Abstract
Active recombinant proteins are used for studying the biological functions of genes and for the development of therapeutic drugs. Overexpression of recombinant proteins in bacteria often results in the formation of inclusion bodies, which are protein aggregates with non-native conformations. Protein refolding is an important process for obtaining active recombinant proteins from inclusion bodies. However, the conventional refolding method of dialysis or dilution is time-consuming and recovered active protein yields are often low, and a cumbersome trial-and-error process is required to achieve success. To circumvent these difficulties, we used controllable diffusion through laminar flow in microchannels to regulate the denaturant concentration. This method largely aims at reducing protein aggregation during the refolding procedure. This Commentary introduces the principles of the protein refolding method using microfluidic chips and the advantage of our results as a tool for rapid and efficient recovery of active recombinant proteins from inclusion bodies.
Introduction
Biologically active proteins are used to study the biological functions of genes, for the development of therapeutic drugsCitation1,2 and represent bioelements in various industries.Citation3 The Escherichia coli overexpression system is the most convenient and frequently used approach to produce recombinant proteins.Citation1 In overexpression systems, the rate of target protein aggregation is often much greater than the rate of correct protein folding.Citation4 These inactive and insoluble protein aggregates, called inclusion bodies, are a drawback in the use of E. coli overexpression systems.Citation3,4 Because inclusion bodies contain relatively pure and intact recombinant proteins, several approaches have been reported to refold the aggregated protein into a biologically active form. In standard procedures, aggregated proteins are denatured and solubilized with high concentrations of denaturant such as urea or guanidinium chloride. The refolding procedure from denatured proteins (unfolded form) to active proteins (folded form) involves the gradual removal of denaturant.
The procedure for removing the denaturant from denatured proteins is a key step in the efficient recovery (or refold) of proteins. The dilution method is often selected for this purpose because the procedure is simple: the denatured protein solution is diluted directly with the refolding buffer that contains no denaturants. However, this method requires a large volume of buffer to dilute the denaturant to a concentration that does not disturb refolding, and there are difficulties encountered in uniform mixing of these large volumes, wherein aggregates reform.
The one-step dialysis (high denaturant concentration to the refolding buffer) is another simple method. Because the concentration of the denaturant decreases as the dialysis period increases, the rate of refolding of the protein into the native structure increases. However, the rate of misfolding and/or aggregation will also increase, possibly because of contact between exposed hydrophobic surfaces.Citation3,5 This suggests that a quick decrease in denaturant concentration initiates the reformation of aggregates or misfolded species, as observed in the dilution method. The step-wise dialysis approach has been used to overcome this problem. The denatured protein is first brought to equilibrium with a high denaturant concentration, then with a mid concentration, and finally a low concentration is used. This shows that gradual removal of denaturant from denatured proteins can achieve high refolding efficiency.Citation6 Although step-wise dialysis may provide refolded (active) proteins, it is a time-consuming procedure (i.e., multiple days) and the proteins often take the inactive form during the refolding process because of aggregates forming and the presence of other misfolded species.Citation3,6 Recent studies show that protein aggregation predominantly occurs during the mid concentration denaturant step (1 to 2 M),Citation7,8 suggesting that a refolding procedure that takes place over a short period of time may reduce the formation of protein aggregation. However, it is difficult to refold proteins in a short time using either dilution or dialysis methods.
In this commentary, we introduce our protein refolding method using laminar flow in microfluidic chips for the effective recovery of active proteins from inclusion bodies.Citation9,10 The strategy aims to inhibit the formation of protein aggregates during the refolding process, as observed in dilution or dialysis methods. With this technique, controllable diffusion by laminar flow in microchannels is used to control the denaturant concentration over a short time frame.
Microfluidic Chips for Initial Folding Events
Microfluidics systems are widely used in chemistry and biotechnology fields.Citation11-13 A laminar flow in microchannels can create a well-defined and predictable interfacial region among streams.Citation13 Hydrodynamic focusing is the steering of the central stream with a secondary boundary stream, where under the proper conditions these fluids do not mix (). Thus, hydrodynamic focusing generates a large surface-area-to-volume ratio, which creates an interface between fluids that can be controlled by changing flow rates. Additionally, a diffusional mass-transfer property is enhanced in the microchannel. These characteristics of microfluidics inspired us to control the gradual removal of denaturants from chemically denatured proteins.
