Abstract
Genome integrity is achieved and maintained by the sum of all of the processes in the cell that ensure the faithful duplication and repair of DNA, as well as its genetic transmission from one cell division to the next. As central players in virtually all of the DNA transactions that occur in vivo, DNA helicases (molecular motors that unwind double-stranded DNA to produce single-stranded substrates) represent a crucial enzyme family that is necessary for genomic stability. Indeed, mutations in many human helicase genes are linked to a variety of diseases with symptoms that can be generally described as genomic instability, such as predispositions to cancers. This review focuses on the roles of both DNA replication helicases and recombination/repair helicases in maintaining genome integrity and provides a brief overview of the diseases related to defects in these enzymes.
Abbreviations
| BVP | = | bovine papilloma virus |
| DSB | = | double-strand break |
| dsDNA | = | double-stranded DNA |
| HR | = | homologous recombination |
| ICL | = | interstrand crosslink |
| MCM or Mcm | = | mini-chromosome maintenance |
| mtDNA | = | mitochondrial DNA |
| PEO | = | progressive external opthalmoplegia |
| ssDNA | = | single-stranded DNA |
| SF | = | superfamily |
| ts | = | temperature sensitive |
| TAg | = | T-antigen |
Introduction
It has been written that “The human body can achieve many things, but perhaps its greatest role is to act as a storage mechanism for the genetic information of the species.”Citation1 However, an organism does not merely store genetic information; the integrity of the genome is also safeguarded through high-fidelity replication, recombination, and repair of genetic information. Indeed, breakdown of genome integrity, a state known as genomic instability, is a characteristic of many diseases such as cancer. Thus, maintaining genomic stability in the face of the approximately 10,000 DNA damaging events that every cell in the human body experiences every dayCitation2 is essential for the faithful propagation of genetic material from one generation to the next.
Although a multitude of diverse proteins are involved in DNA replication, recombination, and repair, members of only 2 enzymatic families—DNA helicases and DNA polymerases—play roles in virtually all aspects of these processes. This review focuses on the former, but interested readers are directed to several recent excellent reviews on DNA polymerases and their roles in maintaining genome integrity.Citation3-5
DNA helicases are molecular motors that in most cases use the power of ATP hydrolysis to unwind double-stranded (ds) DNA and RNA-DNA hybrids into single-stranded (ss) DNA templates. The genomes of all organisms encode a variety of DNA and RNA helicases and helicase-like proteins, from ∼30 in model bacteria like Escherichia coli and Bacillus subtilis, to ∼100 in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, to 163 in humans.Citation6 Based on conserved sequence motifs, helicases have been bioinformatically classified into one of 7 superfamilies (SF-I to SF-VII).Citation7 They have also been further categorized by their polarity of unwinding (e.g., 3′-5′ [SF-IA] vs. 5′-3′ [SF-IB]) and placed into 18 subfamilies (DnaB/MCM, DEAD-box, DEAH-box, SWI2/SNF2, SKI1, RecD/UPF1, PIF1, MPH1, DinG/RAD3, RECQ, Lhr/HRQ1, UvrD/SRS2, RuvB/RVB, KU, YRF1, HsdR/IRC3/SSL2, PhoH/Rho/SecA/HerA/UvrB/PriA/YgcB, and unclassified) using a variety of bioinformatics techniques (reviewed inCitation6). Much has also been written about the various mechanisms used by helicases to unwind dsDNACitation8-10 and the biochemical details underpinning this activity.Citation11-13
Attempting to address all of these details with respect to the roles of DNA helicases in maintaining genome integrity is beyond the scope of this review. Similarly, each of the multitude of helicases described above cannot be adequately addressed. Instead, this review focuses on well-known members of the replicative and recombination/repair helicases, as well as helicase-linked diseases that result from genomic instability.
Replicative helicases
Replicative helicases are the enzymes responsible for the bulk dsDNA unwinding necessary for genome replication during every cell cycle. These enzymes share several features that together distinguish them from all other helicases: (1) they are essential for viability, (2) they are required for both the initiation and elongation steps of DNA replication, (3) they function at the point of the replication fork, and (4) nearly all of them function as ring-shaped hexamers. Evolution has led to at least 5 distinct families of replicative helicases used by bacteria, archaea, eukaryotes, viruses, and mitochondria (see below).
As enzymes that interact with every base pair of DNA in the genome, replicative helicases are also critical for the maintenance of genome integrity (). They are the first portion of the replication fork to encounter DNA lesions and proteins bound to the DNA, both of which can stall DNA replication and lead to genomic instability.Citation14 DnaB-like helicases, those in the minichromosome maintenance (MCM) family, viral replicative helicases, and mitochondrial replicative helicases are briefly introduced below, and their connections to genome maintenance are described.
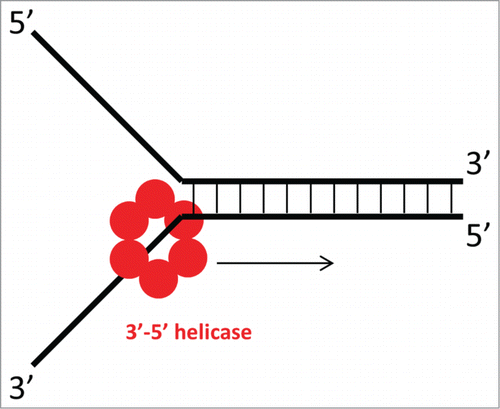
Figure 1. Steric exclusion model of DNA unwinding by a ring-shaped helicase. One strand of ssDNA passes through the central channel of the helicase, while the other is excluded. Unidirectional movement of the helicase (in this case, 3′-5′ as indicated by the arrow) toward the dsDNA and exclusion of the other strand aid in unwinding the DNA duplex.
