond pre-leptotene spermatocytes suggests that transcription and translation of Fanci mRNA were uncoupled whereby translation was shut down after the leptotene stage of meiosis.
FANCJ localized in the nucleus of spermatogonia through to pre-leptotene spermatocytes, after which the protein was not detected during the leptotene phase of meiosis (, Supplementary Figs. 1H and I, and ). FANCJ protein, however, re-appeared in elongating spermatids whereby nuclear localization was observed in steps 9–11 spermatids (Supplementary ). Fancj mRNA was expressed throughout spermatogenesis and its expression peaked in the adult testis (). qPCR analysis on isolated spermatocytes and round spermatids showed that Fancj mRNA was detected in both germ cell types (), indicating that the lack of FANCJ during meiosis and early spermiogenesis was due to translational repression. This phenomenon was similar with translation repression/reactivation observed of the complex I member FANCG.
Sertoli cells contained all of the FANC protein examined. Of note, FANCD2 showed stage specific expression whereby nuclear localization was observed in stages VII and VIII. By contrast, peritubular myoid and Leydig cells contained a subset of FANC protein examined ().
Discussion
In this study we focus on the FANC pathway and its role during spermatogenesis. The current molecular model on how FANC proteins co-operate in DNA interstrand cross-link repair in mitotic cells is based on a stepwise recruitment of FANC complexes on the DNA damage foci.Citation19,30 However, little is known about expression profiles and function of the FANC complexes in male germ cells. This study is the first to define expression profiles of FANC complexes at both mRNA and protein levels in the testis.
Spermatogonia and pre-leptotene spermatocytes contained the majority of the FANC components examined i.e., complex I members FANCB, FANCG and FANCM, complex II members FANCD2 and FANCI, and complex III member FANCJ. These data raise the possibility that these complexes play a role in DNA repair during DNA synthesis i.e. in mitotically active spermatogonia, and in pre-leptotene spermatocytes, which are synthesising DNA. Leptotene, zygotene and early pachytene spermatocytes contained complex I members FANCB, FANCG and FANCM, and complex II member FANCD2. None of the proteins showed nuclear localization in zygotene-leptotene spermatocytes when DSB repair is occurring via homologous recombination. These findings raised 2 possibilities: (i) the FANC proteins examined do not participate in DSB repair during meiosis; or (ii) nuclear localization may occur in a highly dynamic manner and once DSB repair is completed, the repaired protein complexes are being disassembled and re-distributed quickly in the cytoplasm, and/or being rapidly degraded. Given a high incidence of meiotic defects in a number of Fanc knockout mouse lines, the latter is more likely, thus the use of sensitive methods such as meiotic chromosome spreads coupled with immunofluorescence should be conducted to further define nuclear localization of these FANC proteins.
Of note, complex I members FANCB and FANCM, and complex II member FANCD2 were found localized in the cytoplasm and the Golgi complex of spermatogonia through to early pachytene spermatocyte (). The Golgi complex is a protein (and lipid) processing, packaging and distribution center of the cell as it regulates post-translational modifications of proteins synthesized from the ER, sorting and transporting proteins to their ultimate destinations i.e., lysosomes, the plasma membrane, or secretion. Citation31 Recently, a study demonstrated that, in mammalian mitotic cells, DNA damage triggers the Golgi to fragment and disperse throughout the cytoplasm leading to the amplification of the DNA damage response via the DNA damage protein kinase, DNA-PK. Citation32 This study highlights direct links between the DNA damage response and the Golgi. Thus, the presence of FANCB, FANCM and FANCD2 proteins in the Golgi raised the possibility that the newly synthesized FANC proteins may undergo post-translational modification and store for a period of time prior to being distributed to sites of DNA damage foci at appropriate time points of germ cell development.
Of note, spermatogonia and spermatocytes did not contain the catalytic subunit of the FANC core complex (complex I), FANCL. These findings raise the possibility that (i) the component and the assembly of FANC core complex in these germ cells is different from the current proposed models in mitotic cells, and (ii) these germ cells may contain additional members of this pathway.