Figure 1. Hydrodynamic focusing in a microchannel. Confocal fluorescence microscope image at the junction in MR1 showing the laminar flow of the urea stream by the diluting buffer streams.
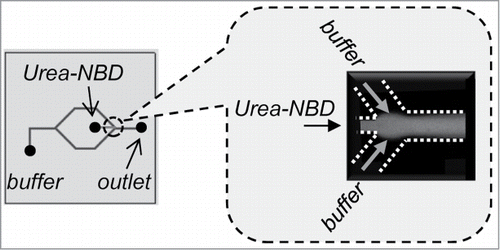
Previous microfluidic chips for protein refolding were designed to study initial folding events by rapid mixing of denatured protein with the dilution buffer in a microchannel. The widely used laminar flow mixers in protein refolding have employed hydrodynamic focusing to achieve a rapid mixing of denatured protein with dilution buffer.Citation14-16 In these studies, the microfluidics have been designed to study the folding kinetics of proteins that have measured folding rates that occur in <30 μs.Citation14-16 Because these microfluidic chips were designed for the analytical chemistry field, the throughput of denatured proteins is significantly smaller than those used for the recovery of bioactive proteins from inclusion bodies. In addition, these kinetic studies used easy-to-fold proteins such as cytochrome c, whereas target recombinant proteins in inclusion bodies are generally difficult-to-fold proteins. These points suggest that microfluidic chips with rapid mixing are not applicable for refolding difficult-to-fold proteins. Moreover, inclusion bodies lead to protein aggregation in a microchannel by rapid mixing because of the quick removal of the denaturant from denatured proteins.Citation17
Protein Refolding from Inclusion Bodies Using a Microfluidic Chip with Step-Wise Dilution
As described above, protein aggregation predominantly occurs during the mid concentration of denaturant. This suggests that reducing the refolding procedure to a short period may reduce the formation of protein aggregates and achieve efficient protein refolding. However, the rapid removal of denaturant from denatured proteins may also lead to protein re-aggregation and/or misfolding. As seen in step-wise dialysis, a gradual decrease of the denaturant concentration from the denatured protein over a short period may lead to efficient protein refolding. Therefore, we have designed microfluidic chips with step-wise dilution.
In a laminar flow, the fluid stream to be mixed flows along the central stream (denatured protein) meeting 2 buffer streams at a junction (). The mean square displacement (<x2>) of molecules in solution has been shown to be proportional to a mixing time t, <x2> = 2Dt. The diffusion coefficients (D) are of the order of 10–7 cm2 s–1 and 10–5 cm2 s–1 for proteins and small molecule denaturants, respectively. This indicates that proteins diffuse 2 orders of magnitude slower than denaturants. The width of the focused stream is also dependent on the flow rate of the dilution buffer.Citation18,19 The denaturant in the central stream of the denatured protein then enables mixing with the buffer by diffusion and the denaturant concentration decreases; thus, the ratio of the flow rates of the dilution buffer can control the denaturant concentration in the microchannel. Based on these concepts, microfluidic chips were designed ().Citation9 In MR1, the denaturant concentration around the protein rapidly decreases because of diffusion, which is expected to have a similar mechanism to the one-step dialysis and dilution approach. In MR2, the denaturant concentration shows a step-wise decrease, which is a similar mechanism to the step-wise dialysis method. To confirm whether the laminar flow in the designed chips can control the distribution of the denaturant concentration, a urea stream in the microchannel was studied by confocal fluorescence microscopy (). As expected, the results showed that the designed chips control the distribution of the denaturant concentration by the ratio of the flow rates.Citation9
Figure 2. Designed microfluidic chip used for protein refolding. (a) In MR1, the denaturant concentration around the protein rapidly decreases because of diffusion, which is expected to have a similar mechanism to one-step dialysis or dilution. (b) In MR2, the denaturant concentration shows a step-wise decrease, which is similar to step-wise dialysis. The denatured protein was injected into channel a. The dilution buffer was injected into channels b and c. The distributions of denaturant concentrations were measured by the relative fluorescence intensities of the fluorophore in the urea stream as a function of the distance from the inlet.
