Abstract
Nuclear protein 95 KDa (Np95, also known as UHRF1 or ICBP90) plays an important role in maintaining DNA methylation of newly synthesized DNA strands by recruiting DNA methyltransferase 1 (DNMT1) during cell division. In addition, Np95 participates in chromatin remodeling by interacting with histone modification enzymes such as histone deacetylases. However, its expression pattern and function in the brain have not been analyzed extensively. We here investigated the expression pattern of Np95 in the mouse brain, from developmental to adult stages. In the fetal brain, Np95 is abundantly expressed at the midgestational stage, when a large number of neural stem/precursor cells (NS/PCs) exist. Interestingly, Np95 is expressed specifically in NS/PCs but not in differentiated cells such as neurons or glial cells. Furthermore, we demonstrate that Np95 is preferentially expressed in type 2a cells, which are highly proliferative NS/PCs in the dentate gyrus of the adult hippocampus. Moreover, the number of Np95-expressing cells increases in response to kainic acid administration or to voluntary running, which are known to enhance the proliferation of adult NS/PCs. These results suggest that Np95 participates in the process of proliferation and differentiation of NS/PCs, and that it should be a useful novel marker for proliferating NS/PCs, facilitating the analysis of the complex behavior of NS/PCs in the brain.
Abbreviations
| Np95 | = | nuclear protein 95 KDa |
| UHRF1, ubiquitin-like | = | containing PHD and RING finger domains 1 |
| ICBP90 | = | inverted CCAAT box-binding protein of 90 KDa |
| NS/PCs | = | neural stem/precursor cells |
| CNS | = | central nervous system |
| SVZ | = | subventricular zone |
| SGZ | = | subgranular zone |
| DG | = | dentate gyrus |
| DNMT1 | = | DNA methyltransferase 1 |
| HDAC | = | histone deacetylase |
| KA | = | kainic acid |
| Sox2 | = | SRY (sex determining region Y)-box 2 |
| VZ/SVZ | = | ventricular and subventricular zone |
| Tbr2 | = | T-box brain 2 |
| IPCs | = | intermediate progenitor cells |
| IZ | = | intermediate zone |
| CP | = | cortical plate |
| OPCs | = | oligodendrocyte progenitor cells |
| GFAP | = | glial fibrillary acidic protein |
| APC | = | adenomatous polyposis coli CC-1 |
| PCNA | = | proliferating cell nuclear antigen |
Introduction
Accumulating studies have indicated that epigenetic regulation plays crucial roles in various biological events such as the development and function of the central nervous system (CNS), including adult neurogenesis.Citation1-5 Mammalian embryonic brain development occurs through the proliferation of multipotent neural stem/precursor cells (NS/PCs) and their differentiation into the 3 major cell types, neurons, astrocytes and oligodendrocytes.Citation6,7 The fate specification of NS/PCs as development progresses is tightly regulated by epigenetic modifications such as DNA methylation and histone modification.Citation8,9 Neurogenesis persists throughout life in 2 restricted adult brain regions: the subventricular zone (SVZ) of the lateral ventricle and the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampus. Adult neurogenesis in the hippocampal DG plays an important role in learning and memory formation, and is also associated with several neurological disorders (e.g., depression, seizure, schizophrenia, and Alzheimer's disease).Citation10-13
DNA methylation is an important epigenetic mark which makes essential contributions to many biological processes, including genomic imprinting, X-inactivation, embryonic development, differentiation,Citation14-16 maintenance of chromosomal stabilityCitation17 and gene transcription control in most eukaryotes.Citation18 Consequently, abnormal patterns of DNA methylation are implicated in various pathologies, from neurodegenerative diseasesCitation19 to cancers.Citation20 During cell division and DNA replication, transferring the DNA methylation pattern of the parent strand onto the daughter strand is essential for the precise regulation of gene expression. Nuclear protein 95 KDa (Np95, also known as Ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1) or inverted CCAAT box-binding protein of 90 KDa (ICBP90)) is a pleiotropic protein with various functional domains (Ubiquitin-like, Tudor, PHD, SRA and RING finger domains), and plays a central role in this methylation pattern transferral. Np95 binds to hemimethylated DNA via its SRA domain and recruits DNA methyltransferase 1 (DNMT1), which methylates the unmethylated strand of hemimethylated DNA.Citation21-25 Mouse embryonic stem cells deficient in Np95 fail to recruit DNMT1 to replicating pericentromeric heterochromatin regions in mid-S-phase, leading to a dramatic reduction of DNA methylation in the genome.Citation22 Furthermore, recent reports have indicated that Np95 also participates in histone modification and chromatin structure by associating with histone deacetylase1 (HDAC1), histone H3K9 methyltransferase G9a and histone H3 di- and tri-methylated at lysine 9 (H3K9me2 and H3K9me3).Citation26-29 In particular, it was reported that both SRA domain-mediated binding of Np95 to hemimethylated DNA strands and Tudor domain-mediated association of Np95 with H3K9me2/3 are necessary for the inheritance of DNA methylation.Citation30,31 Thus, Np95 plays important roles in the epigenetic regulation of the mammalian genome, implying that Np95 also has roles in the CNS. However, the function and even the expression pattern of Np95 in the brain remain elusive.
In this study, as a first step toward understanding the role of Np95 in the nervous system, we investigated the spatiotemporal expression pattern of Np95 and identified Np95-expressing cells in the embryonic and adult mouse brain. We found that expression of Np95 was restricted to NS/PCs in the embryonic brain and to rapidly dividing neural progenitor cells in both the SGZ and the SVZ in the adult brain. Furthermore, the numbers of Np95-expressing NS/PCs in the DG increased in response to kainic acid (KA) administration and voluntary running, both of which are known to stimulate NS/PC proliferation. These findings imply that Np95 plays a role in the epigenetic regulation of NS/PCs under both physiological and pathological conditions in the brain throughout life.
