Abstract
Background. Glioblastoma (GBM) is the most common malignant brain tumor in adults and is nearly always fatal. Emerging evidence suggests that human Cytomegalovirus (HCMV) is present in 90–100% of GBMs and that add-on antiviral treatment for HCMV show promise to improve survival.
Methods. In a randomized, placebo-controlled trial of valganciclovir in 42 GBM patients, blood samples were collected for analyses of HCMV DNA, RNA, reactivity against HCMV peptides, IgG, and IgM at baseline and at 3, 12, and 24 weeks of treatment.
Results. All 42 tumors were positive for HCMV protein. All patients examined had at least one blood sample positive for HCMV DNA, 63% were HCMV RNA positive, and 21% were IgM positive. However, 29% of GBM patients were IgG negative for HCMV. Five of these samples were positive in an enzyme-linked immunosorbent assay (ELISA) that used antigens derived from a clinical isolate. Blood T cells from 11 of 13 (85%) HCMV IgG-negative GBM patients reacted against HCMV peptides. Valganciclovir did not affect IgG titers, DNA, or RNA levels of the HCMV immediate early (HCMV IE) gene in blood.
Conclusion. In GBM patients, HCMV activity is higher than in healthy controls and serology is a poor test to define previous or active HCMV infection in these patients.
Abbreviations:
- ELISA, enzyme-linked immunosorbent assay
- FACS, flow cytometry analyses
- FITC, fluorescein isothiocyanate
- GBM, glioblastoma
- HCMV, human Cytomegalovirus
- HCMV IE, human Cytomegalovirus-immediate early
- HIV, human immunodeficiency virus
- HSV, herpes simplex virus
- PBMC, Peripheral blood mononuclear cells
- PBS, Phosphate buffered saline
- PCR, polymerase chain reaction
- VIGAS study, Efficacy and Safety of Valcyte® as an Add-on Therapy in Patients with Malignant Glioblastoma and cytomegalovirus infection
- SEB, staphylococcal snterotoxin B
- Valcyte
- valganciclovir
Introduction
GBM is the most aggressive malignant primary brain tumor in adults and is nearly always fatal. Despite advances in diagnostic imaging, surgery, and radiation therapy and the development of new antineoplastic agents, the survival rate for patients with GBM remains dismal. With current standard therapy, median overall survival (OS) for GBM patients is less than 15 mo after diagnosis.Citation1,2
An active HCMV infection, as determined by HCMV protein expression, can be detected in tumor cells in 90–100% of GBM patients;Citation3-8 however, the infection is absent in healthy surrounding tissues, indicating a potential oncomodulatory or oncogenic role of HCMV. We found that the median OS of GBM patients correlates with the grade of HCMV infection in tumor tissue.Citation7,9 Patients with a low-grade infection (<25% infected cells) at diagnosis survived significantly longer than those with a high-grade infection and had a higher 2 y survival rate (63.6% vs. 17.2%, p = 0.003).Citation7 These findings imply that HCMV affects tumor progression, consistent with substantial evidence that HCMV has an oncomodulatory or even a direct oncogenic role in tumor development.Citation10-14
To determine whether treatment for HCMV improves the prognosis for GBM patients, we performed an exploratory clinical study, Valcyte Treatment of Glioblastoma Patients in Sweden (VIGAS), to assess the safety and potential efficacy of valganciclovir as an add-on therapy.Citation15 Forty-two patients with GBM were enrolled and received valganciclovir or placebo for 24 weeks in combination with temozolomide and/or radiation therapy.Citation15 We observed trends but no statistically significant differences in tumor growth at 12 and 24 weeks between the placebo and valganciclovir groups; however, survival was significantly higher among patients who received long-term Valcyte treatment. Therefore, several patients were prescribed valganciclovir for compassionate use at our hospital. In 50 GBM patients who received valganciclovir in addition to standard therapy, survival was remarkably improved, especially in those receiving continuous valganciclovir treatment.Citation16
In this study, plasma samples collected from 42 VIGAS patients at baseline and after 3, 12, and 24 weeks of treatment were analyzed for IgG and IgM antibodies against HCMV and for HCMV IE DNA and RNA; T-cell reactivity against HCMV peptides (IE and pp65) were examined at baseline. Plasma samples from 130 healthy blood donors (controls) were tested for HCMV IgG and IgM. We found evidence of a higher activity of HCMV IgM in GBM patients and a substantial proportion of HCMV-seronegative patients with evidence of HCMV infection in their tumor and blood cells. These observations have important implications for the currently ongoing immunotherapy trials for GBM patients.
