Abstract
Research on innate immune signaling and regulation has recently focused on pathogen recognition receptors (PRRs) and their signaling pathways. Members of PRRs sense diverse microbial invasions or danger signals, and initiate innate immune signaling pathways, leading to proinflammatory cytokines production, which, in turn, instructs adaptive immune response development. Despite the diverse functions employed by innate immune signaling to respond to a variety of different pathogens, the innate immune response must be tightly regulated. Otherwise, aberrant, uncontrolled immune responses will lead to harmful, or even fatal, consequences. Therefore, it is essential to better discern innate immune signaling and many regulators, controlling various signaling pathways, have been identified. In this review, we focus on the recent advances in our understanding of the activation and regulation of innate immune signaling in the host response to pathogens and cancer.
Abbreviations
| PRRs | = | pathogen recognition receptors |
| PAMPs | = | pathogen-associated molecular patterns |
| DAMPs | = | danger-associated molecular patterns |
| TLRs | = | Toll-like receptors |
| RLRs | = | RIG-I-like receptors |
| NLRs | = | Nod- like receptors |
| ALRs | = | AIM2-like receptors |
| CLRs | = | C-type lectin receptors |
| IFN | = | interferon |
| DCs | = | dendritic cells |
| NOD | = | nucleotide-binding oligomerization domain |
| LRR | = | leucine-rich repeat |
| RIG-I | = | retinoic acid-inducible gene 1 |
| LGP 2 | = | laboratory of genetics and physiology 2 |
| MDA5 | = | melanoma differentiation-associated protein 5 |
| AIM2 | = | absent in melanoma 2 |
| IFI16 | = | interferon, gamma-inducible protein 16 |
| DDX41 | = | DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 |
| cGAS | = | cyclic GMP-AMP synthase |
| TIR | = | Toll IL-1 receptor |
| LPS | = | lipopolysaccharide |
| MyD88 | = | myeloid differentiation factor 88 |
| TIRAP | = | TIR domain-containing adapter protein |
| TRIF | = | TIR domain-containing adaptor inducing IFN-b |
| TRAM | = | TRIF-related adaptor molecule |
| SARM | = | sterile a- and armadillo motif-containing protein |
| IRAK | = | interleukin-1 receptor-associated kinase |
| TRAF | = | TNFR-associated factor |
| NEMO | = | NF-kB essential modulator |
| IKK | = | IkB kinase |
| TAK1 | = | TGF-b-activating kinase 1 |
| IRF | = | interferon regulatory factor |
| RIP | = | receptor-interacting protein |
| TANK | = | TRAF family-member-associated NF-kB activator |
| TBK1 | = | TANK binding kinase 1 |
| IKKi | = | inducible IkB kinase |
| iE-DAP | = | g-D-glutamyl-meso-diaminopimelic acid |
| MDP | = | muramyl dipeptide |
| Atg16L | = | autophagy related 16-like |
| MAVS | = | mitochondrial antiviral signaling protein |
| ASC | = | apoptosis-associated speck-like protein containing a CARD |
| CARD | = | caspase recruitment domain |
| LT | = | lethal toxin |
| NAIPs | = | neuronal apoptosis inhibitory proteins |
| STING | = | stimulator of interferon gene |
| CMV | = | cytomegalovirus |
| ER | = | endoplasmic reticulum |
| CDNs | = | cyclic dinucleotides |
| CYLD | = | the familial cylindromatosis tumor suppressor gene |
| Nrdp1 | = | neuregulin receptor degradation protein 1 |
| TRIMs | = | tripartite motif containing proteins |
| LBP | = | LPS-binding protein |
| SIGIRR | = | single Ig IL-1-related receptor |
| TRAILR | = | tumor-necrosis factor-related apoptosis-inducing ligand receptor |
| cIAP | = | cellular inhibitor of apoptosis protein |
| ROS | = | reactive oxygen species |
| BMM | = | bone marrow-derived macrophage |
| SOCS | = | suppressor of cytokine signaling |
| GBP5 | = | guanylate-binding protein 5 |
| PKC-d | = | protein kinase C delta |
| PKR | = | dsRNA-dependent protein kinase |
| ULK1 | = | autophagy related serine threonine UNC-51- like kinase |
| AMPK | = | AMP activated protein kinase |
| LUBAC | = | linear ubiquitin assembly complex |
| GSK3β | = | Glycogen synthase kinase 3β |
| MIB | = | mind bomb |
| TRIP | = | TRAF-interacting protein |
| cDC | = | conventional dendritic cell |
| pDC | = | plasmacytoid dendritic cell |
| KSHV | = | Kaposi's sarcoma-associated herpesvirus |
| RAUL | = | RTA-associated E3 ligase |
| RACK1 | = | receptor for activated C kinase 1 |
| HCC | = | hepatocellular carcinoma |
Introduction
The innate immune system plays an important role as the first line of defense against invading microbes by sensing pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs). The initiation and regulation of innate immune responses are orchestrated by several classes of germline-encoded pattern-recognition receptors (PRRs), including Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), Nod-like receptors (NLRs), AIM2-like receptors (ALRs), C-type lectin receptors (CLRs) and other DNA sensors.Citation1-3 Upon pathogen invasion, these PRRs trigger the activation of NF-κB, type I interferon (IFN), or other inflammasome signaling pathways, which, in turn, leads to the production of a variety of proinflammatory and antiviral cytokines and chemokines, subsequently inducing adaptive immune responses.Citation4,5
TLRs belong to a class of type I integral membrane glycoproteins that play an important role in host defense against pathogens by recognizing a wide variety of PAMPs. To date, 10 and 12 functional TLRs have been identified in humans and mice, respectively. These receptors are expressed on the cell surface (TLR1, TLR2, TLR4, TLR5, and TLR6) or in the endosome (TLR3, TLR4, TLR7, TLR8 and TLR9). TLRs are expressed on various immune cells including macrophages, monocytes, neutrophils, master cells, eosinophils, dendritic cells (DCs), and T cells.Citation4 Since first being described in the fruit fly, TLRs have been identified as a major component in inflammatory conditions, antiviral responses, and several immune-related diseases.Citation6
CLRs comprise a large family of receptors, consisting of one or more carbohydrate recognition domains (CRDs), which recognize various carbohydrate ligands. CLRs play a major role in shaping immune responses to microbial pathogens, especially fungi.Citation7 They can function directly as PRRs to induce specific gene expression in macrophages and DCs, or to modulate the functions of TLRs through signaling crosstalk.Citation8,9
NLRs constitute a large protein family of intracellular sensors, the members of which share a conserved central nucleotide-binding, oligomerization domain (NOD), a leucine-rich repeat (LRR) region and a variable N-terminal effector domain.Citation10 NLRs can be classified as receptors (Nod1 and Nod2), adaptors (NLRP3, NLRC4, and NLRP6) and regulators (NLRX1, NLRC3, NLRC5 and NLRP4) by their diverse functions. Much work has been done to reveal the functions of NLRs as receptors of PAMPs to sense microbial structures (e.g. peptidoglycan and muramyl dipeptide for Nod1 and Nod2), and activate NF-κB signaling, or as the adaptors to form different types of inflammasomes to regulate IL-1β and IL-18 secretion.Citation11 However, accumulating evidence suggests that NLRs can also function as the negative regulators of innate immune responses, including inflammation, antiviral immunity and autophagy.
Besides TLRs and NLRs, RLRs including retinoic acid-inducible gene 1 (RIG-I), laboratory of genetics and physiology 2 (LGP2) and melanoma differentiation-associated protein 5 (MDA5) function as cytoplasmic receptors for RNA viruses.Citation12 Recently, several DNA sensors, including absent in melanoma 2 (AIM2), interferon, gamma-inducible protein 16 (IFI16), DNA-dependent activator of IRFs/Z-DNA binding protein 1 (DAI/ZBP1), DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (DDX41), RNA polymerase III and the cyclic GMP-AMP synthase (cGAS) have been identified to recognize viral DNA and activate inflammasome or type I IFN signaling pathways to establish the antiviral state in the cell.Citation3 Thus, TLRs, CLRs, NLRs, RLRs, ALRs and other DNA sensors are critical in bridging innate and adaptive immune responses by activating several key signaling pathways and producing many important cytokines and chemokines to mediate the immunological state of the host.
Innate Immune Signaling Pathways
Toll like receptor signaling pathways
TLRs are conserved membrane-associated receptors, which contain a variable number of LRR motifs, a transmembrane region and a Toll/IL-1 receptor (TIR) domain.Citation13 Their LRR region may recognize and bind to PAMPs as foreign ligands. TLR4, the first identified TLR, recognizes lipopolysaccharide (LPS), a unique cell wall component of gram-negative bacteria. Specific members of the TLR family, including TLR1, TLR2, TLR5 and TLR6, sense the protein, flagellin, or lipid components on infectious microbes.Citation14 On the other hand, TLR3, TLR7, TLR8 and TLR9 detect single or double-strand RNA (ssRNA or dsRNA) as well as unmethylated CpG DNA.Citation5,Citation15-17
Upon ligand binding, TLRs have the ability to dimerize and to undergo conformation changes, which lead to recruitment of TLR domain-containing adaptor proteins. To date, 5 adaptors with TLR domains have been identified, including myeloid differentiation factor 88 (MyD88), TIR domain-containing adapter protein (TIRAP)/MyD88 adaptor-like (Mal), TIR domain-containing adaptor inducing interferon (IFN)-β (TRIF), TRIF-related adaptor molecule (TRAM), and sterile α- and armadillo motif-containing protein (SARM).Citation18 Most TLRs, including TLR1, 2, 4, 5, 7, 8 and 9 and excluding TLR3, use MyD88 as the adaptor protein to activate the canonical NF-κB pathway.Citation19,20 Once the MyD88-interleukin-1 receptor-associated kinase (IRAK) 1-IRAK4 complex is activated, it will recruit and activate TNFR-associated factor (TRAF) 6. Next, TRAF6, together with 2 ubiquitination E2 enzymes, UBC13 and URV1A, catalyze the formation of a K63-linked polyubiquitin chain on TRAF6, itself, and on NF-κB essential modulator (NEMO), a regulatory subunit of IκB kinase (IKK) complex. The polyubiquitin chain acts as a scaffold, recruiting TGF-β-activating kinase 1 (TAK1) and its binding proteins, TAB1, TAB2, TAB3, which leads to the phosphorylation and activation of IKKα/β.Citation21-23 Activated IKKα/β specifically phosphorylates the inhibitory IκB protein, which, in turn, directly interacts with and inactivates NF-κB. This phosphorylation results in the degradation of IκB, allowing NF-κB nuclear localization, and subsequent synthesis of proinflammatory cytokines, such as TNF-α, IL-1β and IL-6, which are critical mediators of inflammatory responses.Citation24 Interestingly, MyD88 may also directly interact with interferon regulatory factor (IRF) 5 and IRF7 to induce IFN-α production through TLR7/9 signaling.Citation25,26 In addition, TLR4 and TLR3 can use another adaptor protein TRIF, to recruit TRAF3, TRAF6 and receptor-interacting protein (RIP) 1, which activates IKKα/β complex and TRAF family-member-associated NF-κB activator (TANK) binding kinase 1(TBK1)/inducible IκB kinase (IKKi), respectively. TBK1/IKKi directly phosphorylates IRF3 and IRF7 to activate type I IFN signaling pathways.Citation14,27 These TLRs and their signaling pathways are extremely important for initiating and propagating innate immune responses, and for maintaining organism homeostasis.
