Abstract
Inheritable epigenetic regulation is integral to the dynamic control of gene expression under different stimuli for cellular homeostasis and disease progression. Histone methylation is a common and important type of chromatin modification. LSD1, the first known histone lysine-specific demethylase, operates as a key component of several corepressor complexes during development and in disease states. In this review, we focus on the regulation of LSD1 in mammary carcinogenesis. LSD1 plays a role in promoting mammary tumor metastasis and proliferation and in maintaining mammary cancer stem cells. Therefore, LSD1 represents a viable therapeutic target for effective treatment of mammary carcinogenesis.
Keywords:
Abbreviations
| ATM | = | ataxia telangiectasia mutated |
| BRAF35 | = | BRCA2-associated factor 35 |
| BRCA1 | = | breast cancer type 1 susceptibility protein 1 |
| CSC | = | cancer stem cell |
| CtBP | = | C-terminal-binding protein |
| HDAC | = | histone deacetylases |
| MMTV | = | mouse mammary tumor virus |
| PHF8 | = | PHD finger protein 8 |
| RNF | = | RING finger protein |
| Sirt1 | = | NAD-dependent protein deacetylase sirtuin-1 |
| TAL1 | = | T-cell acute lymphoblastic leukemia 1 |
| TLX | = | tailless-like |
Introduction
The basic unit of chromatin, the nucleosome, is composed of DNA and histone proteins. Histone post-translational modifications include phosphorylation, acetylation, ubiquitylation, and methylation, among many others.Citation1 The balance between the absence and presence of histone modifications influences chromatin compaction and signaling protein complexes, and thus contributes to accurate gene expression.
During the last few years histone methylation has attracted increasing attention following the discovery that methylation markers are not static but under dynamic regulation. Histone methylation occurs on all basic residues: arginines, lysines, and histidines. The most extensively studied histone lysine methylation sites include lysine 4 on histone H3 (H3K4), H3K9, H3K27, H3K37, H3K36, H3K39, and H4K20.Citation1 In general, methylation sites of H3K9, H3K27, H3K37, or H4K20 are linked to the formation of tightly packed chromatin and gene silencing, whereas methylation on H3K4, H3K36, and H3K39 is associated with actively transcribed regions and gene activation.Citation2 Lysine can be monomethylated, dimethylated, or trimethylated. Histone methylation is reversible and can be modulated by methyltransferases and demethylases, which respectively catalyze the addition and removal of methyl groups on Lys and Arg residues of histone tails. To date, two evolutionarily conserved families of demethylases have been identified that utilize different reaction mechanisms to demethylate methyl-lysines: amine oxidases and jumonji c (JmjC)-domain-containing, iron-dependent dioxygenases.Citation3 The amine oxidase lysine-specific demethylase (LSD)1 was the first H3K4 lysine-specific demethylase to be identified. Recent studies have shed light on the role of LSD1 in the maintenance of pluripotency in stem cells and cancer progression.
LSD1 Structure and Complexes
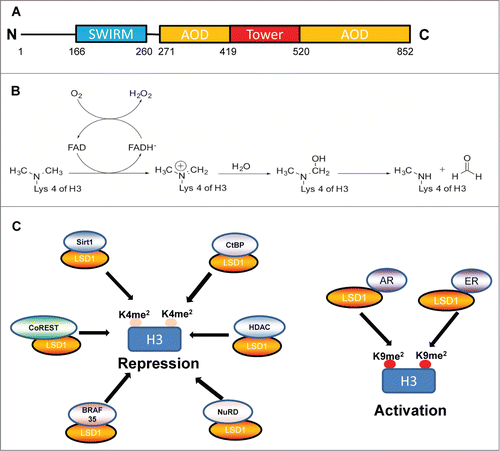
LSD1 contains 2 protein domains, a Swi3p, Rsc8p and Moira (SWIRM) domain and an amine oxidase (AO) domainCitation4 (). The SWIRM domain is present in several proteins that interact with histones whereas the AO domain mediates FAD-dependent demethylation.Citation5 The SWIRM and AO domains are closely associated with each other through extensive hydrophobic interactions to form a core structure.Citation6 The AO domain, which constitutes the main catalytic center, is divided by the tower domain into 2 functional lobes, the FAD-binding lobe and the substrate binding and recognition lobe (). The catalytic center is located within the substrate-binding lobe of the AO domain and is substantially more spacious and open in comparison with other FAD-dependent amine oxidases for accurate substrate placement.Citation6 Inside the active site cavity, 4 major invaginations with distinct chemical properties are used for sensing not only the histone tail sequence but also its chemical modifications.Citation6 Furthermore, there is a cleft between the SWIRM and the AO domains that functions as an additional recognition platform for substrate specificity.Citation6,7 LSD1 also contains a putative nuclear localization signal at its N-terminus and an insertion sequence protruding from the AO domain (the tower domain) that provides a surface platform for interaction with partners.Citation6 This tower domain mediates the interaction between LSD1 and REST corepressor 1 (CoREST). CoREST increases LSD1 stability by maintaining the conformation of LSD1 within the larger complex.Citation8 Recently, LSD1 has also been identified as a subunit of several complexes, including the CtBP, NRD, BRAF35, HDACCitation9 and Sirt1Citation10 complexes, in which it functions as a corepressor or coactivator (). The combination of different histone-modifying enzymes in one complex facilitates concurrent modifications of multiple sites on the histone and associated proteins.