Results
Np95 is expressed abundantly in NS/PCs in the embryonic brain
First, to verify the specificity of antibodies against mouse Np95, HEK293T human embryonic kidney-derived cells were transfected with an Np95-expressing plasmid (BOSE-Np95-Flag), and western blotting was performed using anti-FLAG (Sigma, mouse), -Np95 (ref. 32, rat) and -UHRF1 (Santa Cruz, rabbit) antibodies. A specific band at approximately 95 kDa (the deduced molecular mass of Np95) was detected with all antibodies (). In addition, proteins immunoprecipitated with anti-FLAG and rat anti-Np95 antibodies were also detected by the 3 antibodies (Fig. S1). Having thus validated its specificity, we decided to use the rabbit anti-UHRF1 antibody, which is commercially available, to detect Np95 in the following experiments.
Figure 1. Np95 is expressed abundantly in the brain at midgestation, when a large number of NS/PCs exist. (A) Confirmation of antibody reactivity to mouse Np95. HEK293T cells were transfected with FLAG-tagged Np95 expression plasmid and immunoblotted with anti-FLAG, -Np95 and -UHRF1 antibodies. (B) Western blot analysis of mouse whole brain at various stages of development, from E11 to postnatal day 7 (P7) and in 8-week-old adults, using anti-DNMT1, -Np95 and -actin antibodies. Np95 and DNMT1 bands were strongest in E11 brains. (C) Representative immunofluorescence images for Np95, Ki67 and Sox2 in E14 mouse forebrain sections. Scale bar: 250 μm. (D) Confocal immunofluorescence images for Np95, Ki67 and Sox2 in E14 mouse forebrain sections. Scale bars: 10 μm. (E–G) Representative immunofluorescence images of E14 mouse forebrain sections. Immunostaining of Np95 (red), Nestin (cyan) and Ki67 (E), DCX (F) or MAP2ab (G) (green). The insets in Merge images show Hoechst staining. Np95 expression was observed only in Ki67+ or Nestin+ proliferating NS/PCs. Scale bars: 100 μm. VZ: ventricular zone; SVZ: subventricular zone; IZ: intermediate zone; CP: cortical plate.
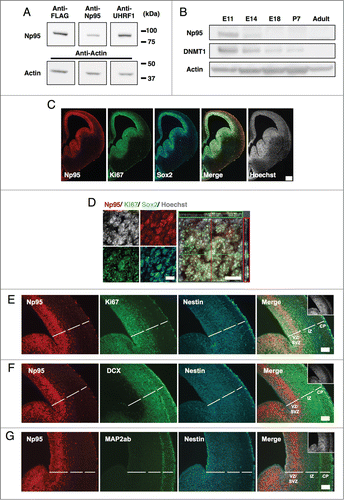
Next, to examine the expression level of Np95 protein at each developmental stage, we subjected total proteins from embryonic, post-natal and adult mouse brains to western blot analysis. In the fetal brain, we found that Np95 was abundantly expressed at the midgestational stage, when a large number of NS/PCs exist (), and decreased as embryonic development proceeds. DNMT1 expression was similar to that of Np95 (), probably because Np95 functions in concert with DNMT1 to maintain DNA methylation patterns in the developing brain.
To determine precisely where Np95 is expressed in the embryonic brain, immunohistochemistry was performed on embryonic day (E) 14 brain sections using antibodies for the proliferating cell marker Ki67 and the NS/PC marker SRY (sex determining region Y)-box 2 (Sox2) (). We observed that Np95 was restricted to the ventricular and subventricular zone (VZ/SVZ) where Sox2+ NS/PCs reside, and co-localized almost exclusively with Ki67. Furthermore, when we stained the brain sections with anti-Np95 antibody together with antibodies for Ki67, for another NS/PC marker (Nestin), and for markers of immature neurons (doublecortin (DCX)) and mature neurons (Map2ab) (), Np95 was detected only in Ki67+ or Nestin+ NS/PCs. In addition, Np95 staining was observed in a region very similar to that displaying T-box brain 2 (Tbr2)+ neurogenic basal progenitors (intermediate progenitor cells (IPCs)), which have proliferation potential and exist in the upper VZ and SVZ (Fig. S2A). In contrast, no Np95 was observed in the intermediate zone (IZ) and or the cortical plate (CP), where DCX+ and MAP2ab+ neurons were distributed (). This Np95 expression pattern also holds true for E11 and E18 brains: Np95 is specifically expressed in Sox2+ NS/PCs but not in Map2ab+ mature neurons (Fig. S2). In , the western blot data appeared to indicate that Np95 is not expressed in the brain at E18 or thereafter, but this is not the case. As is evident in and S2, Np95 expression is sustained in Sox2+ NS/PCs, while the ratio of NS/PCs to whole brain cells gradually decreases. This explains the apparent but deceptive reduction of Np95 expression in the western blot. In addition, although the ratio of proliferative oligodendrocyte progenitor cells (OPCs) (Olig2 and Ki67 double-positive) to whole brain cells was low, Np95 signal was detected in the proliferative OPCs in postnatal day (P) 10 mouse brain (Fig. S5).
Similar to Np95, DNMT1 expression was observed in Sox2+ NS/PCs at each developmental stage in the embryonic cortex (Fig. S2). However, in contrast to Np95, weak DNMT1 signals were also seen in Map2ab+ mature neurons at E18 (Fig. S2C). Therefore, it is conceivable that Np95 collaborates with DNMT1 for the maintenance of DNA methylation, whereas DNMT1 functions independently of Np95 in differentiated neurons.
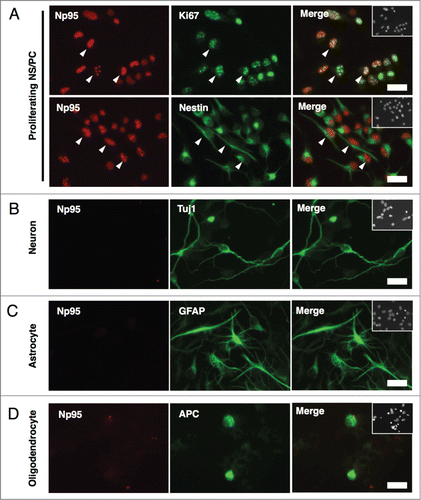
To further characterize Np95 expression in neural cell types, NS/PCs derived from E14 telencephalon were cultured under conditions that induced their differentiation into neurons or glial cells, and subjected to immunocytochemistry (). Np95 signal was detected predominantly in Ki67+ and Nestin+ proliferating NS/PCs (), as it was in the brain sections, and we did not observe any positive signals for Np95 in βIII-tubulin (Tuj1)+ neurons, glial fibrillary acidic protein (GFAP)+ astrocytes or adenomatous polyposis coli CC-1 (APC)+ mature oligodendrocytes (). Taken together, these results demonstrate that Np95 expression in embryonic stages is restricted to NS/PCs.