Signaling pathways of Nod1 and Nod2
Nod1 and Nod2, the 2 most widely studied members of NLRs, recognize bacteria-derived g-D-glutamyl-meso-diaminopimelic acid (iE-DAP) and muramyl dipeptide (MDP), respectively.Citation28 Upon ligand recognition, Nod1 and Nod2 oligomerize and interact with RIP2, subsequently leading to the activation of the canonical NF-κB and MAPK pathways.Citation29,30 Nod1 and Nod2 are also involved in the autophagic response to invasive bacteria. Upon bacteria invasion, Nod1 and Nod2 recruit the autophagy related 16-like (Atg16L) to induce autophagy in a RIP2/NF-κB independent manner.Citation31 Furthermore, Nod2 also regulates antibacterial immunity through its interaction with Atg16L, Atg5 and Atg7 to induce autophagy and to generate MHC class II antigen-specific CD4 T cell responses.Citation32 In addition to their role in antibacterial immunity, a recent study showed that Nod2 senses viral ssRNA, triggering the activation of type I IFN signaling pathway through interaction with mitochondrial antiviral signaling protein (MAVS). However, it is not known whether Nod2 directly binds to viral ssRNA or acts as an adaptor protein.Citation33 Regardless, it is clear that Nod1 and Nod2 function in response to microbial structures, and lead to the activation of NF-κB signaling for cytokine production.
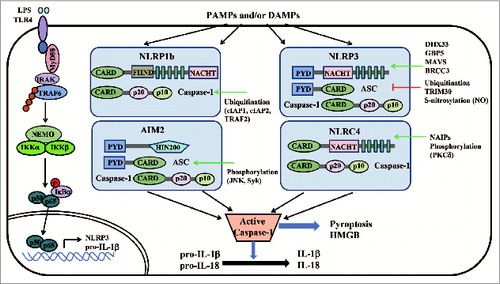
Inflammasome activation by NLRs and ALRs
Several NLRs and ALRs, including NLRP1, NLRP3, NLRP6, NLRP7, NLRC4 and AIM2, function as sensors or adaptors, forming the 'inflammasome', which regulates interleukin-1β (IL-1β) and IL-18 production and induces cell death via pyroptosis in response to diverse microbial and endogenous danger signals.Citation11,34 Inflammasomes are large protein complexes, consisting of the central NLR or ALR protein, the adaptor protein apoptosis-associated speck-like protein containing a CARD (ASC) and caspase-1. Activation of the inflammasome usually requires 2 signals: (1) transcriptional induction of pro-IL-1β, and NLRs or ALRs; (2) specific, agonist-driven oligomerization of NLR/ALRs to initiate inflammasome assembly. Recent studies demonstrated that NLRP3 and AIM2 activation induces prion-like polymerization of the adaptor, ASC, to activate caspase-1. This self-activation modality of ASC, initiated by NLRP3 or AIM2, may represent a unified mechanism for inflammasome assembly.Citation35,36
NLRP1 inflammasome is the first complex identified in the activation of caspase-1. Human NLRP1 contains a C-terminal caspase recruitment domain (CARD) domain, which is thought to be the key component interacting with the CARD domain of procaspase-1.Citation37 Although several studies have shown that MDP activates human NLRP1 inflammasome, the direct activator of human NLRP1 is still unknown.Citation38,39 There are highly polymorphic paralogs (Nlrp1a, Nlrp1b, and Nlrp1c) for mouse NLRP1, which do not have a PYD domain. Mouse Nlrp1b mediates the response to the lethal toxin (LT) of Bacillus anthracis,Citation40 whereas human NLRP1 does not. Like NLRP1, NLRC4 binds to caspase-1 with its own CARD domain. NLRC4 may act as the adaptor protein, downstream of neuronal apoptosis inhibitory proteins (NAIPs) receptor. NAIPs appear to directly bind to flagellin, T3SS rod, or T3SS needle, thereby, activating NLRC4.Citation41-44 NLRP3 is the most extensively studied inflammasome and is composed of NLRP3, ASC, and caspase-1. Human, but not mouse, NLRP3 inflammasome also contains the cardinal protein, whose function remains unclear.Citation45 Notably, NLRP3 inflammasome is responsive to a wide array of stimuli, ranging from microorganisms of both endogenous and exogenous origin to inorganic entities including alum asbestos and silica.Citation46 Whereas specific ligands have been identified for NLRP1, NLRC4, and AIM2, little is known about the direct ligands of NLRP3.Citation47 A recent study reveals that a RNA helicase, DHX33, senses cytosolic RNA and interacts with NLRP3 to trigger NLRP3 inflammasome activation.Citation48 This result indicates that NLRP3 may act as an adaptor protein rather than a direct sensor in innate immune signaling.
AIM2 is the most recently identified DNA sensor that interacts with ASC and capase-1 to regulate IL-1β and IL-18 maturation upon synthetic dsDNA stimulation or DNA virus infection in a NLRP3-independent manner.Citation49-52 Maturation of IL-1β and IL-18 is essentially absent in AIM2-deficient macrophages in response to Poly(dA:dT) or F. tularensis infection.Citation53,54 Collectively, these data demonstrate that these receptors act as nucleotide sensors, mediating the innate immune response in an inflammasome-dependent or -independent manner.
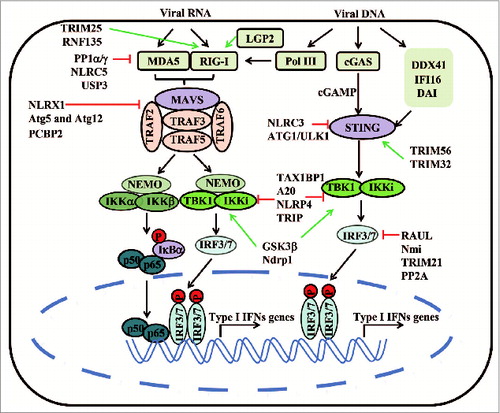
Type I interferon signaling pathways induced by RNA and DNA viruses
Upon RNA virus infection, RLRs, including RIG-I (also known as DDX58) and MDA5 (also known as IFIH1), recognize cytoplasmic viral components and activate the mitochondrial signaling adaptor, MAVS (also known as VISA, IPS-1 or Cardif).Citation55 MAVS is a mitochondrial-associated adaptor protein, which provides the platform to recruit downstream kinases, such as TBK1 and IKKi, and leads to IRF3/IRF7 phosphorylation and to activation of type I IFN signaling pathways.Citation56-59 Recent evidence suggests that viral infection induces the formation of very large MAVS aggregates, which are capable of activating IRF3. The MAVS fibrils operate like prions, converting inactive MAVS into its functional aggregates.Citation60 These results suggest that a prion-like conformational switch of MAVS may be a critical check- point in antiviral signaling cascade initiation. Another member of RLRs, LGP2, was originally believed to negatively regulate RIG-I and MDA5, due to its lack of a CARD domain.Citation61 However, a recent study, using LGP2 knockout mice, indicates that LGP2 positively regulates type I IFN signaling in response to a variety of RNA viruses,Citation62 suggesting that LGP2 may enhance RIG-I/MDA5-mediated recognition of viral RNA.
DAI (also known as ZBP1) was the first viral DNA sensor identified. It interacts with dsDNA and activates TBK1/IRF3 pathway.Citation63 Subsequent work has confirmed a role for DAI in type I IFN activation against cytomegalovirus (CMV) infection in human foreskin fibroblasts.Citation64 However, it has been difficult to discern whether DAI is essential for DNA sensing in vivo.Citation65 Recent studies have shown that RNA polymerase III can also serve as an intracellular sensor of viral DNA by transcribing AT-rich double-stranded DNA into double-stranded RNA, which, in turn, activates RIG-I and initiates the MAVS-dependent signaling cascade.Citation66,67 Furthermore, IFI16 and DDX41 function as cytosolic DNA sensors and interact with another adaptor protein, stimulator of interferon gene (STING, also known as TMEM173 or MITA), to activate TBK1 and downstream signaling.Citation68,69
STING is a newly discovered adaptor protein, which plays a central role in viral dsDNA responses by mediating TBK1-dependent IRF3 activation.Citation70,71 Inactive STING resides in the endoplasmic reticulum (ER), but forms discrete punctate foci with TBK1 in the perinuclear region of the cytosol after dsDNA stimulation.Citation71 Because several DNA sensors, such as DDX41 and IFI16, have been demonstrated to activate STING, the STING-TBK1-IRF3 signaling axis is now known as the major pathway of cytosolic DNA-induced type I IFN signaling. In addition, STING, itself, may work as a sensor, directly recognizing bacterial second messenger molecules called cyclic dinucleotides (CDNs), such as cyclic di-GMP (c-di-GMP), and activating type I IFN signaling.Citation72,73 Notably, Z. Chen's group demonstrated that STING also senses cyclic-di-GMP-AMP (cGAMP), leading to cytosolic DNA-induced type I IFN signaling. Furthermore, cGAMP can be generated in the presence of cytosolic DNA by a cyclase enzyme called cGAS.Citation74,75 The identification of cGAS may provide the missing link between DNA sensing and STING activation. Despite the fact that the cGAS-cGAMP system is essential for type I IFN induction by DNA viruses in fibroblasts and monocytes,Citation74,75 whether the cGAS-cGAMP system operates similarly in in other cell types, as well as in vivo, needs urgent resolution.
Regulation of Innate Immune Signaling Pathways by Diverse Regulators
Because uncontrolled innate immune responses can be harmful, even fatal, to the host,Citation76 stringent regulation of these signaling pathways is essential to maintain immune balance in the host. In the last few years, many positive and negative regulators have been identified to control innate immune signaling pathways at multiple levels through different mechanisms.
Regulation of TLR signaling pathways
Many molecules, ranging from membrane to cytosol, such as helper cofactors, decoy receptors, or specific enzymes, regulate TLR signaling. These molecules include co-receptors (CD14), soluble receptors (sTLR), transmembrane proteins (ST2L, SIGIRR, TRAILR), and intracellular regulators (SOCS-1, MyD88s, TOLLIP, IRAK-M, A20, CYLD (the familial cylindromatosis tumor suppressor gene), Nrdp1, regulatory Nod proteins, TRIAD3A, tripartite motif containing proteins (TRIMs)). These molecules maintain the balance between activation and inhibition of TLR signaling in response to diverse PAMPs ( and Table S1).Citation76,77
Figure 1. A schematic representation of TLR signaling pathways. TLRs are activated by ligand binding, which leads to dimerization of TLRs and to recruitment of TLR domain-containing adaptor proteins. Next, MyD88/IRAK1/IRAK4 or TRIF activates TRAF6, which, in turn, catalyzes the formation of a K63-linked polyubiquitin chain on TRAF6, itself. The polyubiquitin chain acts as the scaffold, recruiting TAK1 and its binding proteins, which leads to IKK-α/β activation. Activated IKKα/β specifically phosphorylates IkBα, resulting in IkBα degradation and NF-kB translocation into the nucleus. TRIF can also recruit TRAF3 to activate TBK1 and IKKi. TBK1/IKKi directly phosphorylates IRF3/7 to activate type I IFN signaling pathways. Various molecules positively (green arrow) or negatively (red blunt arrow) regulate TLR-induced signaling pathways.