Substrates of LSD1
Initially, LSD1 was shown to directly bind and demethylate monomethyl and dimethyl, but not trimethyl, H3K4. LSD1 functions as a flavoenzyme of the oxidoreductase/oxidase class that catalyzes 2-electron oxidation of its substrate to remove the methyl groups from H3K4. Recent emerging evidence shows that LSD1 functions to modulate other cellular processes through actions on non-histone proteins, such as p53 and DNA methyltransferase. Specifically, LSD1 demethylates dimethylated K370 of p53Citation11 and K1096 of DNA methyltransferase 1 (DNMT1). Demethylation of lysine 185 of E2F1 by LSD1 drives E2F degradation.Citation12 Interestingly, LSD1 is also directly recruited to sites of DNA damage in a manner dependent on RNF168 but independent of H2AX and ATM signaling.Citation13 These studies suggested that LSD1 can regulate both global and specific gene expression patterns through demethylation of histone and non-histone substrates. However, the specific functions of LSD1 acting as an “editor” can be specified, in part, by the associated proteins; LSD1 can interact with different transcription factors to facilitate distinct cellular functions. For example, interaction of LSD1 with retinoblastoma (Rb) results in binding of Rb to specific DNA regions in a cell-cycle dependent manner.Citation14 LSD1 is also recruited by nuclear receptor TLX to repress the expression of a TLX target gene and regulate neural stem cell proliferation.Citation15 In the context of repression-associated H3K9me2 marks, LSD1 facilitates androgen- or estrogen-dependent transcriptional activation in prostate or breast cancer cells by demethylation of H3K9 when associated with androgen receptor (AR) or estrogen receptor (ER) ().Citation4,16 Although isolated LSD1 does not act on Lys 9 directly in vitro,Citation17,18 the demethylation activity of LSD1 toward H3K9me2 is inhibited by LSD1 inhibitors such as pargyline and deprenyl in leukemia stem cells (LSCs) and other cell lines.Citation19,20 Likewise, LSD1 is required for activation of Gh transcription in pituitary precursor cells.Citation21 Therefore, LSD1 may contribute directly or indirectly to repress or activate transcription through interactions with various transcription factors and chromatin modifying enzymes.
Regulation of LSD1
LSD1 can be regulated at the transcriptional and post-transcriptional level. The microRNA miR-137 directly targets LSD1 mRNA to repress LSD1 expression in colon cancer and neuroblastoma cells.Citation22,23 Interestingly, TLX, an essential regulator of neural stem cell self-renewal, represses miR-137 expression by recruiting LSD1 to the genomic region of miR-137. Thus, miR-137 forms a feedback regulatory loop with TLX and LSD1 to control the dynamics between neural stem cell proliferation and differentiation during neural development.Citation24 LSD1 can be phosphorylated by protein kinase Cα (PKCα) in a circadian manner and the phosphorylated LSD1 forms a complex with CLOCK:BMAL1 to facilitate E-box-mediated transcriptional activation.Citation25 Interestingly, LSD1 is hyperphosphorylated upon nocodazole synchronization;Citation26 however, whether this phosphorylation is related to cell cycle-dependent association and the dissociation of LSD1 from chromatin is unknown.Citation27 In addition, LSD1 itself is regulated in a cell cycle-dependent manner. LSD1 is localized in the nuclei of cells at G1, S, and G2 phases of the cell cycle, consistent with its chromatin-modifying activity, but is displaced from the chromatin and mainly present in the cytoplasm of cells at mitosis.Citation27
Recently, several studies have been shown that the abundance of LSD1 is under the control of the ubiquitin-proteasome system.Citation8,28 CoREST is required for LSD1 stability in vivo and LSD1 is prone to proteasomal degradation in the absence of CoREST. Our group demonstrated that formation of a Snail-LSD1-CoREST ternary complex is critical for the stability and function of these proteins.Citation28 We further demonstrated that USP28 is a deubiquitinase for LSD1 and stabilizes LSD1 through deubiquitination.Citation29 It will be revealing to identify the E3 ligase that mediates LSD1 proteasomal degradation and the upstream signals that trigger this degradation.