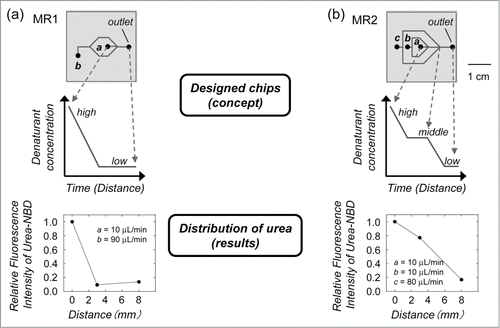
The refolding performance of the designed microfluidic chips was studied using urea-denatured citrate synthase (CS). CS has a low refolding yield when dialysis and dilution methods are used.Citation20 Therefore, CS has been used as a test case for refolding strategies.Citation21,22 The refolding efficiency of the protein was estimated by a CS enzyme activity assay. In our refolding procedure, the urea-denatured proteins were diluted 10-fold in microchannel by buffer within 10 min. Under this experimental condition, we did not observe any protein aggregation during refolding process.Citation9 The refolded CS by MR1 with rapid dilution showed a similar level of recovered enzyme activity (49%) when compared with the batch sample (47%) that was prepared by dilution, suggesting that rapid diffusion of urea from the denatured CS leads to misfolding and/or protein aggregation. In contrast, the recovered activity (76%) was enhanced by MR2 with step-wise dilution. These results indicate that the denatured CS was refolded efficiently by MR2 when compared with samples prepared by dilution and MR1. We also tested microfluidic chips with 3-junctions (MR3) or 4-junctions (MR4) for CS refolding.Citation9 The results showed that the gradual decrease in urea concentration is correlated with the recovery of enzyme activity. The recovered activities by MR3 (73%) and MR4 (74%) were similar to the result obtained by MR2 refolded CS (76%). Recently, the addition of small chemical molecules such as arginine hydrochloride (ArgHCl) has frequently been used to prevent protein aggregation.Citation10 The addition of ArgHCl to the dilution buffer is expected to suppress protein aggregation at the mixing point of the denatured protein and the diluting buffer in the microchannel, leading to improved refolding efficiency. This concept suggests that combining different techniques is a promising approach that may further improve protein refolding by microfluidic chips.
The recovered enzymatic activity of CS by our method is a similar value to those reported using an artificial chaperone-assisted system, which is a successful technique to recover active proteins from denatured forms.Citation22 The refolding by microfluidic chips was achieved within a short period (10 min) at room temperature.Citation9 The estimated throughput of our CS refolding by MR2 was 150 μg/h. This value is one-order higher than those of the artificial chaperone-assisted system (9–14 μg/h in ref. 22) and step-wise dialysis (10 μg/h).Citation9 Rapid and effective refolding is a superior advantage of our refolding procedure over other methods.
Refolding of Recombinant Proteins from Inclusion Bodies Using Microfluidic Chips with Step-Wise Dilution
The microfluidic chips have been evaluated for their efficiency in refolding proteins from inclusion bodies. ζ-Associated protein 70 kDa (ZAP-70) is a tyrosine kinase.Citation23 Because bacterially overexpressed ZAP-70 forms inclusion bodies, this protein is usually expressed in mammalian or insect expression systems.Citation23 However, these expression systems are expensive compared with the cost of expression using an E. coli system and the recovered protein yield is generally low.
In the refolding experiments, the urea-denatured ZAP-70 protein kinase domain, which was purified from E. coli inclusion bodies, was applied to the microfluidic chips to evaluate protein refolding. The circular dichroism (CD) spectrum of refolded ZAP-70 using MR1 showed a similar spectrum to the batch sample by dilution. In contrast, the CD spectrum of ZAP-70 prepared using MR2 was similar to that of folded ZAP-70 prepared by step-wise dialysis over 2 days. The estimated helical content of ZAP-70 using MR2 was higher than that of ZAP-70 prepared using MR1 and dilution.Citation9 Although we could not measure the kinase activity of ZAP-70 due to the fact that the kinase domain involved in this study does not have the enzymatic activity,Citation23 these CD results show that protein refolding by microfluidic chips can be applied to recover folded protein from inclusion bodies.