Results
Serology is a poor test to define HCMV infection status in GBM patients
The VIGAS patients provided blood samples at the indicated time points as long as they were in the study; however, patients whose tumors recurred and exited the study did not provide blood samples, and one patient was lost to follow up at 4.5 mo. Therefore, the numbers of total blood samples tested at the indicated time points change. The number of available samples analyzed is indicated in each analysis. All 42 GBMs were positive for HCMV proteins by immunohistochemistry, as required for entry into the study, and blood cells from all patients were positive for HCMV IE DNA at least once during the 24-week study phase (). Sequencing of polymerase chain reaction (PCR) products confirmed the presence of the HCMV IE gene in blood cells. Nevertheless, only 30 of the 42 patients (71%) were HCMV IgG positive by a commercial ELISA test utilizing HCMV AD169 antigens (Dade Behring) (). One patient who was negative at base line seroconverted at week 12. A similar rate (70%) of CMV IgG positivity was observed among the 50 healthy age-matched controls. Thus, even though HCMV proteins in the tumor specimens were detected with two different antibodies (one to IE and one to a late HCMV protein) and HCMV IE DNA was present in blood cells of all these patients, 13 of 42 patients (31%) were IgG negative at baseline, as shown with this serology test. One of these patients was positive for HCMV IgG (and HCMV DNA) at 12 weeks.
Table 1. HCMV IgG, IgM, DNA, and RNA in serum and tumor samples from glioblastoma patients treated with valganciclovir or placebo.
Figure 1. Levels of HCMV IgG and IgM antibodies at baseline (T0W) and after treatment for 3 weeks (T3W) and 24 weeks (T24W) in VIGAS patients and healthy controls (HC). (A) IgM levels and HC (n = 130). T0 vs. HC, p < 0.0001; T3W vs. HC, p <0 .0001; T12W vs. HC, p = 0.0008; T24W vs. HC, p < 0.0001; H0 vs. T12W, p = 0.009. (B) IgG levels in VIGAS patients (n = 42) and HC (n = 50). (C) IgG levels in placebo-treated VIGAS patients (n = 20) and in HC (n = 50). (D) IgG levels in valganciclovir-treated VIGAS patients (n = 22) and in HC (n = 50). T0 vs. HC, p = 0.005; T0 vs. T3W, p = 0.023.
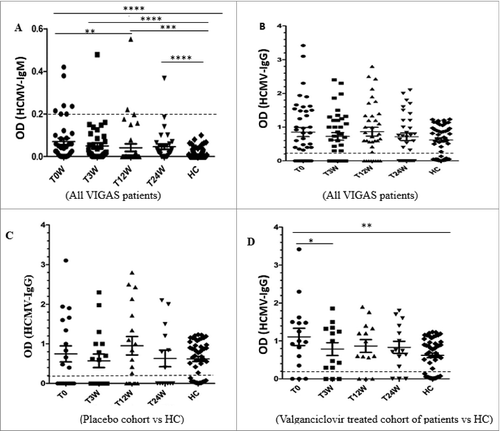
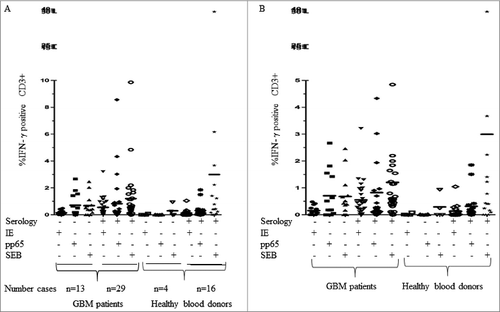
To further determine whether this result was due to the use of one specific kit or whether the patients lacked detectable IgG antibodies against HCMV, we examined the 12 HCMV-negative sera with three additional tests: a commercial kit utilizing HCMV AD169 antigens (BioSite), an in-house test also utilizing HCMV AD169 antigens (used for routine clinical diagnostics for many years at the clinical virology laboratory at the Karolinska University hospital), and an in-house test that utilize antigens of a clinical HCMV isolate in the ELISA assay. All three ELISA tests using HCMV AD169 antigens gave the same results; all 12 HCMV IgG-negative patients remained negative (). However, five serum samples (42%) were HCMV IgG positive (), when we used an ELISA test with antigens from a clinical HCMV isolate.Citation17 Furthermore, of the 12 patients who were HCMV seronegative by ELISA utilizing AD169 antigen, 10 (83%) had T cells in the blood that reacted against HCMV IE and/or pp65 peptides (), while 96.6% of the HCMV-seropositive GBM patients responded to HCMV peptides. Among the healthy blood donors, CD3+ T cells in peripheral blood from 75% of HCMV-seronegative donors and 81% of HCMV-seropositive donors responded to HCMV peptides in vitro (). These observations confirm a discrepancy of the results of ELISA tests vs. other methods to detect a previous or active HCMV infection in GBM patients and healthy controls. CD3-positive blood cells obtained from five patients in the VIGAS cohort (two HCMV-IgG positive and three HCMV-IgG negative) and five healthy donors (three HCMV-IgG-positive and two HCMV-IgG negative) were not activated by SEB peptides ().
Figure 2. Results of serology tests with 4 ELISAs utilizing AD169 as antigen (AD169-ag in-house ELISA, CMV-IgG serology kit from Dade Behring and BioSite) and one ELISA using antigen from a clinical isolate (CI-ag).