Bacterial DnaB-like helicases
All bacteria with sequenced genomes encode a homolog of the well-studied E. coli DnaB (R. Ramalho, unpublished), the prototypical bacterial replicative helicase. In vivo, the E. coli genome is replicated bidirectionally from a single origin of replication, (reviewed inCitation15) where homohexameric rings of DnaB are opened and clamped around the ssDNA by the DnaC loader protein.Citation16 One DnaB hexamer is loaded onto the ssDNA on each side of the origin, and DNA unwinding proceeds in opposite directions around both halves of the circular E. coli chromosome.
The importance of DnaB to the integrity of the E. coli genome is exemplified by experiments performed with temperature sensitive (ts) alleles of the dnaB gene. At the restrictive temperature, DNA replication elongation is blocked in these cells and newly replicated DNA is extensively degraded.Citation17 This is in contrast to types of damage that arrest DNA replication without directly targeting DnaB (e.g., UV damage), in which the replication forks are stabilized and protectedCitation18 until the damage can be repaired.Citation19,20 In any event, DnaB(ts)-mediated replication fork stalling and the associated nascent DNA degradation are disastrous to the cell. Therefore, in E. coli (and probably also other bacteria), a properly functioning replicative helicase is essential to maintain genome integrity. As such, small molecules that target and inactivate DnaB-like helicases should function as potent antibiotics.
MCM helicases
The replicative helicases of all prokaryotes studied to date are homohexamers, including those found in archaea. However, unlike the bacterial DnaB helicases, archaeal genomes encode MCM replicative helicases. Although they are functional homologs (i.e., they are both localized at replication forks to unwind genomic DNA during replication), DnaB and MCM helicases are not orthologous. Further, archaeal MCMs translocate along DNA with an opposite polarity to DnaB (3′-5′ vs. 5′-3′; reviewed inCitation21,22). The eukaryotic replicative helicase is also a 3′-5′ MCM family enzyme. However, in contrast to the archaeal MCM, the eukaryotic Mcm2–7 complex is a heterohexamer comprised of 6 distinct subunits (individually numbered Mcm2 through Mcm7).Citation23 As with DnaB in bacteria, though, the MCM/Mcm2–7 helicases are the vanguards of the replication forks in archaea and eukaryotes.
Biochemical studies of the simpler and more stable archaeal MCM complexes, especially those from thermophilic archaea, have yielded a tremendous wealth of structural (e.g.,Citation24-27) and mechanistic (e.g.,Citation28-31) information about this enzyme family. However, most work connecting MCM helicases to genomic stability has been performed with Mcm2–7 and eukaryotic model organisms.
Because of its essential role in DNA replication, which must occur once and only once per cell cycle in eukaryotes, loading and activation of the Mcm2–7 complex at origins of replication are tightly and redundantly controlled processes.Citation32 As stated above, genomic instability is a hallmark of cancers, thus perturbing Mcm2–7 regulation or activity can lead to carcinogenesis. For example, work in S. cerevisiae and mammals indicates that a hypomorphic allele of Mcm4 (Mcm4Chaos3) is linked to increased rates of lossCitation33 and mutationCitation34 of genetic information in yeast and a variety of defects in mice, including mammary adenocarcinomas ().Citation33 Similarly, deregulating Mcm7 expression actively increases tumor formation in a mouse chemical carcinogenesis model.Citation35 Indeed, altering the expression levels of any of the 6 Mcm2–7 subunits renders cells susceptible to chromosome loss, increased recombination rates, altered viability, and/or early-onset cancer.Citation36,37
Table 1. Helicase-linked diseases
The loss of genome integrity in tumor cells allows them to rapidly accumulate mutations that lead to their uncontrolled replication and can also lead to the development of resistance to chemotherapeutic agents. However, as a vital player in DNA replication, targeting the Mcm2–7 complex with drugs is hypothesized to be a viable method to fight cancer.Citation38 If a drug is able to inhibit all 6 Mcm2–7 subunits, 6 mutational events would be needed to develop resistance if drug resistance is even possible (as these are essential proteins, many mutations will simply be lethal). Schwacha and colleagues are screening small molecules to uncover Mcm2–7-specific inhibitors and their effects on yeast and human cells.Citation38
Viral replicative helicases
The replicative helicases from DNA viruses are members of SF-I to SF-III,Citation39 with well-studied examples that include simian virus 40 T-antigen (SV40 TAg)Citation40 and the bovine papilloma virus (BVP) E1 protein.Citation41 Both of these helicases bind to origins of replication in their respective viral genomes in a sequence-dependent manner. However, TAg is unique among replicative helicases in that it associates with origin DNA on its own;Citation40 all of the other helicases require additional DNA replication initiation proteins to help target them to origins of replication (e.g., DnaA and DnaC for E. coli DnaBCitation15,16 and the papillomavirus E2 protein for E1).Citation41 In most other respects, however, TAg and E1are quite similar. They both load at origins, where they undergo an ATP-dependent multistep oligomerization process to form 2 ring-shaped head-to-head hexamers (i.e., double hexamers) with the DNA topologically constrained within the central channels of the hexameric rings. Based on a crystal structures of BVP E1 in the presence and absence of DNA and nucleotides,Citation42,43 it is believed that these ring-shaped helicases contain ssDNA within their central channels and thus move along only one strand of the DNA, melting the double helix by steric exclusion of the unbound strand (). Similar unwinding models have also been proposed for DnaB and the MCM/Mcm2–7 helicases (seeCitation29,44 and references therein).