Conclusions
Recovering biologically active proteins at low cost is an important goal in protein refolding from bacterial inclusion bodies, not only for analysis of the protein structure and function but also for the development of therapeutic drugs and industrial processes. In addition to standard dilution and dialysis methods, several approaches have been reported to refold aggregated or misfolded proteins, such as size-exclusion chromatography, reversed micelle systems, zeolite absorbing systems and the natural chaperone protein system.Citation24-26 Although these methods work well for many proteins located in inclusion bodies and denatured model proteins, in most cases there is a significant amount of protein precipitation during the refold step, leading to low recovery yields. Therefore, these procedures are still performed with a series of cumbersome trial-and-error refolding experiments to optimize recovery yields. In our refolding method, the denaturant concentration was controlled through laminar flow in microchannels. The protein folding efficiency was greater than observed with either the dialysis or dilution methods. Currently, there is no study describing the effective recovery of bioactive proteins from inclusion bodies using a microfluidic chip. Microfluidic devices apparently may allow facile refolding of proteins from inclusion bodies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported in part by the Research and Study Program of the Tokai University Education System General Research Organization.
References
- Swartz JR. Advances in Escherichia coli production of therapeutic proteins. Curr Opin Biotechnol 2001; 12:195-201; PMID:11287237; http://dx.doi.org/10.1016/S0958-1669(00)00199-3
- Chakrabarty AM, Bernardes N, Fialho AM. Bacterial proteins and peptides in cancer therapy. Bioengineered 2014; 5:234-42; PMID:24875003; http://dx.doi.org/10.4161/bioe.29266
- Clark EDB. Protein refolding for industrial processes. Curr Opin Biotechnol 2001; 12:202-7; PMID:11287238; http://dx.doi.org/10.1016/S0958-1669(00)00200-7
- Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 2004; 22:1399-408; PMID:15529165; http://dx.doi.org/10.1038/nbt1029
- Marston FAO. The purification of eukaryotic polypeptides synthesized in Escherichia coli. Biochem J 1986; 240:1-12; PMID:3548705
- Umetsu M, Tsumoto K, Hara M, Ashish K, Goda S, Adschiri T, Kumagai I. How additives influence the refolding of immunoglobulin-folded proteins in a stepwise dialysis system. Spectroscopic evidence for highly efficient refolding of a single-chain Fv fragment. J Biol Chem 2003; 278:8979-87; PMID:12519771; http://dx.doi.org/10.1074/jbc.M212247200
- Ho JGS, Middelberg APJ, Ramage P, Kocher HP. The likelihood of aggregation during protein renaturation can be assessed using the second virial coefficient. Protein Sci 2003; 12:708-16; PMID:12649429; http://dx.doi.org/10.1110/ps.0233703
- Liu W, Cellmer T, Keerl D, Prausnitz JM, Blanch HW. Interactions of lysozyme in guanidinium chloride solutions from static and dynamic light-scattering measurements. Biotchnol Bioeng 2005; 90:482-90; PMID:15778988; http://dx.doi.org/10.1002/bit.20442
- Yamaguchi H, Miyazaki M, Briones-Nagata MP, Maeda H. Refolding of difficult-to-fold proteins by a gradual decrease of denaturant using microfluidic chips. J Biochem 2010; 147:895-903; PMID:20207823; http://dx.doi.org/10.1093/jb/mvq024
- Yamaguchi H, Miyazaki M. Refolding techniques for recovering biologically active recombinant proteins from inclusion bodies. Biomolecules 2014; 4:235-51; PMID:24970214; http://dx.doi.org/10.3390/biom4010235
