ABSTRACT
CTLA-4 inhibition produces durable T cell–driven antitumor responses, but understanding which patients achieve a long-term benefit remains unclear. Deep sequencing of rearranged T cell receptor β (TCRβ) genes can monitor the effects of CTLA-4 inhibition and potentially identify patients with long-term survival.
Inhibition of cytotoxic T lymphocyte-associated antigen-4 (CTLA-4), one of a number of immune checkpoint receptors that negatively regulate T cells, has shown unique activity in melanoma and other solid tumors. As an immune checkpoint receptor that binds to B7, thereby blocking costimulation, CTLA-4 inhibits T cells after dendritic cells or other antigen presenting cells have primed them, preventing immune responses from overextending their welcome. Because CTLA-4 expression on the T cell surface is induced upon T cell activation, higher levels of CTLA-4 may represent pre-activated T cells in the periphery. CTLA-4 may also provide an important function for regulatory T cells (Treg), where constitutively high levels of CTLA-4 have also been observed.
Treatment with monoclonal antibodies (mAbs) that target CTLA-4, such as ipilimumab and tremelimumab, produce unique results that defy the conventions noted with directly cytotoxic therapies.Citation1 Approximately one of every 5 patients with advanced melanoma treated with ipilimumab, an IgG1 human mAb, are alive at 3 years or more, yet objective responses are relatively uncommon. Given the discrepancy between survival and response rates, it is conceivable that the reported response rates may underestimate clinical benefit, as patients treated with anti-CTLA-4 mAbs may develop delayed responses, or even early progression of disease prior to regression. Anti-CTLA-4 mAbs also are notable for the development of immune-mediated toxicities that underscore the risk of nonspecific T cell activation. Previous analyses reported dynamic immune responses to individual pre-defined cancer antigens,Citation2 or have linked an elevated lymphocyte count to overall survival,Citation3 but it remained unclear if clinical activity in humans was associated with pre-existing antitumor immunity or generation of new immune response. To understand who may obtain long-lasting benefit from treatment and who are at risk for self-reactive immunity, a deeper, systemic analysis of the entire circulating pool of unique T cells would be required.
Next generation sequencing of rearranged CDR3 sequences in the T cell receptor β (TCRβ) genes allows determination of the T cell repertoire at an unprecedented depth, ordering on more than 106 in individual clonotypes that can be identified.Citation4 To address how CTLA-4 blockade can affect T cell responses, next generation sequencing was performed on blood samples collected before and during dosing from patients who received anti-CTLA-4 treatments for metastatic melanoma and castration-resistant prostate cancer (CRPC).Citation5 Metastatic CRPC patients were treated with ipilimumab, an IgG1 human antibody, every 4 weeks in combination with granulocyte macrophage-colony stimulating factor.Citation6 Melanoma patients were dosed with tremelimumab, an IgG2 human antibody, every 3 months.Citation7
Regardless of antibody isotype or tumor histology, the study demonstrated rapid turnover and increased diversity of circulating T cell clones, to the point where in a few patients, there was minimal resemblance in the detectable T cell population before and after a single dose of anti-CTLA-4 treatment. These changes were amplified with multiple doses of anti-CTLA-4 mAbs. This pattern of repertoire evolution was also nearly universally noted in every patient analyzed in the study, suggesting that the increased diversity and global turnover of T cell clones reflected the systemic effects of indiscriminate T cell activation, expansion and mobilization ().
Figure 1. Anti-CTLA-4 monoclonal antibodies (mAbs) block CTLA-4 from inhibiting T cell activation. The resultant expansion and mobilization of activated T cells is reflected in the rapid turnover of the entire T cell repertoire in the periphery. In this representative example, each color represents a unique T cell clone as determined by next generation sequencing of rearranged TCRB genes and ranked by abundance.
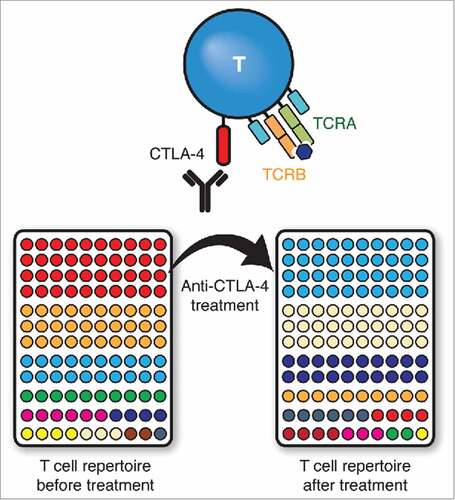
Despite the overall increased churn and diversity across the board, the study demonstrated that less turnover of the most common T cell clonotypes, at frequencies ≥ 10−3, is associated with longer survival. Worse outcomes were noted when patients lost more high frequency T cells, all of which largely consisted of effector cells. Immune responses to known tumor-associated antigens such as MART1 and gp100 for melanoma,Citation8 and PSA for CRPC were transient or difficult to detect. In contrast, cytomegalovirus-reactive clones, which typically carry high affinity TCR, were highly abundant and stable over time. These findings have important implications: that more favorable outcomes arise from a pre-existing, presumably high affinity T cell response, and that this response is likely to antigens not represented well by non-mutant self-proteins.