Initially, the replicative helicases from eukaryotic viruses served as models to begin delineating the similarities and differences between bacterial and eukaryotic DNA replication in vitro. This is because the Mcm2–7 helicase has only recently been found to be amenable to biochemical investigations through the use of buffer conditions that more closely resemble the nuclear milieuCitation45 and the discovery of associated factors that stimulate its activity.Citation46 However, like Mcm2–7, both SV40 TAg and the E1 proteins of papillomaviruses are linked to genome stability. For example, SV40 (a non-human primate virus that was widely introduced into the human population through polio vaccines contaminated with the virus),Citation47 and related polyomaviruses induce malignant transformation of cells.Citation40 This process occurs when TAg binds to and suppresses the activity of the tumor suppressor proteins p53 and Rb, inducing uncontrolled cellular proliferation and rendering the genome susceptible to damage. Papillomaviruses are similarly linked to tumorigenesis.Citation48 As the most conserved protein encoded by papillomavirus genomes and the only one with enzymatic activity, E1 is vital for the virus to commandeer the normal DNA replication machinery of the cell.Citation41 Thus, although the TAg and E1 helicases may help to ensure the integrity of their viral genomes, they also lead to genetic instability in host cells.
Mitochondrial replicative helicases
It is widely believed that mitochondria arose as the result of an ancient endosymbiosis between a eukaryotic cell and an α-proteobacterium.Citation49 As such, these organelles contain a separate genome (mtDNA) from the nuclear DNA that encodes genes with homology to bacteria and bacteriophages. It has also become clear in recent years that the replication of mtDNA involves a different repertoire of enzymes than replication of the nuclear genome, including a mitochondrial replicative helicase known as Twinkle in metazoans.Citation50
Twinkle is a 5′-3′ helicaseCitation51,52 that is more similar to bacteriophage and DnaB-like helicases than to MCM proteins, supporting a bacterial origin for mitochondria. Like all of the replicative helicases described above, though, Twinkle forms a hexameric complex to unwind DNA, and its proper function is linked to maintaining genome integrity. For example, in tissues under high oxidative stress, high Twinkle levels are necessary to overcome replication fork stalling and reduce mtDNA mutations caused by damage from reactive oxygen species.Citation53 Mutations in the gene encoding human TWINKLE are causative of autosomal dominant progressive external opthalmoplegia (PEO) as a result of associated deletions in the mtDNA ().Citation54 PEO is a disease characterized by weak or paralyzed eye muscles, drooping eyelids, and general skeletal muscle weakness that can be exacerbated by exercise and results from depletion of mitochondria,Citation55 i.e., from loss of mtDNA as a result of genomic instability.
Although all multicellular and most unicellular eukaryotes have mitochondria (very simple parasitic eukaryotes lack themCitation56), not all of these organisms encode a Twinkle homolog. Such organisms include the well-studied budding yeast S. cerevisiae and kinetoplastid parasites such as Trypanosoma brucei. However, these organisms encode one or more members of the Pif1 family of helicases (reviewed inCitation57), which in S. cerevisiaeCitation58-62 and T. bruceiCitation63-65 are necessary for mtDNA maintenance. It is tempting to speculate that Pif1 helicases may act as replicative helicases in these cases, although unlike the enzymes discussed above, Pif1 proteins are not known to form hexamersCitation66 nor do they display the levels of processivity (the number of base pairs unwound per helicase-DNA binding event) that one would expect to be necessary to unwind the mtDNA genome.Citation67 Speculation aside, the roles of Pif1 family helicases in maintaining the integrity of the nuclear genome are discussed in greater detail below.
Recombination and repair helicases
In addition to replicative helicases, cells encode a cadre of additional helicases that have a variety of functions. For example, accessory helicases such as E. coli Rep, UvrD, and DinG and S. cerevisiae Rrm3 can act in conjunction with their replicative helicases to drive replication fork progression past impediments (e.g., protein-bound DNA) in vivo.Citation68,69 Additionally, helicases can serve more than one role, such as the human XPB and XPD enzymes (S. cerevisiae Ssl2 and Rad3, respectively) that function in both transcription and nucleotide excision repair.Citation70 Many more have niche roles in DNA recombination and repair, both of which are essential for maintaining genome integrity. Examples of such helicases from 2 evolutionarily conserved families and their roles in genome maintenance are discussed below.
RecQ helicase family
RecQ proteins are 3′-5′ helicases that have DNA structure-specific roles in vivo, often functioning at recombination intermediates (reviewed inCitation71). E. coli expresses the founding member of this family, known simply as RecQ, but eukaryotes tend to express several RecQ helicases. Indeed, the human genome encodes 5 RecQs (RECQ1, BLM, WRN, RECQ4, and RECQ5), and even single-celled eukaryotes like yeasts express 2 or 3 RecQs.Citation72-75 Mutations in 3 of the human RecQ helicases (BLM, WRN, and RECQ4) cause diseases characterized by a predisposition to cancers and/or premature aging (), pathologies that are linked to loss of genome integrity.Citation71
Perturbation of the expression levels and biochemical activities of the RecQ helicases have such negative consequences on genome integrity because these enzymes interact with a host of important protein cofactors. Indeed, RecQs affect DNA replication (RECQ1Citation76 and RECQ4Citation77), recombination (all 5 human RecQsCitation71), repair (all 5Citation71), and telomere maintenance (BLM,Citation78 WRN,Citation79 and RECQ4Citation80), as well as transcription (BLM,Citation81 WRN,Citation82 and RECQ5Citation83) and mtDNA maintenance (RECQ4Citation84). In other words, one or more of the RecQ helicases function in virtually all aspects of DNA metabolism.