- Ohno K, Tachikawa K, Manz, A. Microfluidics:applications for analytical purposes in chemistry and biochemistry. Electrophoresis 2008; 29:4443-53; PMID:19035399; http://dx.doi.org/10.1002/elps.200800121
- Song H, Chen DL, Ismagilov RF. Reactions in droplets in microfluidic channels. Angew Chem Int Ed 2006; 45:7336-56; PMID:17086584; http://dx.doi.org/10.1002/anie.200601554
- Golden JP, Justin GA, Nasir M, Ligler FS. Hydrodynamic focusing–a versatile tool. Anal Bioanal Chem 2012; 402:325-35; PMID:21952728; http://dx.doi.org/10.1007/s00216-011-5415-3
- Lapidus LJ, Yao S, McGarrity KS, Hertzog DE, Tubman E, Bakajin O. Protein hydrophobic collapse and early folding steps observed in a microfluidic mixer. Biophys J 2007; 93:218-24; PMID:17416618; http://dx.doi.org/10.1529/biophysj.106.103077
- Gambin Y, Simonnet C, VanDelinder V, Deniz A, Groisman A. Ultrafast microfluidic mixer with three-dimensional flow focusing for studies of biochemical kinetics. Lab Chip 2010; 10:598-609; PMID:20162235; http://dx.doi.org/10.1039/b914174j
- Park HY, Qiu X, Rhoades E, Korlach J, Kwok LW, Zipfel WR, Webb WW, Pollack L. Achieving uniform mixing in a microfluidic device: hydrodynamic focusing prior to mixing. Anal Chem 2006; 78: 4465-73; PMID:16808455; http://dx.doi.org/10.1021/ac060572n
- Yamamoto E, Yamaguchi S, Sasaki N, Kim HB, Kitamori T, Nagamune T. Artificial chaperone-assisted refolding in a microchannel. Bioprocess Biosyst Eng 2010; 33: 171-7; PMID:19727834; http://dx.doi.org/10.1007/s00449-009-0374-1
- Karnik R, Gu F, Basto P, Cannizzaro C, Dean L, Kyei-Manu W, Langer R, Farokhzad OC. Microfluidic platform for controlled synthesis of polymeric nanoparticles. Nano Lett 2008; 8:2906-12; PMID:18656990; http://dx.doi.org/10.1021/nl801736q
- Jahn A, Vreeland WN, deVoe DL, Locascio LE, Gaitan M. Microfluidic directed formation of liposomes of controlled size. Langmuir 2007; 23:6289-93; PMID:17451256; http://dx.doi.org/10.1021/la070051a
- Zhi W, Landry SJ, Gierasch LM, Srere PA. Renaturation of citrate synthase: influence of denaturant and folding assistants. Protein Sci 1992; 1:522-9; PMID:1363914; http://dx.doi.org/10.1002/pro.5560010407
- Daugherty DL, Rozema D, Hanson PE, Gellman SH. Artificial chaperone-assisted refolding of citrate synthase. J Biol Chem 1998; 273:33961-71; PMID:9852049; http://dx.doi.org/10.1074/jbc.273.51.33961
- Machida S, Ogawa S, Xiaohua S, Takaha T, Fujii K, Hayashi K. Cycloamylose as an efficient artificial chaperone for protein refolding. FEBS Lett 2000; 486:131-5; PMID:11113453; http://dx.doi.org/10.1016/S0014-5793(00)02258-4
- Jin L, Pluskey S, Petrella EC, Cantin SM, Gorga JC, Rynkiewicz MJ, Pandey P, Strickler JE, Babine RE, Weaver DT. et al. The three-dimensional structure of the ZAP-70 kinase domain in complex with staurosporine. J Biol Chem 2004; 279:42818-25; PMID:15292186; http://dx.doi.org/10.1074/jbc.M407096200
- Eiberle MK, Jungbauer A. Technical refolding of proteins: do we have freedom to operate? Biotechnol J 2010; 5:547-59; PMID:20518058; http://dx.doi.org/10.1002/biot.201000001
- Gautam S, Dubey P, Rather GM, Gupta MN. Non-chromatographic strategies for protein refolding. Recent Pat Biotechnol 2012; 6:57-68; PMID:22420882; http://dx.doi.org/10.2174/187220812799789172
- Machold C, Schlegl R, Buchinger W, Jungbauer A. Matrix assisted refolding of proteins by ion exchange chromatography. J Biotechnol 2005; 117:83-97; PMID:15831250; http://dx.doi.org/10.1016/j.jbiotec.2005.01.004