Figure 2. Np95 is not expressed in differentiated neural cells. (A–D) Representative immunofluorescence images of cells cultured under various conditions. Neuroepithelial cells from E14 mice were cultured with bFGF (10 ng/ml) for 4 days, and then cultured as described below, for the indicated number of days, either to maintain the undifferentiated condition or to induce specific differentiation. (A) Undifferentiated cells: bFGF (10 ng/ml) for 2 d. (B) Neurons: 0.5% FBS for 4 d. (C) Astrocytes: bFGF (10 ng/ml), leukemia inhibitory factor (LIF) (40 ng/ml) and bone morphogenetic protein 2 (BMP2) (40 ng/ml) for 4 d. (D) Oligodendrocytes: triiodothyronine (T3) (30 ng/ml) and thyroxin (T4) (40 ng/ml) for 7 d. After each differentiation induction period, the cells in (A-D) were stained with antibodies against Np95 (A–D) (red) and either Ki67, Nestin (A), Tuj1 (B), GFAP (C) or APC (D) (green). Np95 expression was not observed in differentiated neural cells. The insets in Merge images show Hoechst staining. White arrowheads indicate representative Np95-expressing cells. Scale bars: 20 μm.
Np95 is expressed in proliferative NS/PCs in the adult hippocampus
In the adult mouse brain, NS/PCs are retained in the SGZ of the DG in the hippocampus and in the SVZ of the lateral ventricle. Our observation that Np95 signal was detected only in NS/PCs and co-localized with Ki67 signal in embryonic stages prompted us to examine whether Np95 is also expressed in NS/PCs in these 2 regions. We first investigated Np95 expression in the SGZ by double-labeling immunohistochemistry using anti-Np95 and -Ki67 antibodies (). We found that although almost all (97%) Np95-expressing cells were positive for Ki67, 30% of Ki67-expressing cells did not exhibit an Np95 signal (), indicating that not all the proliferating cells in the SGZ are expressing Np95 ().
Figure 3. Np95 is preferentially expressed in actively dividing type 2a cells in the adult hippocampus. (A) Schematic diagram of marker proteins that are expressed in the hippocampal granule cell layer during the 5 discernible stages of adult hippocampal neurogenesis. (B) Representative confocal immunofluorescence images for Np95 and Ki67 in the adult mouse hippocampal DG. Scale bars: 20 μm. (C) Ratio of Np95+/Ki67+ cells to total Np95+ cells or to total Ki67+ cells in hippocampal DG. While 97% of Np95-expressing cells exhibited a Ki67 signal, 30% of Ki67-expressing cells did not exhibit an Np95 signal. Error bars represent the mean ± SEM (n = 3). (D) Representative immunofluorescence images of the adult hippocampal DG. Immunostaining for Np95 (red, left panel), Ki67 (green, left middle panel) and Nestin, GFAP or DCX (cyan, right middle panel). Scale bars: 50 μm. The insets in Merge images show Hoechst staining. White arrowheads indicate representative Np95-, Ki67- and specific cell marker (Nestin-, GFAP-, DCX-) expressing cells. (E) Identification of Np95-expressing cell types in the DG of the adult hippocampus. Ki67+ cells, Ki67+/Np95+ cells and Ki67+/Np95- cells were counted using the merged images of (D). The bar graphs indicate the percentages of Nestin+/− (left), GFAP+/− (middle) or DCX+/− (right) cells among Ki67+ cells, Ki67+/Np95+ cells and Ki67+/Np95- cells in the hippocampal DG. The bar colors correspond to the stage colors used in the illustration in (A). Values are given as mean ± SEM. One-way ANOVA (Prism, GraphPad) followed by Tukey test: n = 3; *p < 0.05, **p < 0.01, ***p < 0.001.
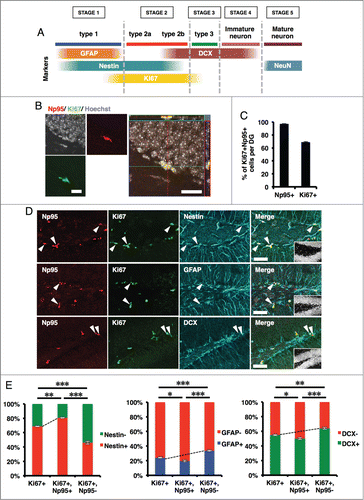
As illustrated in , adult hippocampal neurogenesis has 5 developmental stages characterized by the expression of specific proteins.Citation11,33,34 GFAP is a type 1 NS/PC marker which can be used to distinguish type 1 and type 2 cells. Cells double-positive for GFAP and Ki67 are categorized as actively dividing type 1 cells. In the adult hippocampus, DCX is detected in type 2b and 3 cells and in immature neurons. Furthermore, type 1 and 2 cells are positive for Nestin, and actively dividing type 1 and 2 cells are characterized by the expression of both Nestin and Ki67 (). To identify the type of Np95-expressing cells, we carried out immunohistochemical analyses of the SGZ in the adult mouse hippocampus using antibodies against Ki67 and against marker proteins specific for each cell type (Nestin, GFAP and DCX) (). We found that Nestin+ cells were enriched more in Np95+/Ki67+ cells (80%) than in all Ki67+ cells (69%) (, left panel), suggesting that Np95 is expressed more highly in the type 1 plus 2a population than in type 2b plus 3 cells. In addition, more GFAP+ cells were detected in Np95-/Ki67+ cells (34%) than in all Ki67+ cells (24%) (, middle panel), indicating that Np95 expression is lower in type 1 cells than in the type 2a plus 2b cell population. Furthermore, DCX+ cells were more abundant in Np95-/Ki67+ cells (64%) than in all Ki67+ cells (55%), meaning that Np95 is expressed more weakly in type 3 cells than in the type 2a plus 2b cell population. Considering these results and the fact that DCX is expressed in type 2b but not in type 2a cells, we conclude that Np95 is preferentially expressed in actively dividing type 2a cells in the DG of the adult hippocampus.