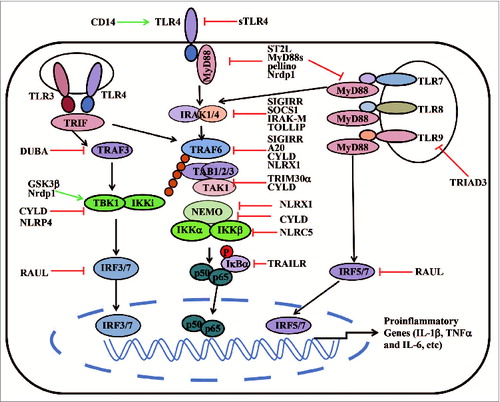
Co-receptors
Several co-receptors have been identified to modulate TLR function. For example, CD14 is a glycosylphosphatidylinositol-anchored protein, which forms a horseshoe-shaped structure similar to TLR ectodomains. CD14 chaperones LPS from the LPS-binding protein (LBP) to the TLR4–MD2 complex at the cell surface and mediates TNF-α and IL-6 production in response to TLR4 and TLR2–TLR6 ligands.Citation78 Furthermore, CD14 is also necessary for induction of proinflammatory cytokines by interacting with TLR7 and TLR9.Citation79
Decoy inhibitors
Some transmembrane proteins serve as the negative regulators of TLR signaling. It is well established that soluble decoy receptors negatively regulate cytokine and chemokine production. In TLR signaling, soluble TLR4 (sTLR4) blocks the interaction between TLR4 and MD2,Citation80 whereas soluble TLR2 (sTLR2) acts as an agonist of TLR2 by binding to CD14.Citation81 Since MyD88 is the major adaptor protein for most TLRs, MyD88s (a short form of MyD88) antagonizes TLR signaling by inhibiting LPS- and IL-1β-induced, but not TNF-induced, NF-κB activation.Citation82 Although MyD88s strongly interacts with IRAK1, it does not directly interact with IRAK4. MyD88s prevents the IRAK4-mediated phosphorylation of IRAK1, which, in turn, inhibits downstream NF-κB activation.Citation83
Transmembrane proteins
Single Ig IL-1-related receptor (SIGIRR) is a membrane protein that is highly expressed in immature DCs and epithelial cells, but not in macrophages. It interacts with IRAK and TRAF6 to negatively regulate TLR4 and TLR9 pathway. SIGIRR−/− mice are more susceptible to LPS-induced septic shock.Citation84,85 ST2L is a type I transmembrane protein that inhibits TLR4-mediated activation and attenuates MyD88 and Mal functions. Macrophages from ST2−/− mice produce higher levels of proinflammatory cytokines in response to a variety of TLR ligands, indicating that ST2L is a potent negative regulator of TLR signaling.Citation86 Another receptor that inhibits TLR signaling is tumor-necrosis factor-related apoptosis-inducing ligand receptor (TRAILR). TRAILR does not have a TIR domain, but belongs to the TNF superfamily. It negatively regulates NF-κB activation by stabilizing IκBα, preventing its degradation.Citation87
Ubiquitin E3 ligases
Protein ubiquitination is a reversible covalent modification, regulating the stability, activity and localization of target proteins. Recent work has shown that many ubiquitin ligases as well as deubiquitinases regulate TLR signaling. In Drosophila, the ubiquitin E3 ligase, Pellino, targets MyD88 for ubiquitination and degradation.Citation88 TRIAD3A is a member of the TRIAD3 family of RING-finger ubiquitin E3 ligase. It binds to the cytoplasmic domain of TLR9 and TLR4, but not to TLR2, to promote the ubiquitination and degradation of TLR4 and TLR9.Citation89 Another RING-finger E3 ligase, TRIM30α, is also a negative regulator of TLR-induced NF-κB signaling. TRIM30α is induced by TLR agonists and interacts with the TAB2-TAB3-TAK1 complex, mediating TAB2 and TAB3 degradation.Citation90 Nrdp1 acts as an E3 ligase and directly binds and polyubiquitinates MyD88, leading to the degradation of MyD88 and subsequent inhibition of NF-κB activation and proinflammatory cytokine production. Interestingly, Nrdp1 also promotes the activation of transcription factor, IRF3, and the production of IFN-β by interacting with TBK1. These results indicate that Nrdp1 may function as a bio-switch to 'preferentially' promote the TLR-type I IFN axis.Citation91
Deubiquitinases
A20 was initially identified as a TNF-induced zinc-finger protein that is rapidly induced by both TNF-α and LPS, and is expressed in many cell types.Citation92,93 A20 has been identified as a key negative regulator, controlling both MyD88-dependent and MyD88-independent TLR-signaling pathways. It acts as a deubiquitinating enzyme, removing K63-specific polyubiquitin chains from TRAF6, thus, blocking NF-κB signaling.Citation94 Recently, it is reported that A20 inhibits the E3 ligase activities of TRAF6, TRAF2, and cellular inhibitor of apoptosis protein 1 (cIAP1) by disrupting their interactions with key E2 enzymes, either Ubc13 or UbcH5c.Citation95 In addition, A20 removes the K63-linked ubiquitin chains on RIP1 (an essential mediator of TNF signaling) and supports E3 ligase activity by facilitating K48-linked ubiquitination of RIP1 for subsequent proteasomal degradation.Citation96 Several A20-interacting proteins, including TAX1BP1, RNF11 and Itch, have been identified to regulate the diverse functions of A20.Citation95,97,98 A20−/− mice are hypersensitive to both TNF-α and LPS, develop severe inflammation, and die prematurely.Citation99 Another deubiquitinase, CYLD, is also important for its inhibitory function in TLR signaling. CYLD removes K63-linked ubiquitin chains from TRAF2, and thus, inhibits NF-κB activation.Citation100,101 It has been demonstrated that CYLD negatively regulates TNF-α or TLR induced-NF-κB activation and type I IFN signaling by targeting a variety of signaling proteins, including TRAF2, TRAF6, RIP1, TAK1, NEMO, RIG-I, and TBK1, in T cells and other immune cells.Citation102-105
Regulatory NLRs
Although NLRs were originally believed to function as the sensors of pathogens and cellular danger signals, recent evidence suggests that several NLRs, known as regulatory NLRs, negatively regulate TLR and RLR signaling. NLRX1 is the first NLR family member shown to be targeted to the mitochondria. It is ubiquitously expressed in a variety of tissues and cells.Citation106 To date, at least 4 distinct functions of NLRX1 have been identified: (1) to negatively modulate RIG-I-mediated antiviral responses by binding to MAVS and disrupting RIG-I-MAVS interaction;Citation106,107 (2) to negatively regulate TLR-induced NF-κB signaling by targeting TRAF6 and IKKα/β-NEMO complex;Citation107,108 (3) to potentiate TNF-α, poly (I:C) and Shigella infection-induced reactive oxygen species (ROS) production, which can further amplify NF-kB and JNK signaling as well as proinflammatory responses;Citation109 (4) to promote virus induced-autophagy in cooperation with mitochondrial Tu translation elongation factor (TUFM).Citation110 NLRC5 is another member of the NLR protein family, recognized as a novel regulator of both adaptive and innate immune responses.Citation111 Unlike NLRX1, NLRC5 expression is upregulated by many cytokines, such as IFNs and LPS.Citation112-114 We, and others, have identified NLRC5 as a negative regulator of both NF-κB and type I IFN signaling. NLRC5 inhibits IKK phosphorylation and NF-κB signaling by interacting with IKKα/β but not NEMO. Whereas, NLRC5 inhibits type I IFN signaling by targeting RIG-I/MDA5 after viral infection and blocking the RIG-I–MAVS interaction. Accordingly, knockdown of NLRC5 by siRNA enhanced the secretion of proinflammatory cytokines (IL-6, TNF-α), and TLR-stimulated antiviral responses in a variety of cell types.Citation112,113 Recently, we, and other groups, generated several lines of NLRC5−/− mice and found that NLRC5 deficiency markedly enhanced proinflammatory and antiviral responses in MEFs and peritoneal macrophages, but not in bone marrow-derived macrophages (BMMs) or BMDCs. This suggests that NLRC5 may regulate innate immune responses in a cell type-specific manner in vivo.Citation114-116 Other regulatory NLRs, such as NLRC3 and NLRP6, have also been identified as negative regulators of TLR-induced NF-κB and MAPK signaling.Citation117,118
Other intracellular inhibitors
SOCS1 belongs to the suppressor of cytokine signaling (SOCS) family and inhibits TLR-induced NF-κB signaling by targeting IRAK1. In accordance, SOCS1−/− mice have increased susceptibly to LPS-induced septic shock.Citation119,120 IRAKM is a member of IRAK kinase family and is predominantly expressed in peripheral blood cells. IRAKM−/− mice produce markedly enhanced levels of proinflammatory cytokines in response to LPS and CpG DNA, suggesting that IRAKM is a negative regulator of TLR-induced NF-κB activation.Citation121 In addition to IRAKM, Toll-interacting protein (TOLLIP) also targets IRAK1 to block TLR2 and TLR4 signaling by inhibiting IRAK1 autophosphorylation.Citation122
Regulation of inflammasomes
Recent studies have reported guanylate-binding protein 5 (GBP5) as a positive regulator of NLRP3 inflammasome. GBP5 stimulates NLRP3-ASC oligomerization by interacting with the pyrin domain of NLRP3 via its GTPase domain, and GBP5 promotes NLRP3 inflammasome activation induced by ATP, nigericin, and bacteria, but not crystalline agents.Citation123 In addition, MAVS, the key regulator of RLR signaling, has also been identified to facilitate NLRP3 inflammasome activation. Similar to GBP5, MAVS selectively promotes NLRP3 inflammasome activation in response to ATP and nigericin, but not to crystalline reagents such as silica and alumCitation124 The mitochondria-dependent NLRP3 inflammasome activation also requires the functional microtubule system and metabolic signals.Citation125
Numerous studies have focused on the role of posttranslational modification of inflammasomes. Several reports reveal that activation of NLRP3 inflammasome is strongly inhibited when NLRP3 is ubiquitinated, indicating that NLRP3 deubiquitination is an essential posttranslational protein modification for NLRP3 activation.Citation126-128 To date, BRCC3, a member of JAMM domain-containing Zn2+ metalloproteases, is the only deubiquitinating enzyme identified to activate NLRP3 inflammasome by deubiquitination.Citation128 Another report demonstrates that an E3 ligase, TRIM30, negatively regulates NLRP3 inflammasome activation by modulating reactive oxygen species production.Citation129 Whether other E3 ligases or deubiquitinases are required for regulating the NLRP3 inflammasome remains to be investigated. Furthermore, the AIM2 or NLRP3 inflammasome in macrophages triggers activation of the G protein, RalB, and formation of autophagosomes to eliminate active, ubiquitinated inflammasomes through autophagy.Citation130 Recently, a study shows that NLRP3, but not AIM2, is negatively regulated by NO-mediated S-nitrosylation.Citation131 Besides NLRP3, the ubiquitination of caspase-1 is also required for optimized inflammasome activity. cIAP1, cIAP2, and the adaptor protein, TRAF2, interact with caspase-1-containing complexes and mediate K63-linked polyubiquitination of caspase-1, thus, activating inflammasomes.Citation132
Phosphorylation is also a critical modification regulating inflammasome activity. Phosphorylation of NLRC4 is critical for the activity of NLRC4 inflammasome, and protein kinase C delta (PKC-δ) phosphorylates NLRC4.Citation133 However, a recent study resolved the structure of NLRC4 and found that inactive NLRC4 is also phosphorylated.Citation134 Thus, the relationship between NLRC4 phosphorylation state and function needs further investigation. The kinase, Syk, controls both pro-IL-1β synthesis and inflammasome activation after cell stimulation with Candida albicans.Citation135 Lyn and Syk kinases also positively regulate malarial hemozoin-induced NLRP3 inflammasome activation.Citation136 In addition, Syk and JNK are required for the activation of caspase-1 via the NLRP3 and AIM2 inflammasomes by phosphorylating ASC.Citation137 Double-stranded, RNA-dependent protein kinase (PKR) has recently been shown to contribute to the activation of the NLRP1, NLRP3, NLRC4, and AIM2 inflammasomes through direct interaction with NLRs and AIM2. Loss of PKR in macrophages results in attenuated IL-1β and IL-18 cleavage and HMGB release in response to various stimuli.Citation138,139 However, the exact role for phosphorylation in PKR-regulated inflammasome activation still needs further clarification ( and Table S1).