LSD1 in Oncogenesis
It has been reported that LSD1 is associated with cancer in leukemia and several types of solid tumors, including non-small cell lung carcinoma (NSCLC), neuroblastoma, pancreatic cancer, prostate cancer, and breast cancer. Inhibition of LSD1 reduces or blocks cell growth in many of these tumors, whereas overexpression of LSD1 contributes to tumorigenesis through chromatin modification.Citation30 The role of LSD1 as essential regulator of LSCs has been demonstrated in mouse and human models of MLL-AF9 leukemia.Citation31 In T-cell acute lymphoblastic leukemia (T-ALL), LSD1 and PHF8 cooperate to epigenetically regulate Notch1 target genes.Citation32 This represents the first example of the dual role of LSD1 as activator and repressor in Notch-mediated target gene regulation. In addition, LSD1 is also associated with the hematopoietic-specific transcription factor TAL1/SCL. This association is disrupted by protein kinase A (PKA)-mediated phosphorylation of serine 172 in TAL1, and the destabilized TAL1–LSD1 interaction leads to H3K4 hypermethylation of the promoter and activation of TAL1 target genes.Citation33,34
LSD1 is significantly upregulated in pancreatic cancer and synergizes with hypoxia inducible factor-1α (HIF1α) to maintain glycolysis; this metabolic process fuels the uncontrolled proliferation in pancreatic cancer.Citation35 LSD1 is also expressed in cancer stem cells (CSCs) that facilitate bladder cancer development, and LSD1-mediated epigenetic modification of developmental genes may play important roles in maintaining pluripotency of these CSCs. Similarly, overexpression of LSD1 enhances cell growth and epithelial–mesenchymal transition (EMT) and correlates with shorter overall survival for patients with NSCLC and hepatocarcinomas.Citation36,37
LSD1 in Mammary Carcinogenesis
Breast cancer is the most common malignancy and the second leading cause of cancer deaths among women in the United States.Citation38 Breast cancer is a heterogeneous disease and has long been recognized to emerge from the progression of atypical ductal hyperplasia to ductal carcinoma in situ (DCIS), with evolution of this preinvasive lesion into invasive breast cancer.Citation39 The development of breast cancer is accompanied by a succession of genetic and epigenetic abnormalities. Epigenetic modifications, including DNA methylation, histone modifications, nucleosome remodeling, and modifications of noncoding RNAs (ncRNAs), are becoming increasingly recognized for their critical roles in normal differentiation and development.Citation40 Deregulation or misinterpretation of these heritable, fine-tuned epigenetic patterns lead to aberrant activation/inhibition of signaling pathways that can culminate in cancer.Citation41,42 Overexpression of LSD1 positively correlates with ER− status in breast carcinoma.Citation43 Additionally, LSD1 expression gradually increases during tumor progression from low-, intermediate- and high-grade DCIS to invasive ductal breast carcinoma.Citation44 These associations implicate a key role for LSD1 in breast cancer development, proliferation, metastasis, and CSC propagation.
LSD1 in Mammary Cancer Initiation
Upregulation of LSD1 may be an early tumor-promoting event in breast carcinoma. Exposure to carcinogens influences the expression of multiple genes critical for early-stage mammary carcinogenesis through the induction of LSD1 expression.Citation45 LSD1 relocates from the periphery to the nucleus after carcinogen treatment of human mammary epithelial cell lines. Demethylation of H3K4 by LSD1 influences the expression of genes such as p57kip2, which encodes a cyclin-dependent kinase inhibitor that is known to be essential for breast tumor formation.