By contrast to pre-meiotic germ cells and germ cells that undergoing meiotic divisions, round spermatids contained one out of 8 FANC proteins examined i.e. FANCL, however FANCL was only observed as round spermatids commenced their final differentiation into elongating spermatids. These data imply that different DNA repair pathway(s) may be responsible for DNA repair in round spermatids. As spermiogenesis proceeded into the spermatid condensation and elongation phase, some FANC proteins re-appeared in elongating and elongated spermatids i.e., complex I members FANCB, FANCG and FANCL, and complex III member FANCJ. These data suggest that FANCB, FANCG, FANCL and FANCJ may play a role in repairing of damage DNA that occurs during chromatin condensation.
Furthermore, our data demonstrate that translational repression exists for some of the FANC components during spermiogenesis. qPCR analysis of Fancg, Fancl, Fancm, Fancd2, Fanci and Fancj on isolated round spermatids and spermatocytes showed that these genes were expressed in both of these germ cell types, however proteins were not observed in round spermatids by immunohistochemistry. Thus the lack of the proteins seems likely to be due to translational repression in round spermatids, a mechanism by which mRNAs are synthesized and stored in a repressed state within ribonucleoprotein complexes (RNPs), which led to delayed translation, is often observed in spermiogenic transcripts Citation33. As spermatids began to elongate, translation of Fancg, Fancl, Fancm, and Fancj mRNAs is activated whereas translation of Fancm, Fancd2 and Fanci remained repressed. As a result, the core complex I members FANCB, FANCG and FANCL, and the complex III member FANCJ were found in elongating spermatids at steps 9–11 (steps 10–16 for FANCB). However, the lack of complex II members FANCD2 and FANCI (ID complex) in elongating and elongated spermatids suggests that DNA repair mediated via FANCB, FANCG and FANCL, and FANCJ during the late phase of spermiogenesis is independent from the ID complex.
In summary, our data demonstrate that specific components of FANC complexes are expressed at particular times during germ cell development and some degree of translational repression during the transition from meiosis to spermiogenesis occurs for some FANC proteins. Our data also raises the possibility that the assembly of FANC protein complexes in each male germ cell type is unique and may be distinct from the proposed model in mitotic cells.
Materials and Methods
Animals and tissue collection
Animal experiments were approved by the Monash University Animal Ethics Committees. Testes were collected from wild-type C57BL/6J mouse strain and fixed in Bouin's fixative solution (Amber Scientific) for 5 hours prior to processing and paraffin embedding using standard methods.
Germ cell isolation
Germ cell sub-populations were isolated from adult mouse testes using the Staput method as previously described.Citation34 Spermatocytes and spermatid fractions were collected at 3.5-hour sedimentation period in 2–4% continuous BSA gradient and were washed in 3 times with PBS buffer to eliminate BSA contamination.
RNA isolation and cDNA conversion
Total RNA from whole testes and isolated cells was isolated using TriZol reagent (Life Technologies). Genomic DNA was eliminated by DNase treatment using DNA-free Kit (Life Technologies). cDNA conversion was performed using SuperScriptIII reverse transcriptase (Life Technologies), and 2 μg of total RNA from the whole testis and 1 μg of total RNA from isolated germ cells.
Quantitative PCR (qPCR)
100–200 ng of cDNA was used as a template in qPCR reactions using TaqMan assays (Life Technologies): Fanca (Assay ID: Mm00516836_m1); Fancb (Mm01167819_m1); Fancc (Mm00514846_m); Fance (Mm00511654_m1); Fancf (Mm03412528_s1); Fancg (Mm00474063_m1), Facnl (Mm00840321_m1); Fancm (Mm00626872_m1); Fancd2 (Mm01184611_m1); Fannci (Mm00524140_m1) and Fancj (Mm01297848_m1). Expression of each Fanc mRNAs in the whole testis was normalized against Hprt (Mm00446968). Due to very low expression levels of Hprt in round spermatids (Ct below 35 cycles), expression of each Fanc mRNAs in the isolated spermatocytes and round spermatids was normalized against Gapdh (Mm99999915). All assays were labeled with FAM and reactions were performed using the Mx3000P qPCR System (Agilent). Data were analyzed using MxPro qPCR software (Agilent). Data were replicates of n = 3 mice per age group and 3 replicates per qPCR reaction. Cycling condition is as follow: 95°C for 10 min (1 cycle), followed by 50 cycles of 95°C for 15 sec and 60°C for 1 min.