We also found that Np95 is expressed in the SVZ, and that a positive signal for Ki67 was detected in almost all Np95-expressing cells (Fig. S3A). Furthermore, an Np95 signal was observed in Mash1+ transit-amplifying cells in the SVZ (Fig. S3B), indicating that Np95 is expressed in proliferating NS/PCs not only in the SGZ but also in the SVZ. However, as with NS/PCs in the SGZ of the hippocampus, not all Ki67+ cells expressed Np95, suggesting that Np95 is expressed in a subpopulation of Ki67+ cells, most likely actively proliferating NS/PCs, in these 2 neurogenic regions in the adult brain.
Physiological and pathological stimuli increase the number of Np95-expressing cells in the SGZ
Since physiological neurogenic stimuli such as voluntary running enhance the proliferation of NS/PCs and facilitate neurogenesis in the DG of the adult mouse hippocampus,Citation35-38 the number of Np95+ cells in this region was expected to increase in response to such stimuli. To confirm this, 8-week adult mice were allowed 14 d of voluntary access to a running disc. As shown in Fig. S4, these running mice displayed an increase of proliferating cell nuclear antigen (PCNA)+ proliferating cells, indicating that voluntary running indeed enhanced NS/PC proliferation in the SGZ of the DG. Likewise, the number of Np95+ cells increased 1.5-fold in running mice compared with sedentary mice (Fig. S4).
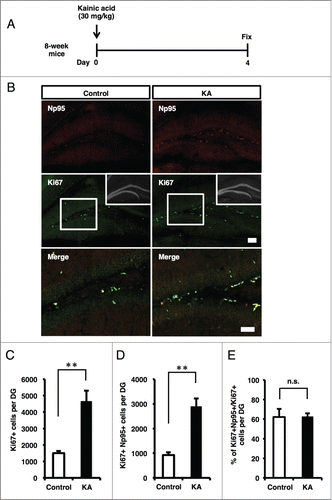
In addition to physiological stimuli, adult neurogenesis increases in response to pathological stimuli. For example, epileptic seizure induced by KA administration increases NS/PC proliferation in the DG of adult mice.Citation39 A marked increase of Ki67+ or PCNA+ proliferating cells at 3 to 4 d after KA treatment has also been reported.Citation40,41 To determine whether the pathological condition induced by KA administration also affects the number of Np95+ cells in the DG, KA was injected into 8-week adult mice, which were sacrificed 4 d later (). Immunohistochemical analysis revealed a significant increase of Ki67+ cells in the KA-injected mice compared with control mice (). Similarly, a marked increase in the number of Np95+/Ki67+ cells was observed in the KA-injected mice (). On the other hand, the proportion of Np95+ cells in Ki67+ cells was unchanged under the epileptic condition (), suggesting that the total number of Ki67+ proliferating cells, including Np95-expressing type 2a cells, expanded in response to KA-induced seizure. Taken together with the results from the running mice, our findings suggest that Np95 participates in the process of NS/PC proliferation and neurogenesis in response to both physiological and pathological stimuli.
Figure 4. KA injection increases the number of Np95-expressing cells. (A) Experimental scheme for inducing seizure. 8-week-old mice were injected intraperitoneally with KA and sacrificed 4 d later. (B) Representative immunofluorescence images of Np95 and Ki67 staining in control and KA-injected mice. Merge images show higher-magnification views of the white boxes in the middle images. Scale bars: 100 μm in Np95 and Ki67 images, 50 μm in Merge images. (C) Quantification of the number of Ki67+ cells in the hippocampal DG of control and KA-injected mice. KA injection increased the number of Ki67+ cells in the DG. Values are given as mean ± SEM. Student's t-test: n = 3; **p < 0.01. (D) Quantification of the number of Ki67+/Np95+ cells in the DG of control and KA-injected mice. KA injection increased the number of Ki67+/Np95+ cells in the DG. Values are given as mean ± SEM. Student's t-test: n = 3; **p < 0.01. (E) Ratio of Np95+/Ki67+ cells to total Ki67+ cells in the DG. Values are given as mean ± SEM. Student's t-test: n = 3; n.s., not significant.
Discussion
Since it has been reported that Np95/UHRF1/ICBP90 is expressed in many tissues (e.g., spleen, lung, testis, thymus) but not in the adult brain,Citation42,43 no extensive study has been undertaken of the expression pattern or the function of Np95 in the CNS. We too were unable to detect Np95 expression in whole brain lysates of adult mice (). However, we report for the first time, to the best of our knowledge, that Np95 is in fact expressed in proliferating NS/PCs, which comprise only a small proportion of the cells in the adult whole brain. This highly restricted expression pattern is probably the reason that Np95 expression in the adult brain has not been detected. On the other hand, several studies have reported that Np95 has an important role in G1 to S phase cell cycle progression, and that it is overexpressed in various cancer cells, which have high proliferation potency.Citation32,44,45 From these observations we speculated that Np95 is expressed in proliferating NS/PCs, and we have shown in this study that Np95 is indeed expressed in cells of both embryonic and adult mouse brains ().
Figure 5. Schematic representation of the expression pattern of Np95 in embryonic brain and adult hippocampal DG. (A) NS/PCs acquire the pluripotency to differentiate into neurons, astrocytes and oligodendrocytes during embryonic development. Np95 is specifically expressed in NS/PCs and not expressed in their completely differentiated neural cell progeny. (B) Schematic diagram illustrating the current view of lineage relationships and marker expression during adult hippocampal neurogenesis. Adult hippocampal neurogenesis occurs in 5 stages: [1] activation of quiescent radial glia-like cells (type 1 (change of morphology from radial to horizontal)) in the SGZ; [2] proliferation of precursor and intermediate progenitors (type 2a, 2b, transit-amplifying cells); [3] generation of neuroblasts (type 3); [4] differentiation into immature neurons; [5] maturation of immature neurons. Np95 is preferentially expressed in type 2a transit-amplifying NS/PCs. Circular arrows indicate the frequency of cell division. Solid arrows show frequently dividing cells compared with dashed arrows. ML: molecular layer; GCL: granule cell layer; SGZ: subgranular zone.