LSD1 in Breast Cancer Proliferation
The estrogen receptors belong to the nuclear receptor superfamily. ERα is predominantly expressed in the majority of breast tumors and has emerged as a major target for breast cancer therapeutics.Citation46 LSD1 enzymatic activity is required for ERα function. Inhibition of LSD1 abrogates recruitment of estrogen-liganded ERα to estrogen-responsive gene promoters, and exhibits a strong antiproliferative effect on breast cancer cells.Citation47 However, CDK2-associated cullin (CAC1), a ligand-independent ERα corepressor, interacts with LSD1 and alleviates the ERα coactivator function. Interestingly, CAC1 and LSD1 reciprocally modulate paclitaxel-mediated cell death in ERα-positive, paclitaxel-resistant MCF7 cells.Citation48 In addition, LSD1 also interacts with β-catenin to regulate the expression of the tumor suppressor Lefty1. The mRNA levels of LSD1 and β-catenin are inversely correlated with expression of Lefty1 in breast tumors. LSD1 also closely interacts with histone deacetylases (HDACs) to control breast cancer cell growth. Combined treatment with LSD1 inhibitor, pargyline, and the HDAC inhibitor, SAHA (Vorinostat), leads to superior growth inhibition and apoptotic death in triple-negative breast cancer (TNBC) cells.Citation49,50
LSD1 in Breast Cancer Metastasis
More than 90% of cancer-related deaths in patients with solid tumors are caused by metastases rather than the primary tumor.Citation51 Metastasis is an exceedingly complex process involving tumor cell invasion, intravasation, survival in the circulation of the blood or lymph system, extravasation, and subsequent outgrowth in new tissues and organs. Initiation of tumor cell invasion is a prerequisite for the metastatic cascade. The increased motility and invasiveness of metastatic tumor cells are reminiscent of the events that occur during the EMT, a characteristic feature of physiologic and pathophysiologic events that occur during embryonic development and morphogenesis, chronic tissue remodeling and fibrosis of mature organs, and cancer progression and metastasis.Citation52-54 Several transcription factors have been implicated in the regulation of EMT, including the zinc finger proteins of the Snail/Slug family, the basic helix-loop-helix factor Twist, and SIP1.Citation52,55,56 We and others have demonstrated that Snail recruits LSD1 to epithelial gene promoters with demethylation of H3K4me2 and subsequent silencing of target genes to enhance tumor metastasis.Citation28,57,58 In addition, Slug, another transcriptional factor of the Snail family, can also interact with LSD1 to facilitate tumor metastasis.Citation59 Both Snail and Slug recruit LSD1 and bind to a series of E-boxes located within the BRCA1 promoter to repress BRCA1 expression.
LSD1 and Breast Cancer Stem Cells
Breast CSCs are characterized by low levels of heat stable antigen (CD24) and high levels of hyaluronan receptor (CD44) and EpCAM expression.Citation60 This subpopulation of cells has the ability to self-renew and initiate tumor formation, and is intrinsically resistant to therapies. It has been hypothesized that the similar features shared by stem and cancer cells are attributed to their shared gene expression patterns. The dedifferentiation process that results in tumor cells is somewhat reminiscent of the acquisition of pluripotency.Citation61 Recently, a series of studies have linked LSD1 function to the control of gene expression during embryonic stem cell (ESC) differentiation. LSD1 is essential to maintain the proper balance between H3k4me2/m3 and H3K27me3 at the lineage-specific genes in human ESCs.Citation62 The Young group demonstrated that LSD1 formed a complex with NuRD (nucleosome remodeling and histone deacetylase) to decommission enhancers of the pluripotency program during differentiation.Citation63 LSD1 functions as a gatekeeper for the onset of differentiation of trophoblast stem cells (TSCs). LSD1-deficient TSCs display features of differentiation initiation, including alterations in cell morphology and increased migration and invasion.Citation64
Recently, we found that LSD1 is also involved in the regulation of CSC self-renewal and differentiation.Citation29 We found that expression of LSD1 was significantly higher in cells isolated from MMTV-Wnt1 tumors than in those from normal mammary glands or tumors from MMTV-HER2 mice. Cells isolated from MMTV-Wnt1 tumors grew as sphere-cluster structures in a monolayer; LSD1 knockdown changed the morphology and the cells became disassociated and scattered, a phenomenon associated with cellular differentiation.Citation65,66 LSD1 knockdown also resulted in fewer and smaller tumorspheres under nonadherent conditions, with a shift in the cytoarchitecture of these colonies from solid organoid to a more acinar-like structure. These morphological changes were concomitant with a reduction in the expression of the basal cell marker CK14 and smooth muscle actin (SMA), and increased expression of the luminal molecules E-cadherin and ELF5. Consistent with these findings, LSD1 knockdown reduced the expression of Sox2 and Oct4. Therefore, LSD1 plays a critical role in controlling CSC-like properties and tumorigenicity in breast cancer. In agreement with these findings, LSD1 inactivation causes selective increased methylation of H3K4 and H3K9 on Sox2 and cell cycle genes, but only induces increased H3K4 methylation on the promoters of differentiation genes, resulting in the downregulation of Sox2 and cyclins and the induction of genes required for differentiation.Citation67
LSD1 Inhibitors
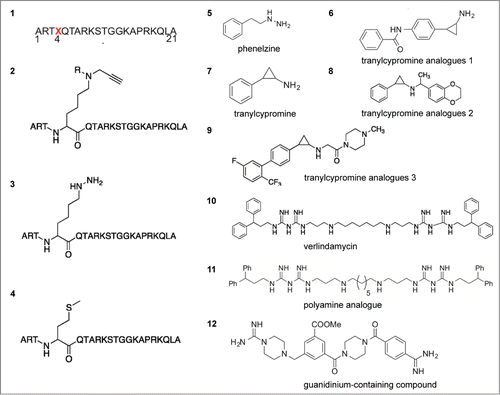