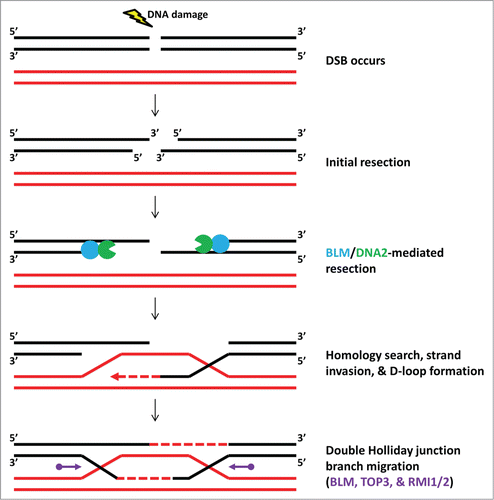
RecQs are perhaps best known for their roles in homologous recombination (HR; reviewed inCitation85). Indeed, they are involved in multiple steps of the HR repair pathway, from beginning to end (). In human cells, when a DNA double-strand break (DSB) occurs, the DNA ends are initially resected in the 5′-3′ direction. The resulting 3′ ssDNA is the perfect substrate for the 3′-5′ BLM helicase, which partners with the nuclease DNA2 to processively unwind dsDNA and degrade the resulting 5′ ssDNA strand, leading to further resection. The remaining 3′ ssDNA is eventually coated by the RAD51 recombinase, which aids in the homology search, strand invasion, and D-loop formation necessary to carry out HR. One pathway used to resolve the D-loop involves the formation of double Holliday junctions, which themselves are resolved by BLM in a complex with TOP3 (a topoisomerase) and RMI1/RMI2 (factors that stimulate TOP3 activity). Furthermore, biochemical experiments suggest that RECQ1,Citation86 BLM,Citation87 and RECQ5Citation88 can inhibit or correct the formation of unproductive recombination intermediates. All of these steps are vital to proper HR, a DSB repair pathway that does not result in loss of genetic information and hence aids in maintaining genomic integrity.
Figure 2. Simplified model of double-strand break (DSB) repair. When a DSB occurs, the DNA surrounding the break is initially resected in the 5′-3′ direction to produce 3′ ssDNA overhangs. The BLM helicase can load onto this 3′ ssDNA and translocate in the 3′-5′ direction, unwinding the double helix to create additional 5′ ssDNA that the DNA2 nuclease degrades to further resect the DNA away from the lesion. The RAD51 recombinase (not pictured) coats the ssDNA to initiate a homology search, strand invasion, and D-loop formation on the undamaged chromosome (red). DNA synthesis (red dashed arrow) serves to copy the missing genetic information. One of the pathways used to resolve these recombination intermediates involves the formation of double Holliday junctions and their branch migration and resolution by a complex composed of BLM, TOP3, RMI1, and RMI2 (purple).
Pif1 helicase family
Pif1 helicases function with a 5′-3′ polarity, and like the RecQs, perform a diverse set of known and hypothesized functions in vivo.Citation57 Although Pif1s were originally thought to be present only in eukaryotes, genes encoding these enzymes have recently been identified in numerous bacteria, bacteriophages, and eukaryotic viruses.Citation89 To date, little is known about the functions of Pif1s in bacteria and viruses, but the roles of Pif1 helicases in genome maintenance in S. cerevisiae, S. pombe, and other eukaryotes have been investigated by several groups (reviewed inCitation57).
Unlike most model eukaryotes (e.g., mice and humans), S. cerevisiae encodes 2 Pif1 family helicases: the founding member Pif1 and its paralog Rrm3.Citation57 Neither protein is essential, nor are cells lacking both the PIF1 and RRM3 genes inviable (N. Ahmad and M. Bochman, unpublished). However, Pif1 and Rrm3 perform multiple (and often opposing) functions to help maintain both nuclear and mitochondrial genome integrity.Citation57 As the best-studied family member, the S. cerevisiae Pif1 is focused on here.
As hypothesized above, Pif1 may be the S. cerevisiae mitochondrial replicative helicase.Citation58-62 In the nucleus, however, Pif1 is a veritable jack-of-all-trades. It acts as a catalytic inhibitor of telomerase by using its helicase activity to physically evict telomerase from chromosome ends and DSBs.Citation90 Additionally, Pif1 is involved in Okazaki fragment maturation, where it probably creates long ssDNA flaps that are degraded by the Dna2 nuclease during Okazaki fragment processing.Citation91,92 Pif1 also helps to oppose DNA replication at rDNA repeats, where it aids in establishing replication fork barriers to prevent the head-on collision of replication and transcription.Citation93 More recently, S. cerevisiae Pif1 was shown to suppress genomic instability at DNA motifs with the potential to form very stable secondary structures that can impede replication fork progression (G-quadruplex motifs)Citation94,95 and to promote break-induced repair by helping to migrate a bubble-like replication fork.Citation96,97
It is unclear how many of these activities are conserved in the human PIF1 helicase, but defects in any of them could explain why mutation of a conserved residue in the PIF1 ATPase/helicase domain is linked to inherited breast cancer ().Citation98 It is also unclear what bacterial Pif1 helicases do in vivo, especially in organisms encoding more than one family member.Citation89 However, if the bacterial Pif1s are as vital to genome integrity as S. cerevisiae Pif1, they may also prove to be useful drug targets in human pathogens.
Helicase-linked diseases
An obvious theme that arises from the above examples of helicases and their roles in maintaining genome integrity is that when the activity of these enzymes is altered (either by mutation or changes in expression level), disease ensues. Many of these pathologies are predispositions to cancer, suggesting that a large number of helicases are tumor suppressors (see ). Additional helicases that are known to be linked to disease have been covered in several excellent reviews.Citation99,100 Some of the best studied include those linked to Fanconi anemia (FANCJ and FANCM in humans; Chl1 and Mph1 in S. cerevisiae), a genetic disease leading to cancer and bone marrow failure in most patients as a result of defects in repairing DNA interstrand crosslinks (ICLs).Citation101
ICLs are covalent linkages between the two strands of the double helix and are particularly dangerous DNA lesions because they block both replication and transcription. Indeed, mutations in many other human helicases are linked to ICL sensitivity, including BLM, CHLR1/DDX11, HELQ, the Mcm8/9 complex, RECQ4, RECQ5, RTEL1, and WRN () (Rogers, van Kessel, and Bochman, in press). Unsurprisingly, there are known and suspected disease links with these enzymes, such as the CHLR1/DDX11 mutations that cause Warsaw breakage syndrome, which is characterized by defects in sister chromatid cohesion and Fanconi anemia-like symptoms.Citation102 Similarly, RTEL1 mutations are associated with dyskeratosis congenitaCitation103 and Hoyeraal-Hreidarsson syndrome,Citation104 related diseases that are characterized by bone marrow and telomere maintenance defects.