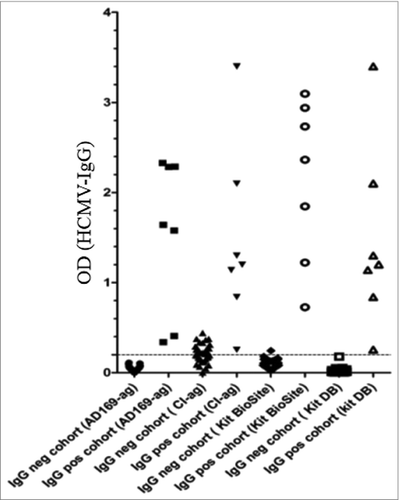
Figure 3. Peripheral blood T cells from glioblastoma patients and healthy controls (HC) were stimulated with HCMV IE or pp65 antigens. Figure B is an enlarged scale of figure A.
HCMV DNA or RNA levels did not change during the treatment phase ( and ). However, HCMV IgG titers were significantly lower at 3 weeks than at baseline (p = 0.023) and was thereafter stable ().
Higher HCMV activity in GBM patients than in healthy controls
Since HCMV remains in a latent state in most HCMV carriers, IgM and HCMV RNA are rarely detected in healthy subjects. At baseline, 7 (17%) of 42 patients were positive for HCMV IgM, compared with none of 50 healthy blood donors (p < 0.0001) ( and ). Two patients became IgM positive later, one at 12 and one at 24 weeks. Thus, 9 (21%) of 42 GBM patients were positive for HCMV IgM during the 24-week study phase. All IgM-positive patients were, as expected during a natural course of infection, negative at 24 weeks ( and ), except for the patient who became HCMV IgM-positive at 24 weeks. This patient was IgG and HCMV DNA and RNA positive at base line, and IgG positive at 3, 12, and 24 weeks and was in the valganciclovir treatment group.
Seven (37%) of 19 patients in the valganciclovir group and 4 (21%) of 19 placebo patients were positive for HCMV RNA in monocyte-enriched blood cell samples at baseline (one sample was missing for one patient; ). Twenty-six (63%) of 41 available RNA samples from VIGAS patients were positive for HCMV RNA in blood monocytes at least once during the 24-week study period: 16 of 22 (73%) in the valganciclovir group and 11 of 19 (58%) in the placebo group (one patient was lost to follow-up; ). HCMV IE RNA levels did not significantly decrease during valganciclovir treatment; 5 (29%) of 17 valganciclovir-treated patients were still positive for HCMV IE RNA in peripheral blood cells at 24 weeks (three patients had exited the study and did not provide blood samples;). Twenty-three (85%) of the RNA-positive patients were IgG positive; thus 15% of HCMV RNA-positive patients were HCMV IgG negative. Furthermore, T cells from one HCMV IgG-negative patient who had a positive HCMV RNA tests responded to HCMV peptides (IE and/or pp65).
Discussion
The results of this study suggest that the activity of HCMV is higher in GBM patients than in healthy blood donors and that serologic tests may be insufficient to define previous or active HCMV infection in these patients as well as in other HCMV-positive tumor patients. In several patients, serum samples were negative for IgG by three serological tests used for clinical diagnosis, even though HCMV proteins were detected in the patients’ tumor samples, their blood cells were positive for HCMV DNA, most of them were also positive for HCMV RNA; the majority of them though had T cells that responded against HCMV peptides. Evidently, a serology test does not appear to be an optimal method to define previous or present HCMV infection in GBM patients. Thus, an alternative to available serology tests may be needed to determine whether or not a GBM patient or a patient with another HCMV-related disease is HCMV positive. This may be true also for patients with other HCMV-positive tumors and needs further investigations, as it would have important clinical implications.