Immnostaining
Immunohistochemistry was performed as previously describedCitation35 using 3 adult wild-type mice. Antibodies were purchased from commercial sources including: FANCB (Abcam, ab86136, rabbit polyclonal) used at 20 μg/ml; FANCD2 (Abcam, ab2187, rabbit polyclonal) used at 2 μg/ml; FANCG (Abcam, ab54645, mouse monoclonal) used at 20 μg/ml; FANCI (Abcam, ab15344, rabbit polyclonal) used at 10 μg/ml; FANCL (Abcam, ab94458, rabbit polyclonal) used at 10 μg/ml, FANCM (Abcam, ab95014, rabbit polyclonal) used at 2 μg/ml and FANCJ (Abcam, ab16608, rabbit polyclonal) used at 1:100 (whole antiserum).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
979061_Supplementary_Materials.zip
Download Zip (13.5 MB)Funding
This work was supported by the National Health and Medical Research Council (NHMRC) and the Australian Research Council (ARC); Grant Numbers: 143786, 384297 and DP110102288.
References
- Hermo L, Pelletier RM, Cyr DG, Smith CE. Surfing the wave, cycle, life history, and genesproteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc Res Tech 2010; 73:241-78; PMID:19941293; http://dx.doi.org/10.1002/jemt.20783
- Skinner M, Griswold M, eds. Sertoli Cell Biology. San Diego: Elsevier Academic Press, 2005.
- Aitken RJ, De Iuliis GN. Origins and consequences of DNA damage in male germ cells. Reprod Biomed Online 2007; 14:727-33; PMID:17579989; http://dx.doi.org/10.1016/S1472-6483(10)60676-1
- Leduc F, Nkoma GB, Boissonneault G. Spermiogenesis and DNA repair: a possible etiology of human infertility and genetic disorders. Syst Biol Reprod Med 2008; 54:3-10; PMID:18543861; http://dx.doi.org/10.1080/19396360701876823
- Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr Rev 2006; 27:398-426; PMID:16543383; http://dx.doi.org/10.1210/er.2005-0017
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 2010; 11:124-36; PMID:20051984
- Ahmed EA, van der Vaart A, Barten A, Kal HB, Chen J, Lou Z, Minter-Dykhouse K, Bartkova J, Bartek J, de Boer P, et al. Differences in DNA double strand breaks repair in male germ cell types: lessons learned from a differential expression of Mdc1 and 53BP1. DNA Repair 2007; 6:1243-54; PMID:17376750; http://dx.doi.org/10.1016/j.dnarep.2007.02.011
- Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma 2003; 111:483-8; PMID:12743712; http://dx.doi.org/10.1007/s00412-002-0227-z
- Marcon L, Boissonneault G. Transient DNA strand breaks during mouse and human spermiogenesis new insights in stage specificity and link to chromatin remodeling. Biol Reprod 2004; 70:910-8; PMID:14645105; http://dx.doi.org/10.1095/biolreprod.103.022541
- Sakkas D, Alvarez JG. Sperm DNA fragmentation: mechanisms of origin, impact on reproductive outcome, and analysis. Fertil Steril 2010; 93:1027-36; PMID:20080235; http://dx.doi.org/10.1016/j.fertnstert.2009.10.046
- Sakkas D, Manicardi G, Bianchi PG, Bizzaro D, Bianchi U. Relationship between the presence of endogenous nicks and sperm chromatin packaging in maturing and fertilizing mouse spermatozoa. Biol Reprod 1995; 52:1149-55; PMID:7626715; http://dx.doi.org/10.1095/biolreprod52.5.1149
- Aitken RJ, De Iuliis GN, McLachlan RI. Biological and clinical significance of DNA damage in the male germ line. Int J Androl 2009; 32:46-56; PMID:19076252; http://dx.doi.org/10.1111/j.1365-2605.2008.00943.x
- Paul C, Povey JE, Lawrence NJ, Selfridge J, Melton DW, Saunders PT. Deletion of genes implicated in protecting the integrity of male germ cells has differential effects on the incidence of DNA breaks and germ cell loss. PLoS One 2007; 2:e989; PMID:17912366; http://dx.doi.org/10.1371/journal.pone.0000989