Adjacent nucleotides in DNA can also be crosslinked (i.e., form intrastrand crosslinks), such as the thymine-thymine dimers caused by UV irradiation. Although not as deleterious as ICLs to cells, intrastrand lesions must still be repaired to maintain genomic integrity, and helicases are involved in this repair. Indeed, mutations in the XPB and XPD helicases mentioned above are linked to xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy – diseases that share the symptom of light sensitivity due to deficiencies in repairing UV damage.Citation70,105
Conclusions
Based on their evolutionary conservation, known and hypothesized in vivo roles, and links to diseases when mutated, it is clear that DNA helicases are essential for maintaining genomic integrity. What is unclear, however, is how defects in these enzymes lead to disease. Indeed, many of the helicases described above are multifunctional, and deficiencies in any one (or more) of the in vivo processes that they take part in could result in a predisposition to cancer. For example, do mutations in RECQ4 alter its activities in DNA replication, recombination, repair, telomere maintenance, or mtDNA maintenance? Furthermore, do different mutations differentially affect RECQ4, accounting for the spectrum of diseases that it is linked to? In the future questions such as these must be addressed, both biochemically using purified protein and in vivo using mutant cell lines and simple model systems (e.g.,Citation72). Such investigations will delineate exactly which of the pathways these helicases are involved in safeguard genome integrity and suggest targets for clinical interventions in helicase-linked diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank Julia van Kessel, David Nickens, Rhakshin Kharwadkar, and Cody Rogers for their insightful comments on the manuscript. I also apologize to those colleagues whose work was not discussed or cited in this review due to space limitations.
References
- Herbert B, Anderson KJ, Herbert F. Hunters of Dune. New York: Tor, 2006.
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993; 362:709-15; PMID: 8469282; http://dx.doi.org/10.1038/362709a0
- Johansson E, Dixon N. Replicative DNA polymerases. Cold Spring Harb Perspect Biol 2013; 5:a012799; PMID:23732474; http://dx.doi.org/10.1101/cshperspect.a012799
- Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol 2013; 5:a010363; PMID:23838442; http://dx.doi.org/10.1101/cshperspect.a010363
- Boyer AS, Grgurevic S, Cazaux C, Hoffmann JS. The human specialized DNA polymerases and non-B DNA: vital relationships to preserve genome integrity. J Mol Biol 2013; 425:4767-81; PMID:24095858; http://dx.doi.org/10.1016/j.jmb.2013.09.022
- Eki T. Genome-wide survey and comparative study of helicase superfamily members in sequenced genomes. In: Urbano KV, ed. Advances in Genetics Research: Nova Science Publishers, Inc., 2010;168-203.
- Berger JM. SnapShot: nucleic acid helicases and translocases. Cell 2008; 134:888- e1; PMID:18775318; http://dx.doi.org/10.1016/j.cell.2008.08.027
- Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem 2006; 281:18265-8; PMID:16670085; http://dx.doi.org/10.1074/jbc.R600008200
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Ann Rev Biophys 2008; 37:317-36; PMID:18573084; http://dx.doi.org/10.1146/annurev.biophys.37.032807.125908
- Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 2009; 73:652-83; PMID:19946136; http://dx.doi.org/10.1128/MMBR.00019-09
- Opresko PL, Cheng WH, Bohr VA. Junction of RecQ helicase biochemistry and human disease. J Biol Chem 2004; 279:18099-102; PMID:15023996; http://dx.doi.org/10.1074/jbc.R300034200
- Brosh RM, Jr, Sharma S. Biochemical assays for the characterization of DNA helicases. Methods Mol Biol 2006; 314:397-415; PMID:16673896; http://dx.doi.org/10.1385/1-59259-973-7:397
- Bell SD, Botchan MR. The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol 2013; 5:a012807; PMID:23881943; http://dx.doi.org/10.1101/cshperspect.a012807
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 2007; 71:13-35; PMID:17347517; http://dx.doi.org/10.1128/MMBR.00030-06
- Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 2007; 5:343-54; PMID:17435790; http://dx.doi.org/10.1038/nrmicro1640
- Davey MJ, O'Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr Biol 2003; 13:R594-6; PMID:12906810; http://dx.doi.org/10.1016/S0960-9822(03)00523-2
- Belle JJ, Casey A, Courcelle CT, Courcelle J. Inactivation of the DnaB helicase leads to the collapse and degradation of the replication fork: a comparison to UV-induced arrest. J Bacteriol 2007; 189:5452-62; PMID:17526695; http://dx.doi.org/10.1128/JB.00408-07
- Chow KH, Courcelle J. RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli. J Biol Chem 2004; 279:3492-6; PMID:14625283; http://dx.doi.org/10.1074/jbc.M311012200
- Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci U S A 2006; 103:9154-9; PMID:16754873; http://dx.doi.org/10.1073/pnas.0600785103
- Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet 1999; 262:543-51; http://dx.doi.org/10.1007/s004380051116
- Beattie TR, Bell SD. Molecular machines in archaeal DNA replication. Curr Opin Chem Biol 2011; 15:614-9; PMID:21852183; http://dx.doi.org/10.1016/j.cbpa.2011.07.017