The fact that some HCMV-positive individuals do not have detectable IgG antibodies to HCMV has been noted before, and it has been questioned whether serologic status is always accurate for HCMV. We and others previously showed that commercial ELISA kits yield negative results for HCMV IgG in some patients who are positive for HCMV DNA.Citation17,18,19 We have also demonstrated that infectious virus could be reactivated from an HCMV DNA-positive but HCMV IgG-negative healthy blood donorCitation18, and we recently also observed that a HCMV-seronegative mother whose breast milk was HCMV DNA positive transferred HCMV to her infant, who developed a HCMV infection (unpublished observation). Bianchi et al. recently presented evidence that HCMV-DNA and proteins are present in brain tumors in the absence of detectable HCMV-specific antibodies against HCMV in the patients serum.Citation20 Sester et al. determined the concordance of serological status and HCMV-specific CD4+ T cells in 388 individuals and found that 2.1% of HCMV IgG-negative individuals had detectable levels of virus-specific CD4 T cells.Citation21 Loeth et. al. further reported that 11% (5 of 44) of seroneagtive blood bank donors had a CMV pp65-specific T cell response ex vivo.Citation22 Furthermore, Zhu et. al. also found a discordant cellular and humoral immune response to HCMV in healthy individuals; 24% of HCMV sronegative blood donors had CMV-specific T cells.Citation23 For herpes simplex virus (HSV) similar observations have been made; Posavad described that 6 of 24 HSV-1- and HSV-2-seronegative immunocompetent individuals without previous history of oral/labial or genital herpes who were HSV negative in oral and genital mucosa secretions, had HSV-specific T cell immunity.Citation24 They discussed whether this was due to an undetected infection or acquired immunity in the absence to infection. They later reported that 17 of 22 (77%) HSV-seronegative individuals who had a sexual relationship with HSV-2 positive partners had an HSV-specific T cell response that was primarily detected to IE proteins and less frequently to peptides of viral components.Citation25 More recently Long et. al. confirmed the presence of CD8+ T cell reactivity to nine peptides of HSV proteins in more than 15% of seronegative individuals.Citation26 The same phenomena is well known in HIV-exposed individuals; T cells reactive to HIV present in some individuals exist in the absence of HIV-specific antibodies.Citation27–34 Similar observations are also made in Hepatitis C- and B-exposed individuals.Citation35–38
It is not known why serology tests for some viruses including HCMV are negative in some virus-exposed individuals. One possibility is that HCMV-positive GBM patients have a high prevalence of a distinct HCMV strain whose antigen profile differs from those of HCMV strains used in commercial kits to detect HCMV-specific IgG and IgM. Indeed, we previously showed that 36% of HCMV IgG-negative blood donors with HCMV DNA-positive blood cells were IgG positive in an ELISA that uses a clinical isolate as antigen.Citation19 Similarly, in the present study, serum from 5 (42%) of 12 patients that were HCMV IgG negative according to three serology tests utilizing AD169-derived antigens, were IgG positive in an ELISA assay using antigens from a clinical isolate. Nevertheless, 9 (58%) of 12 patients were still HCMV IgG negative, although their tumors were positive for HCMV by immunohistochemistry with two different HCMV-specific antibodies, HCMV DNA were detected in their blood cells, and a majority of them had T cells that responded to HCMV peptides.
We speculate that the failure to detect HCMV IgG and IgM in GBM patients may reflect a B-cell tolerance to HCMV, rather than protective immunity and viral clearance or very low levels of virus replication. For example, little is known about the immune response in individuals infected in utero. The prevalence of congenital HCMV infection in newborn babies is 0.5–2.2%.Citation39–40 Some of these individuals may develop HCMV tolerance. Tolerance may also in theory develop during early transmission of the virus to nursing infants. Infections transferred by breast milk are indeed common; over 90% of HCMV-seropositive mothers have reactivated HCMV in their breast milk,Citation41and by 1 y of age, 40% of all children are HCMV positive.Citation42 However, it cannot be excluded that GBM patients may have produced CMV-specific antibodies earlier in life, and that these patients have lost their CMV-specific B cell response during the development of their aggressive tumor, which is associated with immunosuppression of unknown causes. It is also possible that they have an immune defect. Certainly, viral exposure may trigger an immune response controlling further infection in some individuals, but an infection may also persist at a local site in the exposed individuals, in the absence of a proper B cell response to the infection. We provide evidence for the latter hypothesis.
To our knowledge, our report is the first attempt to comprehensively analyze CMV RNA, DNA and/or proteins status in tissue specimens and blood cells and the immune response to the infection in the same individuals. Such analyses have for obvious reasons not been done on tissues samples from healthy individuals, but a discrepancy between HCMV serology data and HCMV DNA detection in blood has been reported as discussed above.Citation17,18,19 Thus, several lines of evidence suggest that HCMV carriers are not always defined by serology, and our findings imply that this is frequently the case in GBM patients. We observed that 29% of GBM patients positive for HCMV DNA in blood cells and HCMV proteins in the tumor, were negative for HCMV IgG by serology, but 85% of them had T-cells that were reactive against HCMV IE and/or pp65 peptides. Thus, we detected a T-cell response to HCMV in HCMV-seronegative GBM patients with positive tests for DNA, RNA, and proteins. We therefore conclude that serology is an insufficient clinical test to identify a previous or active HCMV infection in GBM patients, and that an infection may be present in virus exposed individuals with T cell reactivity in the absence of virus specific antibodies. In HIV it is proposed that the seronegative individuals who have T cells reactive against HIV have either eliminated the virus or carry it in extremely low amounts; these individuals rarely develop AIDS.Citation43 Apparently, this is not the case for HCMV, especially not in GBM patients. These observations are still met with skepticism among many virologists and call for further studies to understand the mechanisms behind these phenomena, and whether HCMV-seronegative but HCMV-positive individuals are more susceptible or resistant to HCMV disease or HCMV-related pathologies, such as cardiovascular diseases or cancer.