![Figure 5. Schematic representation of the expression pattern of Np95 in embryonic brain and adult hippocampal DG. (A) NS/PCs acquire the pluripotency to differentiate into neurons, astrocytes and oligodendrocytes during embryonic development. Np95 is specifically expressed in NS/PCs and not expressed in their completely differentiated neural cell progeny. (B) Schematic diagram illustrating the current view of lineage relationships and marker expression during adult hippocampal neurogenesis. Adult hippocampal neurogenesis occurs in 5 stages: [1] activation of quiescent radial glia-like cells (type 1 (change of morphology from radial to horizontal)) in the SGZ; [2] proliferation of precursor and intermediate progenitors (type 2a, 2b, transit-amplifying cells); [3] generation of neuroblasts (type 3); [4] differentiation into immature neurons; [5] maturation of immature neurons. Np95 is preferentially expressed in type 2a transit-amplifying NS/PCs. Circular arrows indicate the frequency of cell division. Solid arrows show frequently dividing cells compared with dashed arrows. ML: molecular layer; GCL: granule cell layer; SGZ: subgranular zone.](/cms/asset/9b9470db-b0f3-4efd-9edc-453c169b150d/kngs_a_976026_f0005_c.gif)
Np95 is an important factor to maintain DNA methylation of newly synthesized DNA strands by recruiting DNMT1 during cell division.Citation22 Previous studies have ascertained that progressive demethylation of astrocyte-specific gene promoters as gestation proceeds plays a key role in the differentiation of NS/PCs to astrocytes, and that DNMT1 activity in NS/PCs is essential for the maintenance of methylation of these gene promoters.Citation8,Citation46-48
Furthermore, we have previously shown that the demethylation process is triggered by the dissociation of DNMT1 from the promoter regions in response to Notch signal activation.Citation47 In the present study, we revealed that expression of Np95 is restricted to NS/PCs and does not occur in differentiated neural cells in the embryonic brain. This result raises the possibility that Np95 acts together with DNMT1 to maintain the DNA methylation status of astrocyte-specific gene promoters, and may thus take part in the switch from neurogenic to astrocytogenic properties of NS/PCs during the stepwise development of the embryonic brain. In contrast to Np95, DNMT1 was also expressed in differentiated neurons, suggesting that DNMT1 has distinct roles in undifferentiated and differentiated cells. Moreover, an in vitro study has reported very recently that Np95 is a direct target of the transcription factor E2F1 in OPCs and is likely to contribute to the transition from the proliferating to the differentiating state of OPCs.Citation49 Consistent with that study, our in vivo immunohistochemical analysis shows that proliferative OPCs in P10 mouse express Np95, unlike APC+ mature oligodendrocytes (Fig. S5 and ). Further analysis of the expression of Np95 in other proliferating progenitors as well as NS/PCs should provide additional insights into the function of Np95 in the CNS.
The SGZ of the DG in the adult hippocampus contains slowly dividing or relatively quiescent type 1 NSCs, transiently amplifying type 2a and 2b cells, neuronally committed type 3 cells, and immature/mature neurons.Citation11 In this study, we demonstrated that the majority of Np95-expressing cells were actively proliferating type 2a cells in the DG, although a relatively small subpopulation of proliferating type 1, type 2b and type 3 cells also express Np95. We also found that Ki67 is expressed more broadly than Np95 in proliferating cells. This is probably because Ki67 can be detected not only in highly proliferating type 2a cells but also in cells that have just entered the cell cycle from the quiescent state (proliferating type 1 cells) and those that are about to exit the cell cycle to differentiate into neurons (type 2b and type 3 cells). Considering the important function of E2F1 in cell cycle progression and the fact that Np95 is a direct downstream target of E2F1 in OPCs,Citation49 we propose that higher expression of Np95 may be required to maintain the proliferating status of NS/PCs. Moreover, it has been reported that the expression level of Np95 is regulated in a cell cycle-dependent manner, and that deregulating its expression perturbs cell cycle progression.Citation50,51 Therefore, elevated and sustained expression of Np95 may be required for proper cell cycle progression, whereas reduced expression is probably necessary for NS/PCs to exit the cell cycle. It will be of interest to test this hypothesis in future studies.
Several physiological and pathological stimuli are known to facilitate NS/PC proliferation and to promote neurogenesis in the DG of the adult hippocampus.Citation36,38,39 We confirmed that the number of proliferating cells in the DG increased significantly in response to both physiological (voluntary running) and pathological (KA administration) conditions. Concordant with this increase, the number of Np95+ cells also rose. A previous study showed that the reduction of Np95 led to a failure of S phase entry in cell cycle progression of NIH3T3 cells.Citation44 In addition, appropriate DNA methylation is critical for adult neurogenesis by regulating proliferation and/or survival of NS/PCs.Citation52-54 Taking our findings together with these previous ones, we hypothesize that Np95 plays an important role in the regulation of NS/PC proliferation and neurogenesis in the DG in response to physiological and pathological stimuli.
Irrespective of the function of Np95, whose detailed elucidation must await future studies, Np95 promises to be a useful marker to identify proliferating NS/PCs, in particular type 2a cells, in the adult hippocampus. Since Np95 is subject to stricter expression control in type 2a cells than the pan-proliferating cell marker Ki67, it will allow us to conduct more accurate investigations of complex NS/PC behavior in the brain.
Materials and Methods
Animal treatment
8-week-old male C57/BL6 mice were used in this study. The mice were maintained on a 12-h light/dark schedule with free access to food and water. All mice used in this study were handled according to the animal experimentation guidelines of Nara Institute of Science and Technology, which comply with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. To induce epileptic seizure, mice were intraperitoneally injected with 30 mg/kg KA (Enzo Life Sciences, BML-EA123). All mice displayed seizure that started with jerking of the forelimbs, followed by seesaws and involuntary falling. These mice were sacrificed 4 d after the KA injection.