Figure 2. Activation and regulation of inflammasomes. The identified core components of inflammasomes belong to 2 families: (1) the NLR family (such as NLRP1b, NLRP3 and NLRC4); (2) the PYHIN (PYD and HIN200 domain-containing protein) family (such as AIM2). NLRs or AIM2, together with ASC, activate caspase-1, leading to downstream effector functions such as pyroptosis and processing of pro-IL-1β and pro-IL-18. Multiple regulators exert rigorous control on these pathways through their positive (green arrow) or negative (red blunt arrow) regulation. CARD, caspase recruitment domain; FIIND, domain with function to find; LRR, leucine-rich repeat; NACHT, nucleotide binding and oligomerization domain; NLR, Nod-like receptor; PYD, pyrin; HIN200, haematopoietic interferon-inducible nuclear antigens with 200 amino-acid repeats.
Regulation of type I IFN signaling pathway
Regulation of RLRs
RIG-I and MDA5 are key receptors for detecting viral RNAs and inducing type I IFN signaling to establish the antiviral state of the cell. Upon ligand binding, RIG-I undergoes a series of posttranslational modifications, including de-phosphorylation and ubiquitination, to form tetramers and, in turn, to activate MAVS.Citation12 The activity of RIG-I is tightly regulated by many positive and negative regulators. As an IFN stimulator, ZAPS associates with RIG-I to promote the oligomerization and ATPase activity of RIG-I, which leads to robust activation of type I IFN activation.Citation140 K63-linked ubiquitination of the RIG-I CARDs, as well as its CTD, is critical for RIG-I to elicit antiviral activity.Citation141,142 The E3 ligase, TRIM25, interacts with the CARDs of RIG-I and delivers Lys63-linked polyubiquitin chain at Lys172 of the RIG-I CARDs to enhance the interaction between RIG-I and MAVS.Citation141 Moreover, Riplet/RNF135/REUL promotes RIG-I activation, independent of TRIM25, by catalyzing K63linked polyubiquitin chains of RIG-I on its CTD domain.Citation142 A recent study shows that ubiquitin-specific protease 15 (USP15) targets TRIM25 to prevent the LUBAC-dependent degradation of TRIM25 and to promote RIG-I-mediated antiviral signaling.Citation143 It is also reported that an unanchored polyubiquitin chain can also facilitate RIG-I-mediated type I IFN signaling.Citation144 Most recently, a structural-based study demonstrates that both TRIM25-mediated ubiquitination of RIG-I CARD and non-covalent binding of K63-linked polyubiquitin chains induce its tetramer formation, and hence, activate MAVS.Citation145 Ubiquitination is a reversible process. We, and others, have identified 2 deubiquitinases, USP3 and USP21, which negatively regulate RIG-I activity by specifically removing the K63-linked ubiquitin chains on RIG-I.Citation146,147 RIG-I activity is also negatively regulated by RNF125 and Siglec-G-/SHP2/c-Cbl-mediated K48-linked ubiquitination and protein degradation.Citation148,149 Besides ubiquitination, RIG-I is also negatively regulated by Ser-Thr phosphorylation on CARD domain. In unstimulated cells, RIG-I is robustly phosphorylated at S8 and T170 by protein kinase C-α/β (PKCα/β),Citation150-152 whereas the phosphatases, PP1α and PP1γ activate both RIG-I and MDA5 signal pathways by dephosphorylating RIG-I or MDA5 upon viral infection.Citation153 Furthermore, virus-induced upregulation of a short RIG-I splice variant (RIG-I SV) negatively regulates RIG-I signaling in a dominant-negative manner.Citation154 In contrast to the regulatory mechanisms controlling RIG-I signaling activity, mechanisms controlling MDA5 signaling activity are less known. DAK is a dihydroacetone kinase, which associates with MDA5, but not RIG-I, and specifically inhibits MDA5-mediated IFN-β induction.Citation155 MDA5 can also be activated by the phosphatases, PP1α and PP1γ.Citation153 Our group has identified NLRC5 as a negative regulator for both RIG-I and MDA5 via direct interaction upon viral infection.Citation112 Recent evidence shows that K63-linked ubiquitin chains trigger MDA5 oligomerization and activate type I IFN signaling.Citation156 We found that the deubiquitinase, USP3, negatively regulates MDA5-mediated signaling by deubiquitinating its CARD domain.Citation146
Regulation of MAVS
MAVS is a critical adaptor protein for viral RNA-mediated antiviral immunity. NLRX1 was reported to negatively regulate type I IFN signaling by blocking RIG-I-MAVS interaction.Citation106 The autophagic proteins, Atg5 and Atg12, also negatively regulate type I IFN signaling by interacting with RIG-I and MAVS.Citation157 The stability of MAVS protein is regulated by several E3 ligases for 'fine tuning' of antiviral innate immunity. PCBP2 was identified to degrade MAVS in cooperation with a HECT domain-containing E3 ligase AIP4.Citation158 Smurf2 and TRIM25 are also negative regulators of type I IFN signaling by targeting MAVS for K48-linked ubiquitination and degradation.Citation159,160
Regulation of STING
STING has been identified as the most important adaptor protein for DNA-mediated type I IFN signaling. STING is regulated by both K48- and K63-linked ubiquitination. RNF5 ubiquitinates STING with K48 lineage for protein degradation.Citation161 By contrast, TRIM56 and TRIM32 positively regulate STING function by interacting with STING and targeting it for K63-linked ubiquitination. The K63-linked ubiquitination of STING may prompt the interaction between STING and TBK1, and thus, activate type I IFN signaling.Citation162,163 On the contrary, NLRC3 reduces STING-dependent innate immune activation in response to cyclic di-GMP and DNA viruses by impeding STING-TBK1 interaction.Citation164 Recently, the autophagy related serine/threonine UNC-51-like kinase (ULK1/Atg1) has been identified as a novel, negative regulator of STING. After autophagy-dependent delivery of STING to endosomal/lysosomal compartments and activation of IRF3 and NF-κB, ULK1 is free from its repressor, AMP activated protein kinase (AMPK), and subsequently, phosphorylates STING to suppress its function.Citation165
Regulation of TRAF3
The tyrosine kinase, Syk, positively regulates type I IFN production, in response to TLR4 ligand, by interacting with TRAF3 and TBK1. Syk-deficient macrophages exhibited decreased production of type I IFNs.Citation166 A recent study shows that linear, ubiquitinated NEMO, mediated by the linear ubiquitin assembly complex (LUBAC), interacts with TRAF3 and disrupts the MAVS-TRAF3 complex to block type I IFN signaling.Citation167 Deubiquitinating enzyme A (DUBA, also known as OTUD5) was the first identified deubiquitinase to remove K63-linked poly-ubiquitin chains of TRAF3 and negatively regulate type I IFN signaling.Citation168 Recently, HSCARG was found to inhibit TRAF3 ubiquitination via recruiting deubiquitinase OTUB1, therefore negatively regulating RIG-I signaling.Citation169 The protein stability of TRAF3 is controlled by the E3 ubiquitin ligase, TRIAD3A, which leads to K48-linked ubiquitination of TRAF3, and subsquent proteasomal degradation.Citation170 In contrast, USP25 positively regulates TLR4-dependent innate immune responses through deubiquitinating TRAF3. USP25 specifically removes the K48-linked ubiquitination of TRAF3, which is mediated by the E3 ubiquitin ligase, cIAP2, and enables a balanced innate immune response.Citation171
Regulation of TBK1
It is known that TBK1 has several binding partners, including TRAF family member-associated NF-κB activator (TANK), NAK-associated protein (NAP1), and similar to NAP1 TBK1 adaptor (SINTBAD), essential for its activation.Citation172 Recently, GSK3β was shown to interact with TBK1 during virus infection. GSK3 enhances TBK1 self-association and autophosphorylation at Ser 172, which is critical for IRF3 activation.Citation173 The activity of TBK1 is also regulated by a robust, reversible ubiquitination process. TBK1 undergoes ubquitination with K63 lineage by mind bomb (MIB) or Ndrp1 to promote interferon production in response to RLR or TLR ligands, respectively.Citation91,174 However, ABIN1, TAX1BP1 and A20 blocks antiviral signaling by disrupting K63-linked polyubiquitination of TBK1/IKKi.Citation175,176 Recently, we have found that NLRP4 serves as a negative regulator of type I IFN signaling by targeting active TBK1 for degradation. NLRP4-mediated type I IFN signaling is regulated in a TBK1-IRF3-dependent manner, but has no effect on the MyD88-IRF7-dependent pathway. After viral infection or TLR stimulation, NLRP4 specifically interacts with TBK1 and recruits the E3 ubiquitin ligase, DTX4, to promote K48-linked polyubiquitination and degradation of TBK1.Citation177 Another group found the E3 ligase, TRAF-interacting protein (TRIP), also promotes proteasomal degradation of TBK1.Citation178 Interestingly, we observed that TRIP negatively regulates TBK1 in a NLRP4-dependent manner, suggesting that NLRP4 is paramount in mediating TBK1 stability (unpublished data).
Regulation of IRF3/IRF7
IRF3 and IRF7 are master transcriptional factors of the type I IFN pathway. IRF3 and IRF7 can be activated by TLR3, TLR4 or RLRs ligands in macrophages, MEFs and conventional DCs (cDCs). Notably, IRF7 can also be activated by TLR7 and TLR9 ligands through MyD88 signaling, and IRF7 is essential for type I IFN production in plasmacytoid dendritic cells (pDCs).Citation179 Thus, the activity of IRF3/IRF7 must be tightly regulated. It has been reported that IRF7 is targeted for degradation by binding to the RTA immediate-early nuclear transcription factor encoded by Kaposi's sarcoma-associated herpesvirus (KSHV).Citation180 In addition, an RTA-associated E3 ligase RAUL (also known as KIAA10 or UBE3C) negatively regulates type I IFN by targeting both IRF3 and IRF7 for K48-linked ubiquitination and degradation.Citation181 The N-Myc and STATs interactor (Nmi), a Sendai virus-inducible protein, was also found to promote the K48-linked ubiquitination and the proteasome-dependent degradation of IRF7.Citation182 Similarly, the E3 ligase, Ro52 (TRIM21), targets IRF3 for degradation.Citation183 IRF3 can also be deactivated via dephosphorylation mediated by the serine and threonine phosphatase, PP2A, and its adaptor protein, RACK1. PP2A-deficient macrophages show enhanced type I IFN signaling with diverse TLR ligands and viral infections ( and Table S1).Citation184
Figure 3. A schematic representation of cytosolic RNA and DNA-induced type I IFN signaling pathway and its regulators. RIG-I and MDA5 recognize different groups of viral RNAs and initiate signaling cascades that begin with prion-like polymerization of MAVS. MAVS recruits and activates TRAF2, TRAF3, TRAF5, and TRAF6. Polyubiquitin chains on these ligases are sensed by NEMO through its ubiquitin-binding domains, in turn, recruiting IKK and TBK1 complexes to phosphorylate IκBα and IRF3, respectively. LGP2 may function as a regulator to modulate the activity of RIG-I and MDA5. Viral DNA could activate cGAS and other DNA sensors, which are all proposed to transduce signals to the ER-localized adaptor protein STING. STING triggers TBK1‑dependent type I IFN response. RNA polymerase III transcribes the DNA into 5'ppp-RNA, which triggers the RIG-I pathway. Most of the key molecules involved in the RNA- and DNA-sensing pathways can be targeted by E3 ligases, deubiquitinases or regulatory NLRs, which negatively (red blunt arrow) or positively (green arrow) regulate type I IFN responses.