In this study, we also observe that HCMV was clearly more active in GBM patients than in controls. Healthy carriers of latent HCMV rarely show signs of reactivation, active HCMV replication, or clinical signs of disease. Their serum is usually positive for IgG and negative for IgM, and although HCMV DNA is present in some of their cells, some latency associated proteins are produced,Citation44 while the majority of IE, early and late RNAs and proteins are not detectable. If the virus is reactivated from latency, RNA can be detected in blood, which would imply that the virus is active (as a result of reactivation or a new infection). Thus, it is very rare that healthy individuals are HCMV IgM positive or have detectable levels of RNA in their blood cells. In the present study, 50 healthy blood donors were negative for HCMV IgM. In contrast, 21% of GBM patients in the VIGAS cohort were IgM positive for HCMV, and 63% had HCMV RNA in blood monocytes. These observations imply that HCMV is more active in GBM patients than in healthy controls. However, valganciclovir did not significantly change the levels of IE DNA and RNA in blood samples. Since valganciclovir is a DNA polymerase inhibitor that targets late but not IE gene expression, it is not surprising that valganciclovir treatment did not lower the IE RNA levels in blood. Therefore, valganciclovir treatment might not affect the pool of HCMV DNA-positive cells in the blood or the bone marrow, and may have an additional unknown antiviral activity in GBM. However, we observed that IgG titers were significantly lower after surgical debulking at 3 weeks follow up. We also observed highly improved survival in GBM patients receiving continuous valganciclovir treatment as an add-on to their standard therapy.Citation16 Thus, antiviral treatment may benefit GBM patients through a mechanism other than inhibition of HCMV DNA polymerase.
We conclude that several currently used ELISA tests that detect HCMV-specific IgG and IgM in sera from GBM patients do not reliably determine whether a GBM patient is infected with HCMV. More reliable methods were detection of HCMV proteins in the tumor or detection of HCMV DNA in blood cells by PCR. Our observations provide important information about the complex virological and immune status for HCMV in GBM patients that are highly relevant for the current development of immunotherapy protocols for HCMV in cancer patients and encourage further investigations to develop a reliable clinical diagnostic test for HCMV in GBM patients.
Materials and Methods
Study design
VIGAS was a randomized, double-blind, placebo-controlled phase I/II study to assess the efficacy and safety of valganciclovir for 24 weeks in combination with temozolomide and/or radiation therapy in GBM patients. HCMV positivity in the tumor by immunohistochemistry was an inclusion criterion (). Forty-two patients were enrolled between 20 December 2006 and 3 June 2008; 22 were randomized to valganciclovir and 20 to placebo treatment.Citation15 The study was registered at the Swedish Medical Agency (Eudra number 2006–002022–29) and at ClincalTrials.gov and was approved by the Karolinska ethics committee (2006/755–31).
Immunohistochemistry, serology, and PCR
HCMV proteins in paraffin-embedded brain tumors were identified by immunohistochemistry for HCMV IE and late antigens as described.Citation7,9,15 The results were graded from 1–4+ according to the estimated percentage of HCMV-positive tumor cells. Blood samples were collected before and 3, 12, and 24 weeks after surgery and examined for HCMV IgM (Dade Behring, CA# OWBK15) and HCMV-IgG was detected with two commercially available ELISAs utilizing AD169 antigens (Dade Behring, CA# OWBA15 and BioSite, CA# DEIA1437 ) and two in-house ELISA, one utilizing HCMV nuclear antigens of AD169-infected cells (as described beforeCitation45) and one utilizing whole-cell antigens from cells infected with an HCMV clinical isolate.Citation46,47 Internal HCMV-positive and -negative controls were included in both commercial HCMV-ELISA kits (Dade Behring, and BioSite). In the in-house tests, HCMV IgG-negative and -positive samples obtained from healthy blood donors as determined with a commercial HCMV-kit (Dade Behring) served as negative and positive controls, respectively. Plasma samples from 50 aged-matched healthy donors were included as controls. Peripheral blood mononuclear cells were separated with Lymphoprep (Medinor, CA# 1114547 ) and seeded on Primaria dishes (Primaria Falcon, Becton Dickenson, CA# 353801) in RPMI medium including 10% heat inactivated AB serum for at least 4 h in 5% CO2 at 37°C. DNA and RNA were isolated according to the manufacture´s protocol with DNA or RNeasy mini kits (both from Qiagen Sciences, for RNA, CA# 74106 and for DNA CA# 51304). HCMV DNA and RNA in blood monocytes were identified by PCR in triplicate as described before using primer probes targeting HCMV-IE gene.Citation7,47 DNA and RNA from uninfected and HCMV-infected fibroblasts were used as negative and positive controls, respectively.