Cell culture
HEK293T cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (heat inactivated, Biowest, S1820) and gentamicin sulfate solution (100 mg/ml, Nacalai Tesque, 16672-04), under 5% CO2 at 37°C in a cell culture incubator. E14 mouse forebrains were dissected and triturated in calcium- and magnesium-free Hanks’ balanced salts solution (Sigma, H2387) and plated on a poly-ornithine/fibronectin-coated 10-cm dish in proliferating medium (N2-supplemented DMEM/F-12; Invitrogen, 11320-033), containing 10 ng/ml basic fibroblast growth factor (bFGF) (PeproTech, 100-18B) to expand the NS/PCs. Four days later, the cells were re-plated on a poly-ornithine/fibronectin-coated 3.5-cm dish and cultured under specified conditions (see legend).
Plasmids and transfections
Np95 was expressed from the plasmid vector BOSE-Np95-Flag, which allowed the identification of Np95-expressing cells using a specific FLAG antibody (Sigma, F1804) in western blotting. HEK293T cells (2 × 106) were plated on a 6-cm dish one day before transfection. Plasmid vector (3 μg) and 6 μl polyethyleneimine “MAX” (Polysciences, 24765-2) were diluted in 200 μl of OPTI-MEM (Gibco, 22600134), and incubated for 20 min at room temperature. The mixture was then added to confluent (70–80%) HEK293T cells, and the transfected cells were incubated as above for 24 h.
Immunocytochemistry
Cells cultured on coated 3.5-cm dishes were fixed with 4% paraformaldehyde (PFA) in 0.1 M phosphate-buffered saline, pH 7.4 (PBS) for 20 min, and then washed with PBS and incubated in blocking solution (PBS containing 3% FBS and 0.1% Triton X-100). The cells were incubated for 1.5 h at room temperature with the following primary antibodies: rabbit anti-UHRF1 (1:500; Santa Cruz Biotechnology, sc-98817), mouse anti-Ki67 (1:500; BD Biosciences, 550609), mouse anti-βIII-tubulin (Tuj1; 1:500; Sigma, T3952), goat anti-DCX (1:200; Santa Cruz Biotechnology, sc-8066), mouse anti-GFAP (1:500; Sigma, G3893) or mouse anti-APC (1:500; Calbiochem-Merck, OP80). After 3 washes in PBS, the cells were incubated for 1.5 h with the following secondary antibodies: FITC-conjugated donkey anti-chicken/mouse, Cy3-conjugated donkey anti-rabbit or Cy5-conjugated donkey anti-goat (all 1:500; Jackson ImmunoResearch, 705, 606 and 147, respectively). Nuclei were stained using bisbenzimide H33258 fluorochrome trihydrochloride (Hoechst; 1:500; Nacalai Tesque, 04928-92). Samples were washed 3 times with PBS and mounted on glass slides with Immu-Mount (Thermo Scientific, 9990412). Fluorescence images were acquired using a fluorescence microscope (Axiovert 200M, Zeiss) equipped with the appropriate epifluorescence filters. Images were combined for figures using Adobe Photoshop elements 10. All experiments were independently replicated at least 3 times.
Immunohistochemistry
8-week-old male C57/BL6 mice, E18 C57/BL6 mice and P10 C57/BL6 mice were anesthetized and perfused with PBS followed by 4% PFA in PBS. The brains were dissected and post-fixed overnight in the same fixative at 4°C. E11 and E14 C57/BL6 mice were directly preserved in 4% PFA in PBS and incubated overnight at 4°C. For cryosectioning, fixed tissues were cryoprotected in 15% sucrose in PBS overnight at 4°C, and then in 30% sucrose in PBS overnight at 4°C, and were finally embedded in optimal cutting temperature (OCT) compound (Tissue Tek, Sakura Finetek, 25608-930). Cryostat sections (8-week and P10: 40 μm; E11 and E14: 16 μm; E18: 20 μm) were cut and affixed to Matsunami adhesive slide (MAS)-coated glass slides (Matsunami Glass, S9441). Antigen retrieval was performed with autoclave treatment (105°C, 15 min) by soaking the slides in target retrieval solutions (DAKO, S1699). Next, the sections were washed with PBS and incubated in blocking solution (PBS containing 3% FBS and 0.1% Triton X-100) for 1 h, and then incubated overnight at 4°C with one of the following primary antibodies: rabbit anti-UHRF1 (1:500; Santa Cruz Biotechnology, sc-98817), rabbit anti-DNMT1 (1:500; Cosmo Bio, BAM-70-201-EX), mouse anti-Ki67 (1:500; BD Biosciences, 550609), goat anti-Sox2 (1:200; Santa Cruz Biotechnology, sc-17320), chicken anti-Nestin (1:500; Aves Labs, NES), goat anti-DCX (1:200; Santa Cruz Biotechnology, sc-8066), mouse anti-Map2ab (1:500; Sigma, M1406), guinea pig anti-GFAP (1:500; Advanced Immunochemical, 31223), rabbit anti-Tbr2 (1:500; Abcam, ab23345), mouse anti-Mash1 (1:50; BD Biosciences, 556604) or goat anti-Olig2 (1:100; R&D Systems, AF2418). After 3 washes in PBS, cells were incubated for 2 h with the following secondary antibodies: CF488A donkey anti-mouse IgG (H+L), highly cross-adsorbed (1:500; Biotium, 20014), CF543 donkey anti-rabbit IgG (H+L), highly cross-adsorbed (1:500; Biotium, 20308), CF647 donkey anti-goat IgG (H+L), highly cross-adsorbed (1:500; Biotium, 20048), CF647 goat anti-chicken IgY (H+L), highly cross-adsorbed (1:500; Biotium, 20044) or Cy5-conjugated donkey anti-guinea pig (1:500; Chemicon, AP193S). Nuclei were stained using Hoechst (1:500). After a final rinse with PBS, sections were mounted and examined under a fluorescence microscope (Axiovert 200M, Zeiss, or Leica DMI6000 B, Leica) and a scanning laser confocal imaging system (LSM 780 or LSM 700, Zeiss).