LSD1 is a therapeutic target and inhibition of LSD1 is an effective strategy for multiple cancer types. To date, a handful of small molecular inhibitors of LSD1 that are based on different mechanisms have been developed ().Citation68,69 The early LSD1 inhibitors such as tranylcypromine (TCP) and pargyline target its demethylase activity, and are time-dependent and irreversible LSD1 inhibitors. More recently, additional inhibitors with increased selectivity for LSD1 have been developed as derivatives of these scaffolds. These analogs of TCP have been tested preclinically in the treatment of acute myelogenous leukemia (AML).Citation19,70 The polyamine derivatives biguanide and bisguanidine are effective inhibitors of LSD1 in human colon and breast cancer cell lines.Citation20,71 Namoline is another novel selective and reversible inhibitor that is a promising starting compound for the development of effective therapeutics.Citation72 In addition, inhibitors based on LSD1 peptide also have been developed. For example, a 21-mer peptide analog that targets the H3K4 substrate region of LSD1 has been designed, in which Lys4 is replaced by a methionine. This linear peptide inhibits LSD1 activity with a Ki of 0.04 μM, and inhibits LSD1 bound to CoREST with a Ki of 0.05 μM.Citation73 Another novel LSD1 antagonist, SP2509, attenuates the binding of LSD1 with CoREST and increases the permissive H3K4me3 mark on target gene promoters. Cotreatment with the pan-HDAC inhibitor (HDI) panobinostat (PS) and SP2509 is synergistically lethal to cultured and primary AML blasts.Citation74 The LSD1 antagonist GSK2879552 is a potent, selective, mechanism-based inactivator of LSD1/CoREST activity and is in clinical trial. However, currently available small LSD1 inhibitors display poor selectivity, low potency, or in vivo toxicity, limiting further interrogation of the contribution of LSD1 to cancer at the organismal level. Identification of novel LSD1 inhibitors that are potent, selective, and reversible is essential to further elucidate the role of LSD1 in cancer.
Future Perspectives
Extensive studies illustrate the importance of LSD1 in maintaining expression of oncogenic programs and blocking differentiation of multiple cancer types, and highlight the potential therapeutic implications of targeting this important epigenetic regulator in mammary carcinogenesis. Despite these exciting findings, some very important questions still need to be addressed. First, what environmental signals trigger the formation and stability of LSD1 complexes present on different gene targets? Whether these extrinsic signals affect LSD1 activity indirectly through intracellular signaling pathways, which have been known to affect nuclear transcription, or directly through some kinase that activates or suppresses LSD1 activity remains to be determined. Second, what are the molecular signaling mechanisms that determine the inhibitory or stimulatory nature and function of LSD1 complexes? Previous studies indicate that association with AR or ER determines the coactivator versus corepressor function of LSD1. However, more recent results suggest that LSD1 activity is instead determined by properties of the element to which it is being recruited. For example, LSD1 is more likely to demethylate hypoacetylated nucleosomes.Citation8 Moreover, phosphorylation of H3T11 enhances the demethylation of H3K9me by LSD1,Citation75 whereas phosphorylation of H3T6 suppresses the LSD1-mediated demethylation of H3K4me1/2.Citation76 Therefore, it will be interesting to determine the extent to which histone modification or other factors regulate the function of LSD1 as a suppressor versus an activator in distinct cell contexts. Third, how does LSD1 target specific genes and affect chromatin organization during differentiation as opposed to disease progression? LSD1 is the only histone demethylase known to have dual H3K4me2/K9me2 specificities; this unique attribute makes it important for maintaining expression of oncogenic gene programs and blocking differentiation in embryonic stem cells. Its dual functions serve to “decommission” ESC enhancers since pluripotent genes that need to be shut off during the initial steps of differentiation rely on LSD1 for inactivation. Mutant ESCs lacking this enzyme have difficulty progressing through differentiation as a result of unabated transcription of pluripotency genes.Citation63 It would be appealing to properly define the dynamic changes that LSD1 complexes undergo at different stages of embryonic stem cell development. Finally, what long-term effect will LSD1 inhibitors have on normal stem cells and the differentiation program? Considering the plethora of LSD1 inhibitors that have been developed, it is important to undertake a more detailed analysis of the effect of LSD1 inhibitors on normal human stem cells, particularly during human mammary gland development. An understanding of these processes will improve our knowledge of epigenetic modifications in general, and their functional outcome in cancer and stem cell differentiation and reprogramming. Future discoveries will yield more specific and efficient LSD1 inhibitors to suppress tumor growth.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all the investigators that contribute to the LSD1 field and we apologize to those whose work is important but that we are unable to cite here. We also thank Dr. Catherine Anthony for critical reading and editing of this manuscript.
Funding
This work was supported by grants from NIH (CA125454), Mary Kay Ash Foundation, an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103527-06 (to BPZ), and American Cancer Society Research Scholar Award (RSG13187-01 to YW).