Flow cytometry analyses
PBMC were isolated from blood samples, and CD3+ T-cell reactivity against HCMV peptides (IE-1, ID: P13202 and pp65, ID: P06725; Pep Mix; JPT Peptide Technologies) was analyzed with a flow cytometry assay. Briefly, blood mononuclear cells (2 × 106) were isolated with Lymphoprep (Medinor), stimulated for 2 h with HCMV-pp65 or 1 μg of HCMV-IE-1 peptide mix, and incubated over night with 0.02 μg/ul of brefeldin A (Sigma B 6542 ) at 37°C in 5% CO2. Cells were washed with PBS, stained with a CD3-CY antibody (Dako, CA# AC69601), permeabilized with Perm II solution (Becton Dickinson, CA# 340973) according to the manufacturer's instructions, stained with FITC-conjugated antibody against interferon-y (Becton Dickinson, CA# 554551), and analyzed by fluorescence-activated cell sorting. Unstimulated cells and cells stimulated with SEB (Sigma, S 4881) served as negative and positive controls, respectively. Cells were analyzed on a CyAn flow cytometer (Beckman Coulter). The percentage of IFN-interferon gamma positive cells in the negative control stimulation was subtracted from the specific stimulation. Values above zero were counted as positive signals.
Statistical analysis
Categorical variables were analyzed by using frequency tables with numbers and percentages of patients. Unpaired and paired t tests were used to compare antibody levels at different times and in different patient cohorts vs. healthy controls. Statistical analyses were performed with 2-tailed t tests; significance was set at the 5% level. Events with a reasonably high frequency were analyzed with a chi-square test without continuity correction.
One way ANOVA test was used for statistical analysis of peptide stimulation of peripheral blood T-cells. All values above zero were considered as positive signals.
Disclosure of Potential Conflicts of Interest
This work was partly supported by independent grants from Hoffmann La Roche. CSN has received speaker's fees and sponsored travel costs for presentations of data on the indirect molecular effects of HCMV at scientific meetings over 5 y ago.
Acknowledgments
We thank Fredrik Hansson (Norma, Lund) for statistical analyses and comments and Jan Tollemar, Zuzana Lindberg and Mahdi Fahran at F. Hoffman La Roche for input during the design and execution of the VIGAS study.
Funding
This work was supported by BILTEMA Foundation, Stichting af Jochnicks Foundation, Sten A Olssons Foundation for Research and Culture, Familjen Erling-Perssons Stiftelse, RATOS, independent grants from Hoffmann La Roche, Torsten and Ragnar Söderbergs Foundations, Dan och Jane Olssons Foundation, Swedish Cancer Foundation, Swedish Medical Research Council, Swedish Society for Medical Research (SLS), Goljes Memory Foundation, Magnus Bergvalls Foundation, Swedish Society for Medical Research (SSMF), IngaBritt och Arne Lundbergs Foundation and Tore Nilsons Foundation.
References
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009; 10:459-66; PMID:19269895; http://dx.doi.org/10.1016/S1470-2045(09)70025-7
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352:987-96; PMID:15758009
- Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, Nabors LB, Cobbs CG, Britt WJ. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res 2002; 62:3347-50; PMID:12067971
- Mitchell DA, Xie W, Schmittling R, Learn C, Friedman A, McLendon RE, Sampson JH. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro-Oncol 2008; 10:10-18; PMID:17951512
- Ranganathan P, Clark PA, Kuo JS, Salamat MS, Kalejta RF. Significant association of multiple human cytomegalovirus genomic loci with glioblastoma multiforme samples. J Virol 2012; 86:854-64; PMID:22090104; http://dx.doi.org/10.1128/JVI.06097-11
- Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol 2008; 116:79-86; PMID:18351367; http://dx.doi.org/10.1007/s00401-008-0359-1
- Rahbar A, Orrego A, Peredo I, Dzabic M, Wolmer-Solberg N, Straat K et al. Human cytomegalovirus infection levels in glioblastoma multiforme are of prognostic value for survival. J Clin Virol 2013; 57(1):36-42; PMID:23391370; http://dx.doi.org/10.1016/j.jcv.2012.12.018
- Libard S, Popova SN, Amini RM, Karja V, Pietilainen T, Hamalainen KM, Sundström C, Hesselager G, Bergqvist M, Ekman S et al. Human cytomegalovirus tegument protein pp65 is detected in all intra- and extra-axial brain tumours independent of the tumour type or grade. PloS One 2014; 9:e108861; PMID:25268364; http://dx.doi.org/10.1371/journal.pone.0108861
