Abstract
A programmed cell death library based on the Gaussia luciferase protein-fragment complementation assay (GLuc PCA) enables detection of protein–protein interactions (PPI) within the cell death network and quantitative assessments of these interactions. Among future applications for the GLuc PCA cell death library is its potential use as a platform for PPI-targeted drug screening.
Abbreviations
| DAPK2 | = | death-associated protein kinase 2 |
| DAPs | = | death associated proteins |
| GLuc PCA | = | Gaussia luciferase protein-fragment complementation assay |
| PPI | = | protein-protein interaction |
In the past, our conception of programmed cell death was mostly limited to apoptosis. However, it is now becoming clear that other pathways such as programmed necrosis and autophagy are also involved in the cell's life-or-death decisions. Although many points of interface between these pathways have been uncovered in recent years,Citation1 the crosstalk between the different cell death pathways and the backup relationships between them is not completely understood.
Protein–protein interactions play a major role in cell signaling pathways. Environmental and intracellular signals are constantly being collected and converted to a variety of post-translational modifications, including phosphorylations, protein cleavage, and ubiquitinations. These in turn converge on spatial or temporal changes in protein–protein interactions. In the context of programmed cell death, key steps in apoptosis and autophagy and in the crosstalk between these pathways are regulated by protein–protein interactions and are dependent on protein complexes.Citation2
In a recent paper in Cell Reports,Citation3 we describe a programmed cell death GLuc PCA library that enables the discovery and quantitative measurement of protein–protein interactions within the cell death network. The library is based on the Gaussia luciferase protein-fragment complementation assay (PCA) reporter, which enables quantitative measurement of protein–protein interactions in a reversible manner.Citation4 The library encompasses 63 proteins, including apoptosis proteins, autophagy proteins, death-associated proteins (DAPs), and additional cell death regulatory proteins.
By performing an unbiased screen on all possible protein–protein interactions in the library, we managed to detect most of the core machinery interactions of both autophagy and apoptosis and, in addition, uncovered 46 previously unknown interactions. Some of these reflected novel points of interface between apoptosis and autophagy. One of the novel interactions identified in this work, between Death Associated Protein Kinase 2 (DAPK2, also named DRP-1) and 14–3–3τ, was further functionally investigated at both biochemical and cellular levels. We showed that 14–3–3 specifically binds to a Ser-rich region in the C-terminal tail of DAPK2 and thus inhibits its dimerization and activation. Furthermore, we have identified 5 more interacting partners for DAPK2 that may provide clues to its role in autophagy regulation. This proof-of-concept emphasizes the power of the GLuc PCA cell death library for the discovery and characterization of novel protein–protein interactions within the cell death network. Notably, this work was performed under basal growth conditions. One of the future challenges will be to apply different external triggers to this system and test to what extent and how these triggers change the proteomic landscape.
Currently, we forecast several exciting future applications for the GLuc PCA cell death library . As a result of the rapid development of high-throughput functional screening, researchers into cell death face an increasing number of genes that are positively or negatively linked to programmed cell death, and especially autophagy, which is a younger branch of the field.Citation5,6 The bottleneck, however, remains in the annotation of these genes to the pathway's core machinery and identification of their mode of action. Our GLuc PCA library can be used as a complementary tool for functional screening as it enables high-throughput screening of protein–protein interactions. As such, functionally associated genes can be screened against the library in order to identify potential protein partners that mediate their function.
A second application relates to drug screening. The expansion of our understanding of the cell death network and the interconnectivity of the different cell death pathways enables the identification of specific protein–protein interactions as potential targets for drug development. Some examples, such as BH3-mimetic and SMAC-mimetic compounds, already show promising results by triggering apoptosis in cancer cells.Citation7 Specific autophagy inhibitors are also of great interest, as inhibition of autophagy was shown to enhance the cellular response to chemotherapy.Citation8 The GLuc PCA reporters detected in our screen can be easily used as quantitative and cost-effective readouts for novel drug discovery. This approach has several advantages over using a general pathway marker as readout (e.g., caspase activation in apoptosis or MAP1LC3 lipidation in autophagy). First, since a specific pair of protein interactions can be chosen as a target it is easier to understand the drug's mode of action, and the chance for hidden off-target effects is minimized. Second, it enhances the screening sensitivity and enables the detection of drugs with milder effects that might not influence the overall process alone but can be used in combinations with other drugs to achieve a synergistic effect.