Western blot analysis
Western blot analysis was performed as described previously.Citation55 In brief, isolated E11, E14, E18, P7 and adult brains were lysed in lysis buffer (1% Nonidet P-40, 10 mM Tris-HCl pH 7.4, 150 mM NaCl, 100 μM protease inhibitor cocktail (Nacalai Tesque, 03969), 1 mM EDTA). The protein samples were separated in gradient (5–20%) polyacrylamide gels (e-PAGEL; ATTO, 2331830), transferred to a nitrocellulose membrane (GE Healthcare Life Sciences, RPN303F), and probed with anti-FLAG (1:2000; Sigma, F1804), anti-Np95 (1:2000),Citation32 anti-UHRF1 (1:2000; Santa Cruz Biotechnology, sc-98817), anti-DNMT1 (1:2000; Cosmo Bio, BAM-70-201-EX) or anti-actin (1:2000; Abcam, ab3280) antibody. Horseradish peroxidase-conjugated anti-mouse IgG (1:5000; GE Healthcare Life Sciences, NA931), horseradish peroxidase-conjugated anti-rabbit IgG (1:5000; GE Healthcare Life Sciences, NA934) or horseradish peroxidase-conjugated anti-rat IgG (1:5000; Santa Cruz Biotechnology, sc-2006) was used as the secondary antibody. Detection was performed using Chemi-Lumi One L (Nacalai Tesque, 07880).
Immunoprecipitation
HEK293T cells either transduced with BOSE-Np95-Flag or untransduced were lysed in lysis buffer (0.5% Nonidet P-40, 10 mM Tris-HCl pH 7.4, 150 mM NaCl, 100 μM protease inhibitor cocktail (Nacalai Tesque, 03969), 1 mM EDTA). Lysates were sonicated and centrifuged at 20,000 g for 30 min and the supernatant was harvested (cleared lysates). These lysates were divided in half and incubated overnight at 4°C in the presence of animal species-specific antibody-coated beads (Dynabeads M-280 Sheep anti-Mouse IgG or Sheep anti-Rat IgG, Life Technologies, 11202D and 11035, respectively) which were had been pre-incubated for 4 h with anti-FLAG (2 μg; Sigma, F1804) or anti-Np95 (2 μg)Citation32 antibodies. The beads were washed 3 times with lysis buffer and the proteins were eluted with sample buffer (1% β-mercaptoethanol, 10 mM Tris-HCl pH 6.8, 1% sodium dodecyl sulfate (SDS), 8% glycerol, 0.01% Bromophenol blue) for 5 min at 95°C. Each eluate was divided into 3 aliquots and subjected to western blot analysis as described above.
Voluntary running
For the voluntary running experiment, Fast Trac amber with Mouse Igloos (Animec, K-3250) was used as the running disc. 8-week-old mice were housed in a standard cage or voluntary running cage contained a running disc. These mice were sacrificed 2 weeks later.
Statistical analysis
Values are given as the mean ± standard error of the mean (SEM). The Student's t-test (for 2-groups comparison) or one-way ANOVA (Prism, GraphPad) (for multiple groups comparison) followed by the Tukey test was used to evaluate differences. P < 0.05 was considered significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
976026_Supplementary_Materials.zip
Download Zip (11.3 MB)Acknowledgments
We thank Y. Bessho, T. Matsui, Y. Nakahata, T. Imamura, and S. Katada for valuable discussions. We also appreciate the members of our laboratories, in particular Y. Katano, for technical help and suggestions. In addition, we appreciate M. Unoki of the University of Kyushu for technical help with the experiments. Furthermore, we are very grateful to M. Tano and Y. Nakagawa for their excellent secretarial assistance. We thank I. Smith for editing the manuscript.
Funding
This study was supported by the Japan Society for the Promotion of Science (JSPS): Grant-in-Aid for JSPSFellows. This work was also supported by JST, CREST to KN.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev 2004; 14:461-9; PMID:15380235; http://dx.doi.org/10.1016/j.gde.2004.07.006
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediat Res 2007; 61:58R-63R; PMID:17413844; http://dx.doi.org/10.1203/pdr.0b013e3180457635
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci 2007; 8:355-67; PMID:17453016; http://dx.doi.org/10.1038/nrn2132
- Hsieh J, Eisch AJ. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: unraveling the genome to understand the mind. Neurobiol Dis 2010; 39:73-84; PMID:20114075; http://dx.doi.org/10.1016/j.nbd.2010.01.008
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci 2010; 13:1338-44; PMID:20975758; http://dx.doi.org/10.1038/nn.2672
- Gage FH. Mammalian neural stem cells. Science 2000; 287:1433-8; PMID:10688783; http://dx.doi.org/10.1126/science.287.5457.1433
- Temple S. The development of neural stem cells. Nature 2001; 414:112-7; PMID:11689956; http://dx.doi.org/10.1038/35102174
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell 2001; 1:749-58; PMID:11740937; http://dx.doi.org/10.1016/S1534-5807(01)00101-0
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 2009; 63:600-13; PMID:19755104; http://dx.doi.org/10.1016/j.neuron.2009.08.021
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 2007; 130:1146-58; PMID:17825401; http://dx.doi.org/10.1016/j.cell.2007.07.010
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 2011; 70:687-702; PMID:21609825; http://dx.doi.org/10.1016/j.neuron.2011.05.001
- Winner B, Kohl Z, Gage FH. Neurodegenerative disease and adult neurogenesis. Eur J Neurosci 2011; 33:1139-51; PMID:21395858; http://dx.doi.org/10.1111/j.1460-9568.2011.07613.x
- Snyder JS, Soumier A, Brewer M, Pickel J, Cameron HA. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature 2011; 476:458-61; PMID:21814201; http://dx.doi.org/10.1038/nature10287
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 2004; 36:1291-5; PMID:15516931; http://dx.doi.org/10.1038/ng1468
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 2000; 225:294-303; PMID:10985851; http://dx.doi.org/10.1006/dbio.2000.9823
- Meehan RR. DNA methylation in animal development. Semin Cell Dev Biol 2003; 14:53-65; PMID:12524008; http://dx.doi.org/10.1016/S1084-9521(02)00137-4
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet 2000; 9:2395-402; PMID:11005794; http://dx.doi.org/10.1093/hmg/9.16.2395
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 2010; 11:285-96; PMID:20300089; http://dx.doi.org/10.1038/nrg2752
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol 2009; 8:1056-72; PMID:19833297; http://dx.doi.org/10.1016/S1474-4422(09)70262-5