- Rahbar A, Stragliotto G, Orrego A, Peredo I, Taher C, Willems J, Söderberg-Naucler C et al. Low levels of human cytomegalovirus infection in glioblastoma multiforme associates with patient survival; -a case-control study. Herpesviridae 2012; 3:3; PMID:22424569; http://dx.doi.org/10.1186/2042-4280-3-3
- Bongers G, Maussang D, Muniz LR, Noriega VM, Fraile-Ramos A, Barker N, Marchesi F, Thirunarayanan N, Vischer HF, Qin L et al. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J Clin Invest 2010; 120:3969-78; PMID:20978345; http://dx.doi.org/10.1172/JCI42563
- Soderberg-Naucler C. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J Intern Med 2006; 259:219-46; PMID:16476101
- Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res 2011; 157:193-203; PMID:21036194; http://dx.doi.org/10.1016/j.virusres.2010.10.026
- Michaelis M, Doerr HW, Cinatl J, Jr. Oncomodulation by human cytomegalovirus: evidence becomes stronger. Med Microbiol Immunol 2009; 198:79-81; PMID:19198878; http://dx.doi.org/10.1007/s00430-009-0107-8
- John Inge Johnsen, Ninib Baryawno, Söderberg-Nauclér C. Is human cytomegalovirus a target in cancer therapy? Oncotarget 2011; 2(12):1329-38; PMID:2246171
- Stragliotto G, Rahbar A, Solberg NW, Lilja A, Taher C, Orrego A, Bjurman B, Tammik C, Skarman P, Peredo I et al. Effects of valganciclovir as an add-on therapy in patients with cytomegalovirus-positive glioblastoma: a randomized, double-blind, hypothesis-generating study. Int J Cancer 2013; 133(5):1204-13; PMID:23404447; http://dx.doi.org/10.1002/ijc.28111
- Soderberg-Naucler C, Rahbar A, Stragliotto G. Survival in patients with glioblastoma receiving valganciclovir. N Engl J Med 2013; 369:985-6; PMID:24004141; http://dx.doi.org/10.1056/NEJMc1302145
- Rahbar AR, Sundqvist VA, Wirgart BZ, Grillner L, Soderberg-Naucler C. Recognition of cytomegalovirus clinical isolate antigens by sera from cytomegalovirus-negative blood donors. Transfusion 2004; 44:1059-66; PMID:15225248
- Soderberg-Naucler C, Fish KN, Nelson JA. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 1997; 91:119-26; PMID:9335340
- Roback JD, Hillyer CD, Drew WL, Laycock ME, Luka J, Mocarski ES, Slobedman B, Smith JW, Soderberg-Naucler C, Todd DS et al. Multicenter evaluation of PCR methods for detecting CMV DNA in blood donors. Transfusion 2001; 41:1249-57; PMID:11606824
- Bianchi E, Roncarati P, Hougrand O, Guerin-El Khourouj V, Boreux R, Kroonen J, Martin D, Robe P, Rogister B, Delvenne P et al. Human cytomegalovirus and primary intracranial tumors: frequency of tumor infection and lack of correlation with systemic immune anti-viral responses. Neuropathol Appl Neurobiol 2014; PMID:25041908; http://dx.doi.org/10.1111/nan.12172
- Sester M, Gartner BC, Sester U, Girndt M, Mueller-Lantzsch N, Kohler H. Is the cytomegalovirus serologic status always accurate? A comparative analysis of humoral and cellular immunity. Transplantation 2003; 76:1229-30; PMID:14578758
- Loeth N, Assing K, Madsen HO, Vindelov L, Buus S, Stryhn A. Humoral and cellular CMV responses in healthy donors; identification of a frequent population of CMV-specific, CD4+ T cells in seronegative donors. PloS one 2012; 7:e31420; PMID:22347475; http://dx.doi.org/10.1371/journal.pone.0031420
- Zhu J, Shearer GM, Marincola FM, Norman JE, Rott D, Zou JP, Epstein SE. Discordant cellular and humoral immune responses to cytomegalovirus infection in healthy blood donors: existence of a Th1-type dominant response. Int Immunol 2001; 13:785-90; PMID:11369706
- Posavad CM, Wald A, Hosken N, Huang ML, Koelle DM, Ashley RL, Corey L. T cell immunity to herpes simplex viruses in seronegative subjects: silent infection or acquired immunity? J Immunol 2003; 170:4380-8; PMID:12682275
- Posavad CM, Remington M, Mueller DE, Zhao L, Magaret AS, Wald A, Corey L. Detailed characterization of T cell responses to herpes simplex virus-2 in immune seronegative persons. J Immunol 2010; 184:3250-9; PMID:20164419; http://dx.doi.org/10.4049/jimmunol.0900722
- Long D, Skoberne M, Gierahn TM, Larson S, Price JA, Clemens V, Baccari AE, Cohane KP, Garvie D, Siber GR et al. Identification of novel virus-specific antigens by CD4(+) and CD8(+) T cells from asymptomatic HSV-2 seropositive and seronegative donors. Virology 2014; 464–465:296-311; PMID:25108380; http://dx.doi.org/10.1016/j.virol.2014.07.018
- Clerici M, Levin JM, Kessler HA, Harris A, Berzofsky JA, Landay AL, Shearer GM. HIV-specific T-helper activity in seronegative health care workers exposed to contaminated blood. Jama 1994; 271:42-6; PMID:8258885