It is unclear whether these findings are unique for CTLA-4 or emblematic for the variety of T-cell modulating receptors and ligands, but these results do indicate that increased T-cell diversity and presence of a more focused pre-existing immunity are not necessarily unaligned. Indeed, these experiments indicate that for those patients who achieve more favorable outcomes, anti-CTLA4 mAbs are activating pre-existing tumor-reactive T cells and shaping the T cell repertoire, or more accurately, steadying the more abundant T cells of the repertoire to maintain tumor-specific, presumably high affinity T-cell clones that dominate the T cell pool. The effects of diversity on cancer immunity remain unknown; diversity with anti-CTLA-4 treatment may conceivably assist the cohort of patients without strong pre-existing immunity, by shuffling the repertoire to mobilize previously low frequency tumor-reactive clones. However, diversity may also have negative consequences by risking the selection of auto-reactive T cells.
In summary, the findings have important implications not only on the mechanism of action of anti-CTLA-4 mAbs, but on the usage of next generation sequencing for profiling immune response. It will be interesting to see if changes in circulating T cells from peripheral blood, rather than tumor tissue, could potentially have clinical importance with predicting response to immunomodulating agents in prospective studies. In addition, the potential of linking antigen specificity and repertoire sequencing could potentially open the door to novel immunotherapies.Citation9,Citation10
Abbreviations
| CRPC | = | castration-resistant prostate cancer |
| CTLA-4 | = | cytotoxic T lymphocyte-associated antigen-4 |
| mAbs | = | monoclonal antibodies |
| TCRβ | = | T cell receptor β |
| Treg | = | regulatory T cell |
Additional information
Funding
References
- Wolchok JD, Hodi FS, Weber JS, Allison JP, Urba WJ, Robert C, O'Day SJ, Hoos A, Humphrey R, Berman DM, et al. Development of ipilimumab: a novel immunotherapeutic approach for the treatment of advanced melanoma. Ann N Y Acad Sci. 2013;1291:1–13.
- Kwek SS, Cha E, Fong L. Unmasking the immune recognition of prostate cancer with CTLA4 blockade. Nat Rev Cancer. 2012;12:289–97.
- Ku GY, Yuan J, Page DB, Schroeder SEA, Panageas KS, Carvajal RD, Chapman PB, Schwartz GK, Allison JP, Wolchok JD. Single-institution experience with ipilimumab in advanced melanoma patients in the compassionate use setting: lymphocyte count after 2 doses correlates with survival. Cancer. 2010;116:1767–75.
- Klinger M, Kong K, Moorhead M, Weng L, Zheng J, Faham M. Combining next-generation sequencing and immune assays: a novel method for identification of antigen-specific T cells. PloS One. 2013;8:e74231.
- Cha E, Klinger M, Hou Y, Cummings C, Ribas A, Faham M, Fong L. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci Transl Med. 2014;6:238ra70.
- Fong L, Kwek SS, O'Brien S, Kavanagh B, McNeel DG, Weinberg V, Lin AM, Rosenberg J, Ryan CJ, Rini BI, et al. Potentiating endogenous antitumor immunity to prostate cancer through combination immunotherapy with CTLA4 blockade and GM-CSF. Cancer Res. 2009;69:609–15.
- Von Euw E, Chodon T, Attar N, Jalil J, Koya RC, Comin-Anduix B, Ribas A. CTLA4 blockade increases Th17 cells in patients with metastatic melanoma. J Transl Med. 2009;7:35.
- Comin-Anduix B, Lee Y, Jalil J, Algazi A, de la Rocha P, Camacho LH, Bozon VA, Bulanhagui CA, Seja E, Villanueva A, et al. Detailed analysis of immunologic effects of the cytotoxic T lymphocyte-associated antigen 4-blocking monoclonal antibody tremelimumab in peripheral blood of patients with melanoma. J Transl Med. 2008;6:22.
- Van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJA, Behjati S, Hilkmann H, el Atmioui D, et al. Tumor Exome Analysis Reveals Neoantigen-Specific T-Cell Reactivity in an Ipilimumab-Responsive Melanoma. J Clin Oncol. 2013;31:e439–e442.
- Lu Y-C, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, Davis L, Dudley ME, Yang JC, Samuels Y, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20:3401–10.