The GLuc PCA library can help to identify subtle differences between family members. The mammalian apoptosis and autophagy pathways include several protein families whose members display a high degree of similarity in protein structures and functional roles. The Bcl-2 family proteins, inhibitor of apoptosis proteins (IAPs), and the multiple mammalian homologues of the yeastAtg8 (usually referred to as LC3 paralogues) are examples of such protein groups. Understanding the difference/similarity among those family members and quantitative mapping of their interacting partners is highly important in order to understand whether they are fully redundant or display complementary functions in these biological processes. The quantitative information that may result from these experiments can also be used as a powerful resource for computational modeling of the cell death network. Thus, it should be possible to use the GLuc PCA reporters to improve our understanding of the specified roles of each member in these protein families.
Overall, the approach and methodologies discussed above can be extended to other biological processes. Thus, other pathway-based GLuc libraries could also be used as powerful tools in molecular biology.
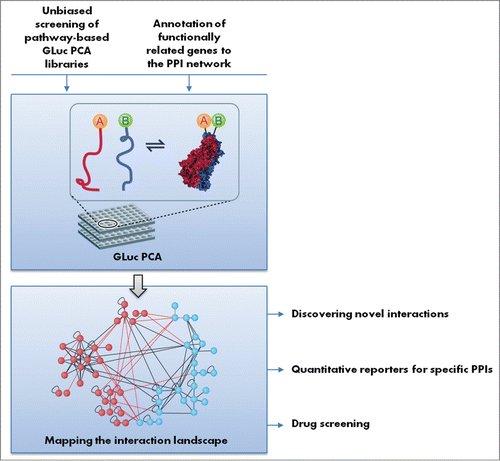
Figure 1. Possible applications of the programmed cell death GLuc PCA library. Functionally related genes or pathway-based libraries can be screened against the library for protein–protein interaction (PPI) mapping. Detected interactions can be further developed as quantitative reporters for specific PPIs or can be used as readouts for drug screening.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
References
- Amaravadi R K, Lippincott-Schwartz J, Yin X M, Weiss W A, Takebe N, Timmer W, Dipaola R S, Lotze M T, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 2011; 17:654-66; PMID:21325294; http://dx.doi.org/10.1158/1078-0432.CCR-10-2634
- Bai L and Wang S. Targeting apoptosis pathways for new cancer therapeutics. Annu Rev Med 2014; 65:139-55; PMID:24188661; http://dx.doi.org/10.1146/annurev-med-010713-141310
- Bialik S, Zalckvar E, Ber Y, Rubinstein A D, Kimchi A. Systems biology analysis of programmed cell death. Trends Biochem Sci 2010; 35:556-64; PMID:20537543; http://dx.doi.org/10.1016/j.tibs.2010.04.008
- Gilad Y, Shiloh R, Ber Y, Bialik S, Kimchi A. Discovering protein-protein interactions within the programmed cell death network using a protein-fragment complementation screen. Cell Rep; 2014; 8:909-21; PMID:25066129; http://dx.doi.org/10.1016/j.celrep.2014.06.049
- Lipinski M M, Hoffman G, NG A, Zhou W, PY B F, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell 2010; 18:1041-52; PMID:20627085; http://dx.doi.org/10.1016/j.devcel.2010.05.005
- Marino G, Niso-santano M, Baehrecke E H, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15:81-94; PMID:24401948; http://dx.doi.org/10.1038/nrm3735
- Orvedahl A, Sumpter R JR, Xiao G, NG, A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 2011; 480:113-7; PMID:22020285; http://dx.doi.org/10.1038/nature10546
- Remy I, Michnick S W. A highly sensitive protein-protein interaction assay based on gaussia luciferase. Nat Methods 2006; 3:977-9; PMID:17099704; http://dx.doi.org/10.1038/nmeth979