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071-8; PMID:19847258; http://dx.doi.org/10.1038/nature08467
- Kastan MB, Lim DS, Kim ST, Xu B, Canman C. Multiple signaling pathways involving ATM. Cold Spring Harbor Symp Quant Biol 2000; 65:521-6; PMID:12760069; http://dx.doi.org/10.1101/sqb.2000.65.521
- Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene 2002; 21:8967-80; PMID:12483513; http://dx.doi.org/10.1038/sj.onc.1206136
- Liu Y, West SC. More complexity to the Bloom's syndrome complex. Genes Dev 2008; 22:2737-42; PMID:18923071; http://dx.doi.org/10.1101/gad.1732808
- D’Andrea AD, Grompe M. The Fanconi anaemiaBRCA pathway. Nat Rev Cancer 2003; 3:23-34; PMID:12509764; http://dx.doi.org/10.1038/nrc970
- Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet 2009; 43:223-49; PMID:19686080; http://dx.doi.org/10.1146/annurev-genet-102108-134222
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 2011; 43:142-6; PMID:21240275; http://dx.doi.org/10.1038/ng.750
- Crossan GP, van der Weyden L, Rosado IV, Langevin F, Gaillard PH, McIntyre RE, Sanger Mouse Genetics P, Gallagher F, Kettunen MI, Lewis DY, et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet 2011; 43:147-52; PMID:21240276; http://dx.doi.org/10.1038/ng.752
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet 2003; 35:165-70; PMID:12973351; http://dx.doi.org/10.1038/ng1241
- Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol 2010; 17:294-8; PMID:20154706; http://dx.doi.org/10.1038/nsmb.1759
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160-6; PMID:17525332; http://dx.doi.org/10.1126/science.1140321
- Agoulnik AI, Lu B, Zhu Q, Truong C, Ty MT, Arango N, Chada KK, Bishop CE. A novel gene, Pog, is necessary for primordial germ cell proliferation in the mouse and underlies the germ cell deficient mutation, gcd. Hum Mol Genet 2002; 11:3047-53; PMID:12417526; http://dx.doi.org/10.1093/hmg/11.24.3047
- Nadler JJ, Braun RE. Fanconi anemia complementation group C is required for proliferation of murine primordial germ cells. Genesis 2000; 27:117-23; PMID:10951504; http://dx.doi.org/10.1002/1526-968X(200007)27:3%3c117::AID-GENE40%3e3.0.CO;2-7
- Wong JC, Alon N, McKerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet 2003; 12:2063-76; PMID:12913077; http://dx.doi.org/10.1093/hmg/ddg219
- Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev 2003; 17:2021-35; PMID:12893777; http://dx.doi.org/10.1101/gad.1103403
- de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21:776-98; PMID:11105904
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 2007; 8:735-48; PMID:17768402; http://dx.doi.org/10.1038/nrg2159
- De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol 2008; 9:273-84; PMID:18354421; http://dx.doi.org/10.1038/nrm2378
- Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM, et al. DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell 2014; 156:413-27; PMID:24485452; http://dx.doi.org/10.1016/j.cell.2013.12.023
- Bettegowda A, Wilkinson MF. Transcription and post-transcriptional regulation of spermatogenesis. Philos Trans Royal Soc London Series B, Biol Sci 2010; 365:1637-51; PMID:20403875; http://dx.doi.org/10.1098/rstb.2009.0196
- Romrell LJ, Bellve AR, Fawcett DW. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol 1976; 49:119-31; PMID:176074; http://dx.doi.org/10.1016/0012-1606(76)90262-1
- O’Bryan MK, Clark BJ, McLaughlin EA, D’Sylva RJ, O’Donnell L, Wilce JA, Sutherland J, O’Connor AE, Whittle B, Goodnow CC, et al. RBM5 is a male germ cell splicing factor and is required for spermatid differentiation and male fertility. PLoS Genet 2013; 9:e1003628; PMID:23935508; http://dx.doi.org/10.1371/journal.pgen.1003628