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128:683-92; PMID:17320506; http://dx.doi.org/10.1016/j.cell.2007.01.029
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007; 317:1760-4; PMID:17673620; http://dx.doi.org/10.1126/science.1147939
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007; 450:908-12; PMID:17994007; http://dx.doi.org/10.1038/nature06397
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 2008; 455:818-21; PMID:18772891; http://dx.doi.org/10.1038/nature07249
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 2008; 455:822-5; PMID:18772889; http://dx.doi.org/10.1038/nature07273
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008; 455:826-9; PMID:18772888; http://dx.doi.org/10.1038/nature07280
- Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 2004; 23:7601-10; PMID:15361834; http://dx.doi.org/10.1038/sj.onc.1208053
- Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol 2008; 28:705-17; PMID:17967883; http://dx.doi.org/10.1128/MCB.01598-07
- Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 2009; 37:493-505; PMID:19056828; http://dx.doi.org/10.1093/nar/gkn961
- Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res 2010; 38:1796-804; PMID:20026581; http://dx.doi.org/10.1093/nar/gkp1152
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 2012; 19:1155-60; PMID:23022729; http://dx.doi.org/10.1038/nsmb.2391
- Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun 2013; 4:1563; PMID:23463006; http://dx.doi.org/10.1038/ncomms2562
- Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J Biol Chem 2002; 277:34549-55; PMID:12084726; http://dx.doi.org/10.1074/jbc.M205189200
- von Bohlen und Halbach O. Immunohistological markers for proliferative events, gliogenesis, and neurogenesis within the adult hippocampus. Cell Tissue Res 2011; 345:1-19; PMID:21647561; http://dx.doi.org/10.1007/s00441-011-1196-4
- Hsieh J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev 2012; 26:1010-21; PMID:22588716; http://dx.doi.org/10.1101/gad.187336.112
- Kempermann G, Kuhn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature 1997; 386:493-5; PMID:9087407; http://dx.doi.org/10.1038/386493a0
- van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci 1999; 2:266-70; PMID:10195220; http://dx.doi.org/10.1038/6368
- Kempermann G, Gast D, Gage FH. Neuroplasticity in old age: sustained fivefold induction of hippocampal neurogenesis by long-term environmental enrichment. Ann Neurol 2002; 52:135-43; PMID:12210782; http://dx.doi.org/10.1002/ana.10262
- Naylor AS, Bull C, Nilsson MK, Zhu C, Bjork-Eriksson T, Eriksson PS, Blomgren K, Kuhn HG. Voluntary running rescues adult hippocampal neurogenesis after irradiation of the young mouse brain. Proc Nat Acad Sci U S A 2008; 105:14632-7; PMID:18765809; http://dx.doi.org/10.1073/pnas.0711128105
- Gray WP, Sundstrom LE. Kainic acid increases the proliferation of granule cell progenitors in the dentate gyrus of the adult rat. Brain Res 1998; 790:52-9; PMID:9593820; http://dx.doi.org/10.1016/S0006-8993(98)00030-4
- Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Gotz M, Haas CA, Kempermann G, Taylor V, Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 2010; 6:445-56; PMID:20452319; http://dx.doi.org/10.1016/j.stem.2010.03.017
- Beukelaers P, Vandenbosch R, Caron N, Nguyen L, Belachew S, Moonen G, Kiyokawa H, Barbacid M, Santamaria D, Malgrange B. Cdk6-dependent regulation of G(1) length controls adult neurogenesis. Stem Cells 2011; 29:713-24; PMID:21319271; http://dx.doi.org/10.1002/stem.616
- Fujimori A, Matsuda Y, Takemoto Y, Hashimoto Y, Kubo E, Araki R, Fukumura R, Mita K, Tatsumi K, Muto M. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm Genome 1998; 9:1032-5; PMID:9880673; http://dx.doi.org/10.1007/s003359900920
- Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIa expression. Cancer Res 2000; 60:121-8; PMID:10646863
- Bonapace IM, Latella L, Papait R, Nicassio F, Sacco A, Muto M, Crescenzi M, Di Fiore PP. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J Cell Biol 2002; 157:909-14; PMID:12058012; http://dx.doi.org/10.1083/jcb.200201025
- Unoki M, Brunet J, Mousli M. Drug discovery targeting epigenetic codes: the great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochem Pharmacol 2009; 78:1279-88; PMID:19501055; http://dx.doi.org/10.1016/j.bcp.2009.05.035
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 2005; 132:3345-56; PMID:16014513; http://dx.doi.org/; http://dx.doi.org/10.1242/dev.01912
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell 2009; 16:245-55; PMID:19217426; http://dx.doi.org/10.1016/j.devcel.2008.12.014
- Mutoh T, Sanosaka T, Ito K, Nakashima K. Oxygen levels epigenetically regulate fate switching of neural precursor cells via hypoxia-inducible factor 1alpha-notch signal interaction in the developing brain. Stem Cells 2012; 30:561-9; PMID:22213097; http://dx.doi.org/10.1002/stem.1019
- Magri L, Swiss VA, Jablonska B, Lei L, Pedre X, Walsh M, Zhang W, Gallo V, Canoll P, Casaccia P. E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. J Neurosci 2014; 34:1481-93; PMID:24453336; http://dx.doi.org/10.1523/JNEUROSCI.2840-13.2014
- Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1S transition during the cell cycle. Oncogene 2005; 24:7337-45; PMID:16007129; http://dx.doi.org/10.1038/sj.onc.1208878
- Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L, Hu S, Zeng P, Guo R, Diao J, et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Nat Acad Sci U S A 2012; 109:4828-33; PMID:22411829; http://dx.doi.org/10.1073/pnas.1116349109
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 2009; 323:1074-7; PMID:19119186; http://dx.doi.org/10.1126/science.1166859
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 2010; 329:444-8; PMID:20651149; http://dx.doi.org/10.1126/science.1190485
- Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 2013; 13:237-45; PMID:23770080; http://dx.doi.org/10.1016/j.stem.2013.05.006
- Kohyama J, Sanosaka T, Tokunaga A, Takatsuka E, Tsujimura K, Okano H, Nakashima K. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J Cell Biol 2010; 189:159-70; PMID:20351065; http://dx.doi.org/10.1083/jcb.200908048