- Rowland-Jones SL, McMichael A. Immune responses in HIV-exposed seronegatives: have they repelled the virus? Curr Opin Immunol 1995; 7:448-55; PMID:7495507
- Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T, HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med 1995; 1:59-64; PMID:7584954
- Pattacini L, Murnane PM, Kahle EM, Bolton MJ, Delrow JJ, Lingappa JR et al. Differential regulatory T cell activity in HIV type 1-exposed seronegative individuals. AIDS Res Hum Retroviruses 2013; 29:1321-9; PMID:23815575; http://dx.doi.org/10.1089/AID.2013.0075
- Murashev BV, Nazarenko OV, Akulova EB, Artemyeva AK, Verevochkin SV, Shaboltas AV, Skochilov RV, Toussova OV, Kozlov AP. The high frequency of HIV type 1-specific cellular immune responses in seronegative individuals with parenteral and/or heterosexual HIV type 1 exposure. AIDS Res Hum Retroviruses 2012; 28:1598-605; PMID:22475222; http://dx.doi.org/10.1089/AID.2011.0335
- Li L, Liu Y, Bao Z, Chen L, Wang Z, Li T, Li H, Zhuang D, Liu S, Wang X et al. Analysis of CD4+CD25+Foxp3+ regulatory T cells in HIV-exposed seronegative persons and HIV-infected persons with different disease progressions. Viral Immunol 2011; 24:57-60; PMID:21319979; http://dx.doi.org/10.1089/vim.2010.0079
- Prodger JL, Hirbod T, Gray R, Kigozi G, Nalugoda F, Galiwango R, Reynolds SJ, Huibner S, Wawer MJ, Serwadda D et al. HIV infection in uncircumcised men is associated with altered CD8 T-cell function but normal CD4 T-cell numbers in the foreskin. J Infect Dis 2014; 209:1185-94; PMID:24277744; http://dx.doi.org/10.1093/infdis/jit644
- Restrepo C, Rallon NI, del Romero J, Rodriguez C, Sempere-Ortells JM, de la Vega E, Soriano V, Benito JM. HIV Gag-specific immune response mediated by double negative (CD3(+)CD4(-)CD8(-)) T cells in HIV-exposed seronegative individuals. J Med Virol 2013; 85:200-9; PMID:23172685; http://dx.doi.org/10.1002/jmv.23447
- Abdelwahab SF, Zakaria Z, Sobhy M, Rewisha E, Mahmoud MA, Amer MA, Del Sorbo M, Capone S, Nicosia A, Folgori A et al. Hepatitis C virus-multispecific T-cell responses without viremia or seroconversion among Egyptian health care workers at high risk of infection. Clin Vaccine Immunol: CVI 2012; 19:780-6; PMID:23172685; http://dx.doi.org/10.1002/jmv.23447
- Cameron B, Galbraith S, Li H, Lloyd A. Correlates and characteristics of hepatitis C virus-specific T-cell immunity in exposed uninfected high-risk prison inmates. J Viral Hepat 2013; 20:e96-106; PMID:23490396; http://dx.doi.org/10.1111/jvh.12016
- Scognamiglio P, Accapezzato D, Casciaro MA, Cacciani A, Artini M, Bruno G, Chircu ML, Sidney J, Southwood S, Abrignani S et al. Presence of effector CD8+ T cells in hepatitis C virus-exposed healthy seronegative donors. J Immunol 1999; 162:6681-9; PMID:10352286
- Wiegand J, Meya S, Schlaphoff V, Manns MP, Mossner J, Wedemeyer H, Tillmann HL. HBV-specific T-cell responses in healthy seronegative sexual partners of patients with chronic HBV infection. J Viral Hepat 2010; 17:631-9; PMID:19889141; http://dx.doi.org/10.1111/j.1365-2893.2009.01220.x
- Elliott SP. Congenital cytomegalovirus infection: an overview. Infect Disord Drug targets 2011; 11:432-6; PMID:21827436
- Malm G, Engman ML. Congenital cytomegalovirus infections. Semin Fetal Neonatal Med 2007; 12:154-9; PMID: 17337260
- Hamprecht K, Goelz R, Maschmann J. Breast milk and cytomegalovirus infection in preterm infants. Early Hum Dev 2005; 81:989-96; PMID:16278059
- Ahlfors K. IgG antibodies to cytomegalovirus in a normal urban Swedish population. Scand J Infect Dis 1984; 16:335-7; PMID:6098960
- Shacklett BL. Understanding the "lucky few:" the conundrum of HIV-exposed, seronegative individuals. Curr HIV/AIDS Rep 2006; 3:26-31; PMID:16522256
- Mason GM, Jackson S, Okecha G, Poole E, Sissons JG, Sinclair J, Wills MR. Human cytomegalovirus latency-associated proteins elicit immune-suppressive IL-10 producing CD4(+) T cells. PLoS pathogens 2013; 9:e1003635; PMID:24130479; http://dx.doi.org/10.1371/journal.ppat.1003635
- Grillner L. Screening of blood donors for cytomegalovirus (CMV) antibodies: an evaluation of different tests. J Virol Methods 1987; 17:133-9; PMID:2822749
- Schmitz H, Doerr HW, Kampa D, Vogt A. Solid-phase enzyme immunoassay for immunoglobulin M antibodies to cytomegalovirus. J Clin Microbiol 1977; 5:629-34; PMID:195977
- Soderberg C, Larsson S, Bergstedt-Lindqvist S, Moller E. Definition of a subset of human peripheral blood mononuclear cells that are permissive to human cytomegalovirus infection. J Virol 1993; 67:3166-75; PMID:7684461