References
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13:343-57; PMID:22473383; http://dx.doi.org/10.1038/nrg3173
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6:838-49; PMID:16261189; http://dx.doi.org/10.1038/nrm1761
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012; 13:297-311; PMID:22473470
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005; 437:436-9; PMID:16079795
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119:941-53; PMID:15620353; http://dx.doi.org/10.1016/j.cell.2004.12.012
- Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol 2006; 13:626-32; PMID:16799558; http://dx.doi.org/10.1038/nsmb1113
- Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol 2007; 14:535-9; PMID:17529991; http://dx.doi.org/10.1038/nsmb1255
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005; 19:857-64; PMID:16140033; http://dx.doi.org/10.1016/j.molcel.2005.08.027
- Nicholson TB, Chen T. LSD1 demethylates histone and non-histone proteins. Epigenetics 2009; 4:129-32; PMID:19395867; http://dx.doi.org/10.4161/epi.4.3.8443
- Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 2011; 42:689-99; PMID: 21596603; http://dx.doi.org/10.1016/j.molcel.2011.04.020
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007; 449:105-8; PMID:17805299; http://dx.doi.org/10.1038/nature06092
- Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell 2010; 39:152-60; PMID:20603083; http://dx.doi.org/10.1016/j.molcel.2010.06.006
- Mosammaparast N, Kim H, Laurent B, Zhao Y, Lim HJ, Majid MC, Dango S, Luo Y, Hempel K, Sowa ME, et al. The histone demethylase LSD1/KDM1A promotes the DNA damage response. J Cell Biol 2013; 203:457-470; PMID:24217620; http://dx.doi.org/10.1083/jcb.201302092
- Chau CM, Deng Z, Kang H, Lieberman PM. Cell cycle association of the retinoblastoma protein Rb and the histone demethylase LSD1 with the Epstein-Barr virus latency promoter Cp. J Virol 2008; 82:3428-37; PMID: 18216119; http://dx.doi.org/10.1128/JVI.01412-07
- Sun, G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol 2010; 30:1997-2005; PMID:20123967; http://dx.doi.org/10.1128/MCB.01116-09
- Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 2007; 128:505-18; PMID:17289570; http://dx.doi.org/10.1016/j.cell.2006.12.038
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119:941-53; PMID:15620353; http://dx.doi.org/10.1016/j.cell.2004.12.012
- Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett 2005; 579:2203-7; PMID:15811342; http://dx.doi.org/10.1016/j.febslet.2005.03.015
- Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012; 21:473-87; PMID:22464800; http://dx.doi.org/10.1016/j.ccr.2012.03.014
- Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA Jr. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci U S A 2007; 4:8023-8; PMID:17463086; http://dx.doi.org/10.1073/pnas.0700720104
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007; 446:882-7; PMID:17392792; http://dx.doi.org/10.1038/nature05671
- Balaguer, F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, Goel A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res 2010; 70:6609-18; PMID:20682795; http://dx.doi.org/10.1158/0008-5472.CAN-10-0622
- Althoff, K, Koopman R, Denisov D, Schall P. MiR-137 functions as a tumor suppressor in neuroblastoma by downregulating KDM1A. Int J Cancer 2013; 133:1064-73; PMID:23323208; http://dx.doi.org/10.1002/ijc.28091
- Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun 2011; 2:529; PMID:22068596; http://dx.doi.org/10.1038/ncomms1532
- Nam HJ, Boo K, Kim D, Han DH, Choe HK, Kim CR, Sun W, Kim H, Kim K, Lee H, et al. Phosphorylation of LSD1 by PKCalpha is crucial for circadian rhythmicity and phase resetting. Mol Cell 2014; 53:791-805; PMID:24582500; http://dx.doi.org/10.1016/j.molcel.2014.01.028
- Lv S, Bu W, Jiao H, Liu B, Zhu L, Zhao H, Liao J, Li J, Xu X. LSD1 is required for chromosome segregation during mitosis. Eur J Cell Biol 2010; 89:557-63; PMID:20189264; http://dx.doi.org/10.1016/j.ejcb.2010.01.004
- Nair VD, Ge Y, Balasubramaniyan N, Kim J, Okawa Y, Chikina M, Troyanskaya O, Sealfon SC. Involvement of histone demethylase LSD1 in short-time-scale gene expression changes during cell cycle progression in embryonic stem cells. Mol Cell Biol 2012; 32:4861-76; PMID:23028048; http://dx.doi.org/10.1128/MCB.00816-12
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 2010; 29, 1803-16; PMID:20389281; http://dx.doi.org/10.1038/emboj.2010.63
- Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C, Evers BM, Zhou BP. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep 2013; 5:224-36; PMID: 24075993; http://dx.doi.org/10.1016/j.celrep.2013.08.030
- Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer 2011; 128:574-86; PMID: 20333681; http://dx.doi.org/10.1002/ijc.25349
- Lokken AA, Zeleznik-Le NJ. Breaking the LSD1/KDM1A addiction: therapeutic targeting of the epigenetic modifier in AML. Cancer Cell 2012; 21:451-3; PMID:22516254; http://dx.doi.org/10.1016/j.ccr.2012.03.027
- Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, Lelievre JD, Levy Y, Benkirane M. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell 2012; 48:445-58; PMID:23022380; http://dx.doi.org/10.1016/j.molcel.2012.08.022
- Hu X, Li X, Valverde K, Fu X, Noguchi C, Qiu Y, Huang S. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc Natl Acad Sci U S A 2009; 106:10141-6; PMID:19497860; http://dx.doi.org/10.1073/pnas.0900437106
- Li Y, Deng C, Hu X, Patel B, Fu X, Qiu Y, Brand M, Zhao K, Huang S. Dynamic interaction between TAL1 oncoprotein and LSD1 regulates TAL1 function in hematopoiesis and leukemogenesis. Oncogene 2012; 31:5007-18; PMID:22310283; http://dx.doi.org/10.1038/onc.2012.8
- Qin Y, Zhu W, Xu W, Zhang B, Shi S, Ji S, Liu J, Long J, Liu C, Liu L, et al. LSD1 sustains pancreatic cancer growth via maintaining HIF1alpha-dependent glycolytic process. Cancer Lett 2014; 347:225-32; PMID: 24561118; http://dx.doi.org/10.1016/j.canlet.2014.02.013
- Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One 2012; 7:e35065; PMID:22493729; http://dx.doi.org/10.1371/journal.pone.0035065
- Zhao ZK, Dong P, Gu J, Chen L, Zhuang M, Lu WJ, Wang DR, Liu YB. Overexpression of LSD1 in hepatocellular carcinoma: a latent target for the diagnosis and therapy of hepatoma. Tumour Biol 2013; 34, 173-80; PMID:23015317; http://dx.doi.org/10.1007/s13277-012-0525-x
- DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin 2011; 61:409-18; PMID:21969133; http://dx.doi.org/10.3322/caac.20134
- Rivenbark AG, Coleman WB. Field cancerization in mammary carcinogenesis - Implications for prevention and treatment of breast cancer. Exp Mol Pathol 2012; 93:391-8; PMID:23142414; http://dx.doi.org/10.1016/j.yexmp.2012.10.018
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell 2010; 19:698-711; PMID:21074720; http://dx.doi.org/10.1016/j.devcel.2010.10.005
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012; 150:12-27; PMID: 22770212; http://dx.doi.org/10.1016/j.cell.2012.06.013
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128:683-92; PMID:17320506; http://dx.doi.org/10.1016/j.cell.2007.01.029
- Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 2010; 31:512-20; PMID:20042638; http://dx.doi.org/10.1093/carcin/bgp324
- Serce N, Gnatzy A, Steiner S, Lorenzen H, Kirfel J, Buettner R. Elevated expression of LSD1 (Lysine-specific demethylase 1) during tumour progression from pre-invasive to invasive ductal carcinoma of the breast. BMC Clin Pathol 2012; 12:13; PMID:22920283; http://dx.doi.org/10.1186/1472-6890-12-13
- Bradley C, van der Meer R, Roodi N, Yan H, Chandrasekharan MB, Sun ZW, Mernaugh RL, Parl FF. Carcinogen-induced histone alteration in normal human mammary epithelial cells. Carcinogenesis 2007; 28:2184-92; PMID:17468514; http://dx.doi.org/10.1093/carcin/bgm100
- Cosman F, Lindsay R. Selective estrogen receptor modulators: clinical spectrum. Endocr Rev 1999; 20:418-34; PMID:10368777
- Pollock JA, Larrea MD, Jasper JS, McDonnell DP, McCafferty DG. Lysine-specific histone demethylase 1 inhibitors control breast cancer proliferation in ERalpha-dependent and -independent manners. ACS Chem Biol 2012; 7:1221-31; PMID:22533360; http://dx.doi.org/10.1021/cb300108c
- Kim J, Park UH, Moon M, Um SJ, Kim EJ. Negative regulation of ERalpha by a novel protein CAC1 through association with histone demethylase LSD1. FEBS Lett 2013; 587:17-22; PMID:23178685; http://dx.doi.org/10.1016/j.febslet.2012.10.054
- Vasilatos SN, Katz TA, Oesterreich S, Wan Y, Davidson NE, Huang Y. Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells. Carcinogenesis 2013; 34:1196-207; PMID:23354309; http://dx.doi.org/10.1093/carcin/bgt033
- Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat 2012; 131:777-89; PMID:21452019; http://dx.doi.org/10.1007/s10549-011-1480-8
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell 2006; 127:679-95; PMID:17110329; http://dx.doi.org/10.1016/j.cell.2006.11.001