Double-edged Sword Roles of PRRs and Innate Immune Regulators in Cancer
Activation of the innate immune response through TLR- and NLR- signaling pathways serves as a link between chronic inflammation and cancer.Citation185-188 It is well-demonstrated that chronic inflammation is a major driving force in cancer development.Citation189,190 Chronic infection or chronic inflammation causes about 20% of human cancers.Citation189,191 Thus, chronic inflammation, primarily caused by abnormal NF-κB or inflammasome activation, is tightly linked to cancer through PRRs-mediated cytokine productionCitation186,Citation192-194 Emerging evidence suggests that PRRs and their regulators have both favorable and unfavorable consequences on cancer cells. On the one hand, certain PRRs induce an anti-tumor immune response to inhibit tumor progression. Conversely, uncontrolled innate immune signaling may provide a microenvironment for cancer cell proliferation and immune surveillance evasion.
The role of TLRs and their regulators in cancer
Several studies have demonstrated a significant association between prostate cancer and TLR sequence variants, including TLR4, TLR1, TLR6 and TLR10. These studies suggest that TLR sequence polymorphisms are an important risk factor for prostate cancer.Citation190,195,196 However, this may be dependent on the specific TLR, the tumor type, and the types of cells expressing the TLR. Functional analyses show that the loss of TLR4, due to polymorphisms, in human prostate and breast tumor cell lines, inhibits tumor cell proliferation and invasion, and induces tumor cell apoptosis.Citation197,198 In contrast, TLR2−/− mice develop significant larger and more colorectal tumors than wild-type mice, suggesting a protective role of TLR2 in colorectal tumors.Citation199 In addition, activation of TLR3 signaling by poly (I:C) inhibits tumor cell proliferation and induces apoptosis.Citation200,201 Wang et al. reported TLR9 activation in non-small cell lung cancer cells sensitizes tumor cells to apoptosis, which, in turn, leads to tumor growth arrest.Citation202 Collectively, these results reveal anti-tumor functions of certain TLRs. Alternatively, another report demonstrates that specific ligand activation of TLR2, 3 and 4 on human melanoma cells induces cell migration and promotes metastatic events, suggesting that the function of TLR signaling may be tumor specific.Citation203 Another possible interpretation of these results is that TLRs are sensors of many immune cells, and the activation of TLRs by PAMPs can stimulate diverse innate immune pathways (e.g., the NF-κB pathway) to secrete proinflammatory cytokines.
Recent studies identify an important role for MyD88, a molecule downstream in TLR-signaling pathways, in tumor development. MyD88 deficiency reduces the incidence of cancer in mice in an AOM-DSS-induced colon cancer model.Citation204 Loss of MyD88 also reduces chemically induced skin cancer or liver cancer development.Citation205,206 These studies suggest that MyD88-mediated signaling plays a key role in promoting tumor development and progression. However, another study shows that loss of MyD88 increases tumor incidence in AOM-DSS-induced model, due to the reduced ability to heal ulcers and repair DNA damage.Citation207 This further reiterates the dual roles of TLR signaling in cancer.
The role of NLRs in cancer
Nod2 has been the focus of many studies because of its implication in Crohn's disease.Citation208 Notably, inactivating polymorphisms of Nod2 have been associated with increased susceptibility to colorectal cancer in several patient cohorts.Citation209 Similar to that observed in patients with Crohn's disease, Nod2 deficiency leads to increased colorectal cancer development in mice. Both Nod2-deficient mice and patients with Nod2 mutations have intestinal dysbiosis.Citation210 A recent report shows that Nod2-deficient mice are susceptible to dysbiosis, and the susceptibility to inflammation-induced colorectal cancer is transmissible to wild-type mice by co-housing.Citation211
Emerging evidence implicates a pivotal role for NLR inflammasomes in tumor development. NLRP3 is highly expressed in haematopoietic cells but not intestinal epithelial cells. NLRP3−/− mice have increased colitis-associated cancer development.Citation212 Similarly, ASC and caspase-1 deficient mice are susceptible to DSS-induced tissue injury.Citation213,214 Further, another study demonstrates that NLRP3-dependent IL-18 production exerts a protective role against colorectal tumorigenesis through IFN-γ production and STAT1 signaling.Citation214 NLRP6 is highly expressed in intestinal tissue and has an important role in maintaining intestinal homeostasis. NLRP6 inflammasome deficiency leads to aberrant inflammation within the colon and to colitis-induced tumorigenesis.Citation215 NLRP6 may function as a negative regulator of colorectal tumorigenesis, partly through controlling epithelial cell self-renewal upon injury.Citation216 Flavell and his colleagues found that the NLRP6 inflammasome in intestinal epithelial cells may play a role in maintaining healthy gut flora.Citation217 In addition, the NLRC4 inflammasome is also related to inflammation-induced colorectal tumor formation in mice. Loss of the NLRC4 inflammasome causes enhanced epithelial cell proliferation and reduced apoptosis.Citation218 Therefore, NLRs and NLR inflammasomes are promising therapeutic targets in inflammation-induced cancers.
Type I IFN regulators and cancer
A recent study demonstrated significant downregulation of RIG-I expression in human hepatocellular carcinoma (HCC) tissues, which was correlated with poorer patient prognosis and diminished response to IFNα therapy. Mechanistically, RIG-I enhances IFNα production by amplifying IFN-JAK-STAT signaling via promoting STAT1 activation.Citation219 It has also been demonstrated that TBK1 plays a critical role in Kras-mediated tumor development. Suppression of TBK1-induced apoptosis, specifically in human lung cancer cell lines, depends on oncogenic KRAS expression.Citation220 In addition, many negative regulators of type I IFN signaling have been shown to be involved in cancer. For example, A20 and CYLD have been identified as tumor suppressors, and mutations in A20 and CYLD have been identified in different tumor types.Citation100,221,222
Conclusions
Since their discovery 20 y ago, TLRs and RLRs have been shown to be essential for efficient innate and adaptive immunity, and the mechanisms regulating TLR-mediated signaling pathways have been extensively studied. In contrast, for many NLRs and other viral DNA sensors, their recognized ligands and their signaling pathways remain to be determined. This review provides an updated overview of the rapidly evolving field of PRR biology and PRR signaling pathways. These innate immune receptors and their regulators are clearly important in host defense against infectious diseases, in autoimmune diseases as well as in cancer. However, challenges still remain in the understanding of how the different activation pathways of PRRs converge or diverge, and how the different innate immune signaling pathways are regulated at the molecular level. The exact roles of these PRRs in cancer immunosurveillance also remain to be determined. Understanding the regulation of innate immune signaling, cytokine production and their relationship to adaptive immune responses in cancer will be critically important for the development of cancer diagnostics and prognostics as well as effective cancer therapeutics.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Table_1.doc
Download MS Word (140 KB)Funding
This work was in part supported by Key Basic Research Program of China (2014CB910800), the National Natural Science Foundation of China (31370869), Guangdong Natural Science Funds for Distinguished Young Scholar (S2013050014772) and the National Guangdong Innovative Research Team Program (2011Y035 and 201001Y0104687244) as well as grants (R01CA101795, R01CA09327 and DA030338) from the National Cancer Institute and National Institute on Drug Abuse, NIH, and Cancer Prevention and Research Institute of Texas (CPRIT RP121048).
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124:783-801; PMID:16497588; http://dx.doi.org/10.1016/j.cell.2006.02.015
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140:805-20; PMID:20303872; http://dx.doi.org/10.1016/j.cell.2010.01.022
- Paludan SR, Bowie AG. Immune sensing of DNA. Immunity 2013; 38:870-80; PMID:23706668; http://dx.doi.org/10.1016/j.immuni.2013.05.004
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol 2004; 5:987-95; PMID:15454922; http://dx.doi.org/10.1038/ni1112
- Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol 2005; 17:1-14; PMID:15585605; http://dx.doi.org/10.1093/intimm/dxh186
- Basith S, Manavalan B, Yoo TH, Kim SG, Choi S. Roles of toll-like receptors in cancer: a double-edged sword for defense and offense. Arch Pharm Res 2012; 35:1297-316; PMID:22941474; http://dx.doi.org/10.1007/s12272-012-0802-7
- Hoving JC, Wilson GJ, Brown GD. Signalling C-type lectin receptors, microbial recognition and immunity. Cell Microbiol 2014; 16:185-94; PMID:24330199; http://dx.doi.org/10.1111/cmi.12249
- Drummond RA, Brown GD. Signalling C-type lectins in antimicrobial immunity. PLoS Pathog 2013; 9:e1003417; PMID:23935480; http://dx.doi.org/10.1371/journal.ppat.1003417
- Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol 2009; 9:465-79; PMID:19521399; http://dx.doi.org/10.1038/nri2569
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature 2006; 442:39-44; PMID:16823444; http://dx.doi.org/10.1038/nature04946
- Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity 2011; 34:665-79; PMID:21616436; http://dx.doi.org/10.1016/j.immuni.2011.05.007
- Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity 2013; 38:855-69; PMID:23706667; http://dx.doi.org/10.1016/j.immuni.2013.05.007
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol 2003; 21:335-76; PMID:12524386; http://dx.doi.org/10.1146/annurev.immunol.21.120601.141126
- Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 2007; 13:460-9; PMID:18029230; http://dx.doi.org/10.1016/j.molmed.2007.09.002
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 2004; 101:5598-603; PMID:15034168; http://dx.doi.org/10.1073/pnas.0400937101
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004; 303:1526-9; PMID:14976262; http://dx.doi.org/10.1126/science.1093620
- Peng G, Guo Z, Kiniwa Y, Voo KS, Peng W, Fu T, Wang DY, Li Y, Wang HY, Wang RF. Toll-like receptor 8-mediated reversal of CD4+ regulatory T cell function. Science 2005; 309:1380-4; PMID:16123302; http://dx.doi.org/10.1126/science.1113401
- O'Neill LA, Fitzgerald KA, Bowie AG. The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol 2003; 24:286-90; PMID:12810098
- Uematsu S, Akira S. Toll-like receptors and innate immunity. J Mol Med (Berl) 2006; 84:712-25; PMID:16924467; http://dx.doi.org/10.1007/s00109-006-0084-y
- West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol 2006; 22:409-37; PMID:16822173; http://dx.doi.org/10.1146/annurev.cellbio.21.122303.115827
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 2000; 103:351-61; PMID:11057907
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 2001; 412:346-51; PMID:11460167; http://dx.doi.org/10.1038/35085597
- Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci U S A 2002; 99:5567-72; PMID:11960013; http://dx.doi.org/10.1073/pnas.082100399