- Brewster AS, Chen XS. Insights into the MCM functional mechanism: lessons learned from the archaeal MCM complex. Crit Rev Biochem Mol Biol 2010; 45:243-56; PMID:20441442; http://dx.doi.org/10.3109/10409238.2010.484836
- Vijayraghavan S, Schwacha A. The eukaryotic Mcm2-7 replicative helicase. Sub-cell Biochem 2012; 62:113-34; PMID:22918583; http://dx.doi.org/10.1007/978-94-007-4572-8_7
- Fu Y, Slaymaker IM, Wang J, Wang G, Chen XS. The 1.8-A crystal structure of the N-terminal domain of an archaeal MCM as a right-handed filament. J Mol Biol 2014; 426:1512-23; PMID:24378617; http://dx.doi.org/10.1016/j.jmb.2013.12.025
- Slaymaker IM, Fu Y, Toso DB, Ranatunga N, Brewster A, Forsburg SL, Zhou ZH, Chen XS. Mini-chromosome maintenance complexes form a filament to remodel DNA structure and topology. Nucleic Acids Res 2013; 41:3446-56; PMID:23361460; http://dx.doi.org/10.1093/nar/gkt022
- Bae B, Chen YH, Costa A, Onesti S, Brunzelle JS, Lin Y, Cann IK, Nair SK. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure 2009; 17:211-22; PMID:19217392; http://dx.doi.org/10.1016/j.str.2008.11.010
- Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, Chen XS. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci USA 2008; 105:20191-6; PMID:19073923; http://dx.doi.org/10.1073/pnas.0808037105
- Kristensen TP, Maria Cherian R, Gray FC, MacNeill SA. The haloarchaeal MCM proteins: bioinformatic analysis and targeted mutagenesis of the beta7-beta8 and beta9-beta10 hairpin loops and conserved zinc binding domain cysteines. Front Microbiol 2014; 5:123; PMID:24723920; http://dx.doi.org/10.3389/fmicb.2014.00123
- Graham BW, Schauer GD, Leuba SH, Trakselis MA. Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res 2011; 39:6585-95; PMID:21576224; http://dx.doi.org/10.1093/nar/gkr345
- Liew LP, Bell SD. The interplay of DNA binding, ATP hydrolysis and helicase activities of the archaeal MCM helicase. Biochem J 2011; 436:409-14; PMID:21361871; http://dx.doi.org/10.1042/BJ20110084
- Sakakibara N, Kasiviswanathan R, Kelman Z. Different residues on the surface of the Methanothermobacter thermautotrophicus MCM helicase interact with single- and double-stranded DNA. Archaea 2010; 2010:505693; PMID:21151660; http://dx.doi.org/10.1155/2010/505693
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem 2002; 71:333-74; PMID:12045100; http://dx.doi.org/10.1146/annurev.biochem.71.110601.135425
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet 2007; 39:93-8; PMID:17143284; http://dx.doi.org/10.1038/ng1936
- Li XC, Schimenti JC, Tye BK. Aneuploidy and improved growth are coincident but not causal in a yeast cancer model. PLoS Biol 2009; 7:e1000161; PMID:19636358; http://dx.doi.org/10.1371/journal.pbio.1000161
- Honeycutt KA, Chen Z, Koster MI, Miers M, Nuchtern J, Hicks J, Roop DR, Shohet JM. Deregulated minichromosomal maintenance protein MCM7 contributes to oncogene driven tumorigenesis. Oncogene 2006; 25:4027-32; PMID:16518415; http://dx.doi.org/10.1038/sj.onc.1209435
- Chuang CH, Wallace MD, Abratte C, Southard T, Schimenti JC. Incremental genetic perturbations to MCM2-7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stress. PLoS Genet 2010; 6:e1001110; PMID:20838603; http://dx.doi.org/10.1371/journal.pgen.1001110
- Liang DT, Hodson JA, Forsburg SL. Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J Cell Sci 1999; 112 (Pt 4):559-67; PMID:9914167
- Simon N, Bochman ML, Seguin S, Brodsky JL, Seibel WL, Schwacha A. Ciprofloxacin is an inhibitor of the Mcm2-7 Replicative Helicase. Biosci Rep 2013; PMID:24001138
- Frick DN, Lam AM. Understanding helicases as a means of virus control. Curr Pharm Des 2006; 12:1315-38; PMID:16611118; http://dx.doi.org/10.2174/138161206776361147
- Topalis D, Andrei G, Snoeck R. The large tumor antigen: a "Swiss Army knife" protein possessing the functions required for the polyomavirus life cycle. Antiviral Res 2013; 97:122-36; PMID:23201316; http://dx.doi.org/10.1016/j.antiviral.2012.11.007
- Bergvall M, Melendy T, Archambault J. The E1 proteins. Virology 2013; 445:35-56; PMID:24029589; http://dx.doi.org/10.1016/j.virol.2013.07.020
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 2006; 442:270-5; PMID:16855583; http://dx.doi.org/10.1038/nature04943
- Sanders CM, Kovalevskiy OV, Sizov D, Lebedev AA, Isupov MN, Antson AA. Papillomavirus E1 helicase assembly maintains an asymmetric state in the absence of DNA and nucleotide cofactors. Nucleic Acids Res 2007; 35:6451-7; PMID:17881379; http://dx.doi.org/10.1093/nar/gkm705
- Kaplan DL, Davey MJ, O'Donnell M. Mcm4,6,7 uses a "pump in ring" mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J Biol Chem 2003; 278:49171-82; PMID:13679365; http://dx.doi.org/10.1074/jbc.M308074200
- Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell 2008; 31:287-93; PMID:18657510; http://dx.doi.org/10.1016/j.molcel.2008.05.020
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 2010; 37:247-58; PMID:20122406; http://dx.doi.org/10.1016/j.molcel.2009.12.030
- Ferber D. Public health. Creeping consensus on SV40 and polio vaccine. Science 2002; 298:725-7; PMID:12399560; http://dx.doi.org/10.1126/science.298.5594.725b
- White MK, Pagano JS, Khalili K. Viruses and human cancers: a long road of discovery of molecular paradigms. Clin Microbiol Rev 2014; 27:463-81; PMID:24982317; http://dx.doi.org/10.1128/CMR.00124-13
- Scheffler IE. Mitochondria. Hoboken: J. Wiley and Sons, Inc., 2008.