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nature reviews. Mol Cell Biol 2002; 3:155-166; PMID:11994736
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007; 7:415-28; PMID:17508028; http://dx.doi.org/10.1038/nrc2131
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nature reviews. Mol Cell Biol 2006; 7:131-42; PMID:16493418
- Hartwell KA, Muir B, Reinhardt F, Carpenter AE, Sgroi DC, Weinberg RA. The Spemann organizer gene, goosecoid, promotes tumor metastasis. Proc Natl Acad Sci U S A 2006; 103:18969-74; PMID:17142318; http://dx.doi.org/10.1073/pnas.0608636103
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117:927-39; PMID:15210113; http://dx.doi.org/10.1016/j.cell.2004.06.006
- Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 2010; 29, 4896-904; PMID:20562920; http://dx.doi.org/10.1038/onc.2010.234
- Lin Y, Kang T, Zhou BP. Doxorubicin enhances Snail/LSD1-mediated PTEN suppression in a PARP1-dependent manner. Cell Cycle 2014; 13:1708-16; PMID:24675890
- Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic breast cancer 1, early onset (BRCA1) repression. Proc Natl Acad Sci U S A 2012; 109:16654-9; PMID: 23011797; http://dx.doi.org/10.1073/pnas.1205822109
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100:3983-8; PMID:12629218; http://dx.doi.org/10.1073/pnas.0530291100
- Kim J, Orkin SH. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med 2011; 3:75; PMID:22126538; http://dx.doi.org/10.1186/gm291
- Adamo A, Sesé B, Boue S, Castaño J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol 2011; 13:652-9; PMID: 21602794; http://dx.doi.org/10.1038/ncb2246
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 2012; 482:221-5; PMID:22297846; http://dx.doi.org/10.1038/nature10805
- Zhu, D, Hölz S, Metzger E, Pavlovic M, Jandausch A, Jilg C, Galgoczy P, Herz C, Moser M, Metzger D, et al. Lysine-specific demethylase 1 regulates differentiation onset and migration of trophoblast stem cells. Nat Commun 2014; 5:3174; PMID:24448552; http://dx.doi.org/10.1038/ncomms4174
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012; 148:1015-28; PMID:22385965; http://dx.doi.org/10.1016/j.cell.2012.02.008
- Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, Hinds PW. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell 2010; 17:65-76; PMID: 20129248; http://dx.doi.org/10.1016/j.ccr.2009.11.024
- Zhang X, Lu F, Wang J, Yin F, Xu Z, Qi D, Wu X, Cao Y, Liang W, Liu Y, et al. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep 2013; 5:445-7; PMID:24139802; http://dx.doi.org/10.1016/j.celrep.2013.09.018
- Kumarasinghe IR, Woster PM. Synthesis and evaluation of novel cyclic Peptide inhibitors of lysine-specific demethylase 1. ACS Med Chem Lett 2014; 5:29-33; PMID:24883177; http://dx.doi.org/10.1021/ml4002997
- Prusevich P, Kalin JH, Ming SA, Basso M, Givens J, Li X, Hu J, Taylor MS, Cieniewicz AM, Hsiao PY, et al. A selective phenelzine analogue inhibitor of histone demethylase LSD1. ACS Chem Biol 2014; 9:1284-93; PMID:24707965; http://dx.doi.org/10.1021/cb500018s
- Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med 2012; 18:605-11; PMID:22406747; http://dx.doi.org/10.1038/nm.2661
- Zhu Q, Huang Y, Marton LJ, Woster PM, Davidson NE, Casero RA Jr. Polyamine analogs modulate gene expression by inhibiting lysine-specific demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids 2012; 42:887-98; PMID:21805138; http://dx.doi.org/10.1007/s00726-011-1004-1
- Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, Jung M, Forne I, Imhof A, Janzer A, et al. Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. Int J Cancer 2012; 131:2704-9; PMID:22447389; http://dx.doi.org/10.1002/ijc.27555
- Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J Biol Chem 2005; 280:41360-5; PMID: 16223729; http://dx.doi.org/10.1074/jbc.M509549200
- Fiskus W, Sharma S, Shah B, Portier BP, Devaraj SG, Liu K, Iyer SP, Bearss D, Bhalla KN. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia 2014; PMID:24699304; http://dx.doi.org/10.1038/leu.2014.119
- Metzger E, Yin N, Wissmann M, Kunowska N, Fischer K, Friedrichs N, Patnaik D, Higgins JM, Potier N, Scheidtmann KH, et al. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat Cell Biol 2008; 10:53-60; PMID:18066052; http://dx.doi.org/10.1038/ncb1668
- Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Müller JM, Greschik H, Kirfel J, Ji S, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature 2010; 464:792-6; PMID:20228790; http://dx.doi.org/10.1038/nature08839