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell 2008; 132:344-62; PMID:18267068; http://dx.doi.org/10.1016/j.cell.2008.01.020
- Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 2005; 434:1035-40; PMID:15815647; http://dx.doi.org/10.1038/nature03547
- Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 2005; 434:243-9; PMID:15665823; http://dx.doi.org/10.1038/nature03308
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 2003; 301:640-3; PMID:12855817; http://dx.doi.org/10.1126/science.1087262
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol 2006; 7:1250-7; PMID:17110941; http://dx.doi.org/10.1038/ni1412
- Magalhaes JG, Sorbara MT, Girardin SE, Philpott DJ. What is new with Nods? Curr Opin Immunol 2011; 23:29-34; PMID:21190821; http://dx.doi.org/10.1016/j.coi.2010.12.003
- Chen G, Shaw MH, Kim YG, Nunez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol 2009; 4:365-98; PMID:18928408; http://dx.doi.org/10.1146/annurev.pathol.4.110807.092239
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010; 11:55-62; PMID:19898471; http://dx.doi.org/10.1038/ni.1823
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med 2010; 16:90-7; PMID:19966812; http://dx.doi.org/10.1038/nm.2069
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol 2009; 10:1073-80; PMID:19701189; http://dx.doi.org/10.1038/ni.1782
- Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol 2012; 13:325-32; PMID:22430785; http://dx.doi.org/10.1038/ni.2231
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 2014; 156:1207-22; PMID:24630723; http://dx.doi.org/10.1016/j.cell.2014.01.063
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schröder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 2014; 156:1193-206; PMID:24630722; http://dx.doi.org/10.1016/j.cell.2014.02.008
- Jin T, Curry J, Smith P, Jiang J, Xiao TS. Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1. Proteins 2013; 81:1266-70; PMID:23508996; http://dx.doi.org/10.1002/prot.24287
- Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I, Reed JC. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell 2007; 25:713-24; PMID:17349957; http://dx.doi.org/10.1016/j.molcel.2007.01.032
- Gregory SM, Davis BK, West JA, Taxman DJ, Matsuzawa S, Reed JC, Ting JP, Damania B. Discovery of a viral NLR homolog that inhibits the inflammasome. Science 2011; 331:330-4; PMID:21252346; http://dx.doi.org/10.1126/science.1199478
- Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet 2006; 38:240-4; PMID:16429160; http://dx.doi.org/10.1038/ng1724
- Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 2011; 477:592-5; PMID:21874021; http://dx.doi.org/10.1038/nature10394
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 2011; 477:596-600; PMID:21918512; http://dx.doi.org/10.1038/nature10510
- Halff EF, Diebolder CA, Versteeg M, Schouten A, Brondijk TH, Huizinga EG. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J Biol Chem 2012; 287:38460-72; PMID:23012363; http://dx.doi.org/10.1074/jbc.M112.393512
- Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A 2013; 110:14408-13; PMID:23940371; http://dx.doi.org/10.1073/pnas.1306376110
- Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 2004; 20:319-25; PMID:15030775
- Wen H, Miao EA, Ting JP. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity 2013; 39:432-41; PMID:24054327; http://dx.doi.org/10.1016/j.immuni.2013.08.037
- Latz E. The inflammasomes: mechanisms of activation and function. Curr Opin Immunol 2010; 22:28-33; PMID:20060699; http://dx.doi.org/10.1016/j.coi.2009.12.004
- Mitoma H, Hanabuchi S, Kim T, Bao M, Zhang Z, Sugimoto N, Liu YJ. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity 2013; 39:123-35; PMID:23871209; http://dx.doi.org/10.1016/j.immuni.2013.07.001
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 2009; 458:509-13; PMID:19158676; http://dx.doi.org/10.1038/nature07710
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009; 458:514-8; PMID:19158675; http://dx.doi.org/10.1038/nature07725
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 2009; 323:1057-60; PMID:19131592; http://dx.doi.org/10.1126/science.1169841
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 2009; 10:266-72; PMID:19158679; http://dx.doi.org/10.1038/ni.1702
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol 2010; 11:385-93; PMID:20351693; http://dx.doi.org/10.1038/ni.1859
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 2010; 11:395-402; PMID:20351692; http://dx.doi.org/10.1038/ni.1864
- Loo YM, Gale M Jr. Immune signaling by RIG-I-like receptors. Immunity 2011; 34:680-92; PMID:21616437; http://dx.doi.org/10.1016/j.immuni.2011.05.003
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 2005; 19:727-40; PMID:16153868; http://dx.doi.org/10.1016/j.molcel.2005.08.014
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 2005; 6:981-8; PMID:16127453; http://dx.doi.org/10.1038/ni1243
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005; 122:669-82; PMID:16125763; http://dx.doi.org/10.1016/j.cell.2005.08.012
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 2005; 437:1167-72; PMID:16177806; http://dx.doi.org/10.1038/nature04193
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011; 146:448-61; PMID:21782231; http://dx.doi.org/10.1016/j.cell.2011.06.041
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A 2007; 104:582-7; PMID:17190814; http://dx.doi.org/10.1073/pnas.0606699104
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A 2010; 107:1512-7; PMID:20080593; http://dx.doi.org/10.1073/pnas.0912986107
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007; 448:501-5; PMID:17618271; http://dx.doi.org/10.1038/nature06013
- DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Fruh K. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J Virol 2010; 84:585-98; PMID:19846511; http://dx.doi.org/10.1128/JVI.01748-09
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 2008; 451:725-9; PMID:18256672; http://dx.doi.org/10.1038/nature06537
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 2009; 138:576-91; PMID:19631370; http://dx.doi.org/10.1016/j.cell.2009.06.015
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol 2009; 10:1065-72; PMID:19609254; http://dx.doi.org/10.1038/ni.1779
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 2011; 12:959-65; PMID:21892174; http://dx.doi.org/10.1038/ni.2091
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 2010; 11:997-1004; PMID:20890285; http://dx.doi.org/10.1038/ni.1932
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008; 455:674-8; PMID:18724357; http://dx.doi.org/10.1038/nature07317
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009; 461:788-92; PMID:19776740; http://dx.doi.org/10.1038/nature08476
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med 2009; 206:1899-911; PMID:19652017; http://dx.doi.org/10.1084/jem.20082874
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011; 478:515-8; PMID:21947006; http://dx.doi.org/10.1038/nature10429
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013; 339:826-30; PMID:23258412; http://dx.doi.org/10.1126/science.1229963
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013; 339:786-91; PMID:23258413; http://dx.doi.org/10.1126/science.1232458
- Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol 2005; 5:446-58; PMID:15928677; http://dx.doi.org/10.1038/nri1630
- Qian C, Cao X. Regulation of Toll-like receptor signaling pathways in innate immune responses. Ann N Y Acad Sci 2013; 1283:67-74; PMID:23163321; http://dx.doi.org/10.1111/j.1749-6632.2012.06786.x
- Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 2011; 147:868-80; PMID:22078883; http://dx.doi.org/10.1016/j.cell.2011.09.051
- Baumann CL, Aspalter IM, Sharif O, Pichlmair A, Bluml S, Grebien F, Bruckner M, Pasierbek P, Aumayr K, Planyavsky M, et al. CD14 is a coreceptor of Toll-like receptors 7 and 9. J Exp Med 2010; 207:2689-701; PMID:21078886; http://dx.doi.org/10.1084/jem.20101111
- Iwami KI, Matsuguchi T, Masuda A, Kikuchi T, Musikacharoen T, Yoshikai Y. Cutting edge: naturally occurring soluble form of mouse Toll-like receptor 4 inhibits lipopolysaccharide signaling. J Immunol 2000; 165:6682-6; PMID:11120784
- LeBouder E, Rey-Nores JE, Rushmere NK, Grigorov M, Lawn SD, Affolter M, Griffin GE, Ferrara P, Schiffrin EJ, Morgan BP, et al. Soluble forms of Toll-like receptor (TLR)2 capable of modulating TLR2 signaling are present in human plasma and breast milk. J Immunol 2003; 171:6680-9; PMID:14662871
- Janssens S, Burns K, Tschopp J, Beyaert R. Regulation of interleukin-1- and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88. Curr Biol 2002; 12:467-71; PMID:11909531
- Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med 2003; 197:263-8; PMID:12538665
- Wald D, Qin J, Zhao Z, Qian Y, Naramura M, Tian L, Towne J, Sims JE, Stark GR, Li X. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol 2003; 4:920-7; PMID:12925853; http://dx.doi.org/10.1038/ni968
- Garlanda C, Riva F, Polentarutti N, Buracchi C, Sironi M, De Bortoli M, Muzio M, Bergottini R, Scanziani E, Vecchi A, et al. Intestinal inflammation in mice deficient in Tir8, an inhibitory member of the IL-1 receptor family. Proc Natl Acad Sci U S A 2004; 101:3522-6; PMID:14993616; http://dx.doi.org/10.1073/pnas.0308680101
- Brint EK, Xu D, Liu H, Dunne A, McKenzie AN, O'Neill LA, Liew FY. ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance. Nat Immunol 2004; 5:373-9; PMID:15004556; http://dx.doi.org/10.1038/ni1050
- Diehl GE, Yue HH, Hsieh K, Kuang AA, Ho M, Morici LA, Lenz LL, Cado D, Riley LW, Winoto A. TRAIL-R as a negative regulator of innate immune cell responses. Immunity 2004; 21:877-89; PMID:15589175; http://dx.doi.org/10.1016/j.immuni.2004.11.008
- Ji S, Sun M, Zheng X, Li L, Sun L, Chen D, Sun Q. Cell-surface localization of Pellino antagonizes Toll-mediated innate immune signalling by controlling MyD88 turnover in Drosophila. Nat Commun 2014; 5:3458; PMID:24632597; http://dx.doi.org/10.1038/ncomms4458
- Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol 2004; 5:495-502; PMID:15107846; http://dx.doi.org/10.1038/ni1066
- Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol 2008; 9:369-77; PMID:18345001; http://dx.doi.org/10.1038/ni1577
- Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. The E3 ubiquitin ligase Nrdp1 'preferentially' promotes TLR-mediated production of type I interferon. Nat Immunol 2009; 10:744-52; PMID:19483718; http://dx.doi.org/10.1038/ni.1742
- Opipari AW, Jr, Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem 1990; 265:14705-8; PMID:2118515
- Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem 1992; 267:17971-6; PMID:1381359
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 2004; 5:1052-60; PMID:15334086; http://dx.doi.org/10.1038/ni1110
- Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 2010; 327:1135-9; PMID:20185725; http://dx.doi.org/10.1126/science.1182364
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 2004; 430:694-9; PMID:15258597; http://dx.doi.org/10.1038/nature02794
- Shembade N, Harhaj NS, Parvatiyar K, Copeland NG, Jenkins NA, Matesic LE, Harhaj EW. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol 2008; 9:254-62; PMID:18246070; http://dx.doi.org/10.1038/ni1563
- Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J 2009; 28:513-22; PMID:19131965; http://dx.doi.org/10.1038/emboj.2008.285
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 2000; 289:2350-4; PMID:11009421
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 2003; 424:801-5; PMID:12917691; http://dx.doi.org/10.1038/nature01802