- McKinney EA, Oliveira MT. Replicating animal mitochondrial DNA. Genet Mol Biol 2013; 36:308-15; PMID:24130435; http://dx.doi.org/10.1590/S1415-47572013000300002
- Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem 2003; 278:48627-32; PMID:12975372; http://dx.doi.org/10.1074/jbc.M306981200
- Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. Embo J 2004; 23:2423-9; PMID:15167897; http://dx.doi.org/10.1038/sj.emboj.7600257
- Pohjoismaki JL, Williams SL, Boettger T, Goffart S, Kim J, Suomalainen A, Moraes CT, Braun T. Overexpression of Twinkle-helicase protects cardiomyocytes from genotoxic stress caused by reactive oxygen species. Proc Natl Acad Sci U S A 2013; 110:19408-13; PMID:24218554; http://dx.doi.org/10.1073/pnas.1303046110
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet 2001; 28:223-31; PMID:11431692; http://dx.doi.org/10.1038/90058
- Finsterer J, Ahting U. Mitochondrial depletion syndromes in children and adults. Can J Neur Sci 2013; 40:635-44; PMID:23968935
- Cavalier-Smith T. Archamoebae: the ancestral eukaryotes? Bio Systems 1991; 25:25-38; PMID:1854912; http://dx.doi.org/10.1016/0303-2647(91)90010-I
- Bochman ML, Sabouri N, Zakian VA. Unwinding the functions of the Pif1 family helicases. DNA Repair (Amst) 2010; 9:237-49; PMID:20097624; http://dx.doi.org/10.1016/j.dnarep.2010.01.008
- Foury F, Dyck EV. A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. Embo J 1985; 4:3525-30; PMID:16453651
- Van Dyck E, Foury F, Stillman B, Brill SJ. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J 1992; 11:3421-30; PMID:1324172
- O'Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol Cell Biol 2002; 22:4086-93; PMID:12024022; http://dx.doi.org/10.1128/MCB.22.12.4086-4093.2002
- O'Rourke TW, Doudican NA, Zhang H, Eaton JS, Doetsch PW, Shadel GS. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene 2005; 354:86-92; PMID:15907372; http://dx.doi.org/10.1016/j.gene.2005.03.031
- Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion 2007; 7:211-22; PMID:17257907; http://dx.doi.org/10.1016/j.mito.2006.11.023
- Li Z, Lindsay ME, Motyka SA, Englund PT, Wang CC. Identification of a bacterial-like HslVU protease in the mitochondria of Trypanosoma brucei and its role in mitochondrial DNA replication. PLoS Pathog 2008; 4:e1000048; PMID:18421378; http://dx.doi.org/10.1371/journal.ppat.1000048
- Liu B, Wang J, Yaffe N, Lindsay ME, Zhao Z, Zick A, Shlomai J, Englund PT. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol Cell 2009; 35:490-501; PMID:19646907; http://dx.doi.org/10.1016/j.molcel.2009.07.004
- Liu B, Wang J, Yildirir G, Englund PT. TbPIF5 is a Trypanosoma brucei mitochondrial DNA helicase involved in processing of minicircle Okazaki fragments. PLoS Pathog 2009; 5:e1000589; PMID:19779567; http://dx.doi.org/10.1371/journal.ppat.1000589
- Galletto R, Tomko EJ. Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res 2013; 41:4613-27; PMID:23446274; http://dx.doi.org/10.1093/nar/gkt117
- Boule JB, Zakian VA. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucl Acids Res 2006; 34:4147-53; PMID:16935874; http://dx.doi.org/10.1093/nar/gkl561
- McGlynn P. Helicases that underpin replication of protein-bound DNA in Escherichia coli. Bioch Soc Trans 2011; 39:606-10; PMID:21428948; http://dx.doi.org/10.1042/BST0390606
- Azvolinsky A, Giresi P, Lieb J, Zakian V. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 2009; 34:722-34; PMID:19560424; http://dx.doi.org/10.1016/j.molcel.2009.05.022
- Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011; 10:697-713; PMID:21571596; http://dx.doi.org/10.1016/j.dnarep.2011.04.028
- Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem 2014; PMID:24606147
- Bochman ML, Paeschke K, Chan A, Zakian VA. Hrq1, a homolog of the human RecQ4 helicase, acts catalytically and structurally to promote genome integrity. Cell Rep 2014; 6:346-56; PMID:24440721; http://dx.doi.org/10.1016/j.celrep.2013.12.037
- Barea F, Tessaro S, Bonatto D. In silico analyses of a new group of fungal and plant RecQ4-homologous proteins. Comput Biol Chem 2008; 32:349-58; PMID:18701350; http://dx.doi.org/10.1016/j.compbiolchem.2008.07.005
- Groocock LM, Prudden J, Perry JJ, Boddy MN. The RecQ4 orthologue Hrq1 is critical for DNA interstrand cross-link repair and genome stability in fission yeast. Mol Cell Biol 2012; 32:276-87; PMID:22064477; http://dx.doi.org/10.1128/MCB.06184-11
- Mandell JG, Goodrich KJ, Bahler J, Cech TR. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J Biol Chem 2005; 280:5249-57; PMID:15591066; http://dx.doi.org/10.1074/jbc.M412756200
- Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr, Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol 2010; 30:1382-96; PMID:20065033; http://dx.doi.org/10.1128/MCB.01290-09
- Capp C, Wu J, Hsieh TS. RecQ4: the second replicative helicase? Crit Rev Biochem Mol Biol 2010; 45:233-42; PMID:20429771; http://dx.doi.org/10.3109/10409231003786086
- Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res 2012; 40:7358-67; PMID:22576367; http://dx.doi.org/10.1093/nar/gks407
- Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner Syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell 2004; 14:763-74; PMID:15200954; http://dx.doi.org/10.1016/j.molcel.2004.05.023
- Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, Liu Y, Bohr VA. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J Biol Chem 2012; 287:196-209; PMID:22039056; http://dx.doi.org/10.1074/jbc.M111.295063
- Grierson PM, Acharya S, Groden J. Collaborating functions of BLM and DNA topoisomerase I in regulating human rDNA transcription. Mutat Res 2013; 743-744:89-96; PMID:23261817
- Shiratori M, Suzuki T, Itoh C, Goto M, Furuichi Y, Matsumoto T. WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex. Oncogene 2002; 21:2447-54; PMID:11971179; http://dx.doi.org/10.1038/sj.onc.1205334
- Islam MN, Fox D, 3rd, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms–interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol 2010; 30:2460-72; PMID:20231364; http://dx.doi.org/10.1128/MCB.01583-09
- Croteau DL, Rossi ML, Canugovi C, Tian J, Sykora P, Ramamoorthy M, Wang ZM, Singh DK, Akbari M, Kasiviswanathan R, et al. RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell 2012; 11:456-66; PMID:22296597; http://dx.doi.org/10.1111/j.1474-9726.2012.00803.x
- Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet 2010; 44:393-417; PMID:21047263; http://dx.doi.org/10.1146/annurev-genet-102209-163602
- Bugreev DV, Brosh RM, Jr, Mazin AV. RECQ1 possesses DNA branch migration activity. J Biol Chem 2008; 283:20231-42; PMID:18495662; http://dx.doi.org/10.1074/jbc.M801582200
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev 2007; 21:3085-94; PMID:18003860; http://dx.doi.org/10.1101/gad.1609007
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 2007; 21:3073-84; PMID:18003859; http://dx.doi.org/10.1101/gad.1609107
- Bochman ML, Judge CP, Zakian VA. The Pif1 family in prokaryotes: what are our helicases doing in your bacteria? Mol Biol Cell 2011; 22:1955-9; PMID:21670310; http://dx.doi.org/10.1091/mbc.E11-01-0045
- Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 1994; 76:145-55; PMID:8287473; http://dx.doi.org/10.1016/0092-8674(94)90179-1
- Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol 2006; 26:2490-500; PMID:16537895; http://dx.doi.org/10.1128/MCB.26.7.2490-2500.2006
- Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem 2009; 284:25170-80; PMID:19605347; http://dx.doi.org/10.1074/jbc.M109.023325
- Ivessa AS, Zhou J-Q, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 2000; 100:479-89; PMID:10693764; http://dx.doi.org/10.1016/S0092-8674(00)80683-2
- Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013; PMID:23657261
- Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife 2014; 3:e02190; PMID:24843019
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 2013; 502:393-6; PMID:24025768; http://dx.doi.org/10.1038/nature12585
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 2013; 502:389-92; PMID:24025772; http://dx.doi.org/10.1038/nature12584
- Chisholm KM, Aubert SD, Freese KP, Zakian VA, King MC, Welcsh PL. A genomewide screen for suppressors of Alu-mediated rearrangements reveals a role for PIF1. PLoS One 2012; 7:e30748; PMID:22347400; http://dx.doi.org/10.1371/journal.pone.0030748
- van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability, and human genetic disease. Annu Rev Genomics Hum Genet 2000; 1:409-59; PMID:11701636; http://dx.doi.org/10.1146/annurev.genom.1.1.409
- Suhasini AN, Brosh RM, Jr. Disease-causing missense mutations in human DNA helicase disorders. Mutat Res 2013; 752:138-52; PMID:23276657; http://dx.doi.org/10.1016/j.mrrev.2012.12.004
- Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 2012; 26:1393-408; PMID:22751496; http://dx.doi.org/10.1101/gad.195248.112
- Capo-Chichi JM, Bharti SK, Sommers JA, Yammine T, Chouery E, Patry L, Rouleau GA, Samuels ME, Hamdan FF, Michaud JL, et al. Identification and biochemical characterization of a novel mutation in DDX11 causing Warsaw breakage syndrome. Hum Mutat 2013; 34:103-7; PMID:23033317; http://dx.doi.org/10.1002/humu.22226
- Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I. Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet 2013; 92:448-53; PMID:23453664; http://dx.doi.org/10.1016/j.ajhg.2013.02.001
- Le Guen T, Jullien L, Touzot F, Schertzer M, Gaillard L, Perderiset M, Carpentier W, Nitschke P, Picard C, Couillault G, et al. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum Mol Genet 2013; 22:3239-49; PMID:23591994; http://dx.doi.org/10.1093/hmg/ddt178
- Egly JM, Coin F. A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair (Amst) 2011; 10:714-21; PMID:21592869; http://dx.doi.org/10.1016/j.dnarep.2011.04.021
- Liu Y. Rothmund-Thomson syndrome helicase, RECQ4: On the crossroad between DNA replication and repair. DNA Repair (Amst) 2010; 9:325-30; PMID:20096650; http://dx.doi.org/10.1016/j.dnarep.2010.01.006
- Wu Y, Shin-ya K, Brosh RM, Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol 2008; 28:4116-28; PMID:18426915; http://dx.doi.org/10.1128/MCB.02210-07
- Deans AJ, West SC. FANCM connects the genome instability disorders Bloom's Syndrome and Fanconi Anemia. Mol Cell 2009; 36:943-53; PMID:20064461; http://dx.doi.org/10.1038/embor.2008.221