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 2003; 424:793-6; PMID:12917689; http://dx.doi.org/10.1038/nature01803
- Reiley WW, Zhang M, Jin W, Losiewicz M, Donohue KB, Norbury CC, Sun SC. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol 2006; 7:411-7; PMID:16501569; http://dx.doi.org/10.1038/ni1315
- Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EF, Leonard TO, Norbury CC, Fitzpatrick L, Zhang M, et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med 2007; 204:1475-85; PMID:17548520; http://dx.doi.org/10.1084/jem.20062694
- Friedman CS, O'Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep 2008; 9:930-6; PMID:18636086; http://dx.doi.org/10.1038/embor.2008.136
- Lee AJ, Zhou X, Chang M, Hunzeker J, Bonneau RH, Zhou D, Sun SC. Regulation of natural killer T-cell development by deubiquitinase CYLD. EMBO J 2010; 29:1600-12; PMID:20224552; http://dx.doi.org/10.1038/emboj.2010.31
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 2008; 451:573-7; PMID:18200010; http://dx.doi.org/10.1038/nature06501
- Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity 2011; 34:854-65; PMID:21703540; http://dx.doi.org/10.1016/j.immuni.2011.03.026
- Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, Yang X, Hong J, Songyang Z, Chen ZJ, et al. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity 2011; 34:843-53; PMID:21703539; http://dx.doi.org/10.1016/j.immuni.2011.02.022
- Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin SE. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep 2008; 9:293-300; PMID:18219313; http://dx.doi.org/10.1038/sj.embor.7401161
- Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, Swanson KV, Wen KW, Damania B, Moore CB, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity 2012; 36:933-46; PMID:22749352; http://dx.doi.org/10.1016/j.immuni.2012.03.025
- Kobayashi KS, van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nat Rev Immunol 2012; 12:813-20; PMID:23175229; http://dx.doi.org/10.1038/nri3339
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell 2010; 141:483-96; PMID:20434986; http://dx.doi.org/10.1016/j.cell.2010.03.040
- Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol 2010; 185:1681-91; PMID:20610642; http://dx.doi.org/10.4049/jimmunol.0903900
- Tong Y, Cui J, Li Q, Zou J, Wang HY, Wang RF. Enhanced TLR-induced NF-kappaB signaling and type I interferon responses in NLRC5 deficient mice. Cell Res 2012; 22:822-35; PMID:22473004; http://dx.doi.org/10.1038/cr.2012.53
- Kumar H, Pandey S, Zou J, Kumagai Y, Takahashi K, Akira S, Kawai T. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J Immunol 2011; 186:994-1000; PMID:21148033; http://dx.doi.org/10.4049/jimmunol.1002094
- Staehli F, Ludigs K, Heinz LX, Seguin-Estevez Q, Ferrero I, Braun M, Schroder K, Rebsamen M, Tardivel A, Mattmann C, et al. NLRC5 deficiency selectively impairs MHC class I- dependent lymphocyte killing by cytotoxic T cells. J Immunol 2012; 188:3820-8; PMID:22412192; http://dx.doi.org/10.4049/jimmunol.1102671
- Schneider M, Zimmermann AG, Roberts RA, Zhang L, Swanson KV, Wen H, Davis BK, Allen IC, Holl EK, Ye Z, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol 2012; 13:823-31; PMID:22863753; http://dx.doi.org/10.1038/ni.2378
- Anand PK, Malireddi RK, Lukens JR, Vogel P, Bertin J, Lamkanfi M, Kanneganti TD. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 2012; 488:389-93; PMID:22763455; http://dx.doi.org/10.1038/nature11250
- Kinjyo I, Hanada T, Inagaki-Ohara K, Mori H, Aki D, Ohishi M, Yoshida H, Kubo M, Yoshimura A, et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 2002; 17:583-91; PMID:12433365
- Nakagawa R, Naka T, Tsutsui H, Fujimoto M, Kimura A, Abe T, Seki E, Sato S, Takeuchi O, Takeda K, et al. SOCS-1 participates in negative regulation of LPS responses. Immunity 2002; 17:677-87; PMID:12433373
- Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr., Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 2002; 110:191-202; PMID:12150927
- Zhang G, Ghosh S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J Biol Chem 2002; 277:7059-65; PMID:11751856; http://dx.doi.org/10.1074/jbc.M109537200
- Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, MacMicking JD. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science 2012; 336:481-5; PMID:22461501; http://dx.doi.org/10.1126/science.1217141
- Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 2013; 153:348-61; PMID:23582325; http://dx.doi.org/10.1016/j.cell.2013.02.054
- Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol 2013; 14:454-60; PMID:23502856; http://dx.doi.org/10.1038/ni.2550
- Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem 2012; 287:36617-22; PMID:22948162; http://dx.doi.org/10.1074/jbc.M112.407130
- Lopez-Castejon G, Luheshi NM, Compan V, High S, Whitehead RC, Flitsch S, Kirov A, Prudovsky I, Swanton E, Brough D. Deubiquitinases regulate the activity of caspase-1 and interleukin-1beta secretion via assembly of the inflammasome. J Biol Chem 2013; 288:2721-33; PMID:23209292; http://dx.doi.org/10.1074/jbc.M112.422238
- Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell 2013; 49:331-8; PMID:23246432; http://dx.doi.org/10.1016/j.molcel.2012.11.009
- Hu Y, Mao K, Zeng Y, Chen S, Tao Z, Yang C, Sun S, Wu X, Meng G, Sun B. Tripartite-motif protein 30 negatively regulates NLRP3 inflammasome activation by modulating reactive oxygen species production. J Immunol 2010; 185:7699-705; PMID:21048113; http://dx.doi.org/10.4049/jimmunol.1001099
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012; 13:255-63; PMID:22286270; http://dx.doi.org/10.1038/ni.2215
- Mishra BB, Rathinam VA, Martens GW, Martinot AJ, Kornfeld H, Fitzgerald KA, Sassetti CM. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nat Immunol 2013; 14:52-60; PMID:23160153; http://dx.doi.org/10.1038/ni.2474
- Labbe K, McIntire CR, Doiron K, Leblanc PM, Saleh M. Cellular inhibitors of apoptosis proteins cIAP1 and cIAP2 are required for efficient caspase-1 activation by the inflammasome. Immunity 2011; 35:897-907; PMID:22195745; http://dx.doi.org/10.1016/j.immuni.2011.10.016
- Qu Y, Misaghi S, Izrael-Tomasevic A, Newton K, Gilmour LL, Lamkanfi M, Louie S, Kayagaki N, Liu J, Kömüves L, et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 2012; 490:539-42; PMID:22885697; http://dx.doi.org/10.1038/nature11429
- Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 2013; 341:172-5; PMID:23765277; http://dx.doi.org/10.1126/science.1236381
- Gross O, Poeck H, Bscheider M, Dostert C, Hannesschlager N, Endres S, Hartmann G, Tardivel A, Schweighoffer E, Tybulewicz V, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature 2009; 459:433-6; PMID:19339971; http://dx.doi.org/10.1038/nature07965
- Shio MT, Eisenbarth SC, SavariaM, Vinet AF, Bellemare MJ, Harder KW, Sutterwala FS, Bohle DS, Descoteaux A, Flavell RA, et al. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog 2009; 5:e1000559; PMID:19696895; http://dx.doi.org/10.1371/journal.ppat.1000559
- Hara H, Tsuchiya K, Kawamura I, Fang R, Hernandez-Cuellar E, Shen Y, et al. Phosphorylation of the adaptor ASC acts as a molecular switch that controls the formation of speck-like aggregates and inflammasome activity. Nat Immunol 2013; 14:1247-55; PMID:24185614; http://dx.doi.org/10.1038/ni.2749
- Lu B, Nakamura T, Inouye K, Li J, Tang Y, Lundback P, Valdes-Ferrer SI, Olofsson PS, Kalb T, Roth J, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature 2012; 488:670-4; PMID:22801494; http://dx.doi.org/10.1038/nature11290
- He Y, Franchi L, Nunez G. The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. Eur J Immunol 2013; 43:1147-52; PMID:23401008; http://dx.doi.org/10.1002/eji.201243187
- Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, Goto S, Kameoka S, Fujikura D, Yamada T, et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol 2011; 12:37-44; PMID:21102435; http://dx.doi.org/10.1038/ni.1963
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007; 446:916-20; PMID:17392790; http://dx.doi.org/10.1038/nature05732
- Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem 2009; 284:807-17; PMID:19017631; http://dx.doi.org/10.1074/jbc.M804259200
- Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, Gack MU. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal 2014; 7:ra3; PMID:24399297; http://dx.doi.org/10.1126/scisignal.2004577
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 2010; 141:315-30; PMID:20403326; http://dx.doi.org/10.1016/j.cell.2010.03.029
- Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 2014; 509:110-4; PMID:24590070; http://dx.doi.org/10.1038/nature13140
- Cui J, Song Y, Li Y, Zhu Q, Tan P, Qin Y, Wang HY, Wang RF. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res 2014; 24:400-16; PMID:24366338; http://dx.doi.org/10.1038/cr.2013.170
- Fan Y, Mao R, Yu Y, Liu S, Shi Z, Cheng J, Zhang H, An L, Zhao Y, Xu X, et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med 2014; 211:313-28; PMID:24493797; http://dx.doi.org/10.1084/jem.20122844
- Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A 2007; 104:7500-5; PMID:17460044; http://dx.doi.org/10.1073/pnas.0611551104
- Chen W, Han C, Xie B, Hu X, Yu Q, Shi L, Wang Q, Li D, Wang J, Zheng P, et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell 2013; 152:467-78; PMID:23374343; http://dx.doi.org/10.1016/j.cell.2013.01.011
- Gack MU, Nistal-Villan E, Inn KS, Garcia-Sastre A, Jung JU. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol 2010; 84:3220-9; PMID:20071582; http://dx.doi.org/10.1128/JVI.02241-09
- Nistal-Villan E, Gack MU, Martinez-Delgado G, Maharaj NP, Inn KS, Yang H, Wang R, Aggarwal AK, Jung JU, García-Sastre A. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem 2010; 285:20252-61; PMID:20406818; http://dx.doi.org/10.1074/jbc.M109.089912
- Maharaj NP, Wies E, Stoll A, Gack MU. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J Virol 2012; 86:1358-71; PMID:22114345; http://dx.doi.org/10.1128/JVI.06543-11
- Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 2013; 38:437-49; PMID:23499489; http://dx.doi.org/10.1016/j.immuni.2012.11.018
- Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A 2008; 105:16743-8; PMID:18948594; http://dx.doi.org/10.1073/pnas.0804947105
- Diao F, Li S, Tian Y, Zhang M, Xu LG, Zhang Y, Wang RP, Chen D, Zhai Z, Zhong B, et al. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci U S A 2007; 104:11706-11; PMID:17600090; http://dx.doi.org/10.1073/pnas.0700544104
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 2012; 36:959-73; PMID:22705106; http://dx.doi.org/10.1016/j.immuni.2012.03.022
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A 2007; 104:14050-5; PMID:17709747; http://dx.doi.org/10.1073/pnas.0704014104
- You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol 2009; 10:1300-8; PMID:19881509; http://dx.doi.org/10.1038/ni.1815
- Castanier C, Zemirli N, Portier A, Garcin D, Bidere N, Vazquez A, Arnoult D. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol 2012; 10:44; PMID:22626058; http://dx.doi.org/10.1186/1741-7007-10-44
- Pan Y, Li R, Meng JL, Mao HT, Zhang Y, Zhang J. Smurf2 Negatively Modulates RIG-I-Dependent Antiviral Response by Targeting VISA/MAVS for Ubiquitination and Degradation. J Immunol 2014; 192:4758-64; PMID:24729608; http://dx.doi.org/10.4049/jimmunol.1302632
- Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 2009; 30:397-407; PMID:19285439; http://dx.doi.org/10.1016/j.immuni.2009.01.008
- Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 2010; 33:765-76; PMID:21074459; http://dx.doi.org/10.1016/j.immuni.2010.10.013
- Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 2012; 287:28646-55; PMID:22745133; http://dx.doi.org/10.1074/jbc.M112.362608
- Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM, Zhang Z, Schneider M, Jiang Y, Fitzgerald KA, et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 2014; 40:329-41; PMID:24560620; http://dx.doi.org/10.1016/j.immuni.2014.01.010
- Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013; 155:688-98; PMID:24119841; http://dx.doi.org/10.1016/j.cell.2013.09.049
- Lin YC, Huang DY, Chu CL, Lin YL, Lin WW. The tyrosine kinase Syk differentially regulates Toll-like receptor signaling downstream of the adaptor molecules TRAF6 and TRAF3. Sci Signal 2013; 6:ra71; PMID:23962979; http://dx.doi.org/10.1126/scisignal.2003973
- Belgnaoui SM, Paz S, Samuel S, Goulet ML, Sun Q, Kikkert M, Iwai K, Dikic I, Hiscott J, Lin R. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe 2012; 12:211-22; PMID:22901541; http://dx.doi.org/10.1016/j.chom.2012.06.009
- Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O'Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, et al. DUBA: a deubiquitinase that regulates type I interferon production. Science 2007; 318:1628-32; PMID:17991829; http://dx.doi.org/10.1126/science.1145918
- Peng Y, Xu R, Zheng X. HSCARG Negatively Regulates the Cellular Antiviral RIG-I Like Receptor Signaling Pathway by Inhibiting TRAF3 Ubiquitination via Recruiting OTUB1. PLoS Pathog 2014; 10:e1004041; PMID:24763515; http://dx.doi.org/10.1371/journal.ppat.1004041
- Nakhaei P, Mesplede T, Solis M, Sun Q, Zhao T, Yang L, Chuang TH, Ware CF, Lin R, Hiscott J. The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog 2009; 5:e1000650; PMID:19893624; http://dx.doi.org/10.1371/journal.ppat.1000650
- Zhong B, Liu X, Wang X, Li H, Darnay BG, Lin X, Sun SC, Dong C. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci Signal 2013; 6:ra35; PMID:23674823; http://dx.doi.org/10.1126/scisignal.2003708
- Chau TL, Gioia R, Gatot JS, Patrascu F, Carpentier I, Chapelle JP, O'Neill L, Beyaert R, Piette J, Chariot A. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem Sci 2008; 33:171-80; PMID:18353649; http://dx.doi.org/10.1016/j.tibs.2008.01.002
- Lei CQ, Zhong B, Zhang Y, Zhang J, Wang S, Shu HB. Glycogen synthase kinase 3beta regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity 2010; 33:878-89; PMID:21145761; http://dx.doi.org/10.1016/j.immuni.2010.11.021
- Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 2011; 35:426-40; PMID:21903422; http://dx.doi.org/10.1016/j.immuni.2011.06.014
- Parvatiyar K, Barber GN, Harhaj EW. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. J Biol Chem 2010; 285:14999-5009; PMID:20304918; http://dx.doi.org/10.1074/jbc.M110.109819
- Gao L, Coope H, Grant S, Ma A, Ley SC, Harhaj EW. ABIN1 protein cooperates with TAX1BP1 and A20 proteins to inhibit antiviral signaling. J Biol Chem 2011; 286:36592-602; PMID:21885437; http://dx.doi.org/10.1074/jbc.M111.283762
- Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, Wang RF. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 2012; 13:387-95; PMID:22388039; http://dx.doi.org/10.1038/ni.2239
- Zhang M, Wang L, Zhao X, Zhao K, Meng H, Zhao W, Gao C. TRAF-interacting protein (TRIP) negatively regulates IFN-beta production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J Exp Med 2012; 209:1703-11; PMID:22945920; http://dx.doi.org/10.1084/jem.20120024
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 2006; 6:644-58; PMID:16932750; http://dx.doi.org/10.1038/nri1900
- Yu Y, Wang SE, Hayward GS. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 2005; 22:59-70; PMID:15664159; http://dx.doi.org/10.1016/j.immuni.2004.11.011
- Yu Y, Hayward GS. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 2010; 33:863-77; PMID:21167755; http://dx.doi.org/10.1016/j.immuni.2010.11.027
- Wang J, Yang B, Hu Y, Zheng Y, Zhou H, Wang Y, Ma Y, Mao K, Yang L, Lin G, et al. Negative regulation of Nmi on virus-triggered type I IFN production by targeting IRF7. J Immunol 2013; 191:3393-9; PMID:23956435; http://dx.doi.org/10.4049/jimmunol.1300740
- Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol 2008; 181:1780-6; PMID:18641315
- Long L, Deng Y, Yao F, Guan D, Feng Y, Jiang H, Li X, Hu P, Lu X, Wang H, et al. Recruitment of Phosphatase PP2A by RACK1 Adaptor Protein Deactivates Transcription Factor IRF3 and Limits Type I Interferon Signaling. Immunity 2014; 40:515-29; PMID:24726876; http://dx.doi.org/10.1016/j.immuni.2014.01.015
- Clevers H. At the crossroads of inflammation and cancer. Cell 2004; 118:671-4; PMID:15369667; http://dx.doi.org/10.1016/j.cell.2004.09.005
- Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 2004; 118:285-96; PMID:15294155; http://dx.doi.org/10.1016/j.cell.2004.07.013
- Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 2005; 5:749-59; PMID:16175180; http://dx.doi.org/10.1038/nri1703
- Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006; 124:263-6; PMID:16439202; http://dx.doi.org/10.1016/j.cell.2006.01.007
- De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat Rev Cancer 2007; 7:256-69; PMID:17384581; http://dx.doi.org/10.1038/nrc2090
- El-Omar EM, Ng MT, Hold GL. Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene 2008; 27:244-52; PMID:18176606; http://dx.doi.org/10.1038/sj.onc.1210912
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420:860-7; PMID:12490959; http://dx.doi.org/10.1038/nature01322
- Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest 2007; 117:1175-83; PMID:17476347; http://dx.doi.org/10.1172/JCI31537
- Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 2005; 121:977-90; PMID:15989949; http://dx.doi.org/10.1016/j.cell.2005.04.014
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004; 431:461-6; PMID:15329734; http://dx.doi.org/10.1038/nature02924
- Sun J, Wiklund F, Zheng SL, Chang B, Balter K, Li L, Johansson JE, Li G, Adami HO, Liu W, et al. Sequence variants in Toll-like receptor gene cluster (TLR6-TLR1-TLR10) and prostate cancer risk. J Natl Cancer Inst 2005; 97:525-32; PMID:15812078; http://dx.doi.org/10.1093/jnci/dji070
- Zheng SL, Augustsson-Balter K, Chang B, Hedelin M, Li L, Adami HO, Bensen J, Li G, Johnasson JE, Turner AR, et al. Sequence variants of toll-like receptor 4 are associated with prostate cancer risk: results from the CAncer Prostate in Sweden Study. Cancer Res 2004; 64:2918-22; PMID:15087412
- Hua D, Liu MY, Cheng ZD, Qin XJ, Zhang HM, Chen Y, Qin GJ, Liang G, Li JN, Han XF, et al. Small interfering RNA-directed targeting of Toll-like receptor 4 inhibits human prostate cancer cell invasion, survival, and tumorigenicity. Mol Immunol 2009; 46:2876-84; PMID:19643479; http://dx.doi.org/10.1016/j.molimm.2009.06.016
- Yang H, Zhou H, Feng P, Zhou X, Wen H, Xie X, Shen H, Zhu X. Reduced expression of Toll-like receptor 4 inhibits human breast cancer cells proliferation and inflammatory cytokines secretion. J Exp Clin Cancer Res 2010; 29:92; PMID:20618976; http://dx.doi.org/10.1186/1756-9966-29-92
- Lowe EL, Crother TR, Rabizadeh S, Hu B, Wang H, Chen S, Shimada K, Wong MH, Michelsen KS, Arditi M. Toll-like receptor 2 signaling protects mice from tumor development in a mouse model of colitis-induced cancer. PLoS One 2010; 5:e13027; PMID:20885960; http://dx.doi.org/10.1371/journal.pone.0013027
- Matijevic T, Marjanovic M, Pavelic J. Functionally active toll-like receptor 3 on human primary and metastatic cancer cells. Scand J Immunol 2009; 70:18-24; PMID:19522763; http://dx.doi.org/10.1111/j.1365-3083.2009.02262.x
- Nomi N, Kodama S, Suzuki M. Toll-like receptor 3 signaling induces apoptosis in human head and neck cancer via survivin associated pathway. Oncol Rep 2010; 24:225-31; PMID:20514466
- Wang H, Rayburn ER, Wang W, Kandimalla ER, Agrawal S, Zhang R. Chemotherapy and chemosensitization of non-small cell lung cancer with a novel immunomodulatory oligonucleotide targeting Toll-like receptor 9. Mol Cancer Ther 2006; 5:1585-92; PMID:16818518; http://dx.doi.org/10.1158/1535-7163.MCT-06-0094
- Goto Y, Arigami T, Kitago M, Nguyen SL, Narita N, Ferrone S, Morton DL, Irie RF, Hoon DS. Activation of Toll-like receptors 2, 3, and 4 on human melanoma cells induces inflammatory factors. Mol Cancer Ther 2008; 7:3642-53; PMID:19001446; http://dx.doi.org/10.1158/1535-7163.MCT-08-0582
- Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science 2007; 317:124-7; PMID:17615359; http://dx.doi.org/10.1126/science.1140488
- Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 2007; 317:121-4; PMID:17615358; http://dx.doi.org/10.1126/science.1140485
- Swann JB, Vesely MD, Silva A, Sharkey J, Akira S, Schreiber RD, Smyth MJ. Demonstration of inflammation-induced cancer and cancer immunoediting during primary tumorigenesis. Proc Natl Acad Sci U S A 2008; 105:652-6; PMID:18178624; http://dx.doi.org/10.1073/pnas.0708594105
- Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, Wang E, Ma W, Haines D, O'hUigin C, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med 2010; 207:1625-36; PMID:20624890; http://dx.doi.org/10.1084/jem.20100199
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 2003; 278:8869-72; PMID:12527755; http://dx.doi.org/10.1074/jbc.C200651200
- Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol 2008; 8:458-66; PMID:18500230; http://dx.doi.org/10.1038/nri2340
- Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut 2011; 60:1354-62; PMID:21421666; http://dx.doi.org/10.1136/gut.2010.216259
- Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest 2013; 123:700-11; PMID:23281400; http://dx.doi.org/10.1172/JCI62236
- Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med 2010; 207:1045-56; PMID:20385749; http://dx.doi.org/10.1084/jem.20100050
- Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 2010; 32:367-78; PMID:20226691; http://dx.doi.org/10.1016/j.immuni.2010.02.012
- Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010; 32:379-91; PMID:20303296; http://dx.doi.org/10.1016/j.immuni.2010.03.003
- Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol 2011; 186:7187-94; PMID:21543645; http://dx.doi.org/10.4049/jimmunol.1100412
- Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, Lemoine Y, Hot D, Chamaillard M. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A 2011; 108:9601-6; PMID:21593405; http://dx.doi.org/10.1073/pnas.1100981108
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 2014; 156:1045-59; PMID:24581500; http://dx.doi.org/10.1016/j.cell.2014.01.026
- Hu B, Elinav E, Huber S, Booth CJ, Strowig T, Jin C, Eisenbarth SC, Flavell RA. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci U S A 2010; 107:21635-40; PMID:21118981; http://dx.doi.org/10.1073/pnas.1016814108
- Hou J, Zhou Y, Zheng Y, Fan J, Zhou W, Ng IO, Sun H, Qin L, Qiu S, Lee JM, et al. Hepatic RIG-I predicts survival and interferon-alpha therapeutic response in hepatocellular carcinoma. Cancer Cell 2014; 25:49-63; PMID:24360797; http://dx.doi.org/10.1016/j.ccr.2013.11.011
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009; 462:108-12; PMID:19847166; http://dx.doi.org/10.1038/nature08460
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature 2009; 459:712-6; PMID:19412163; http://dx.doi.org/10.1038/nature07969
- Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, Klapper W, Vater I, Giefing M, Gesk S, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med 2009; 206:981-9; PMID:19380639; http://dx.doi.org/10.1084/jem.20090528
