Abstract
G6PDH is the rate-limiting enzyme of the pentose phosphate pathway and one of the principal source of NADPH, a major cellular reductant. Importantly, in ruminant's metabolism the aforementioned NADPH provided, is utilized for de novo fatty acid synthesis. Previous work of cloning the ovine (Ovis aries) og6pdh gene has revealed the presence of two cDNA transcripts (og6pda and og6pdb), og6pdb being a product of alternative splicing not similar to any other previously reported.1 In the current study the effect of energy balance in the ovine G6PDH protein expression was investigated, shedding light on the biochemical features and potential physiological role of the oG6PDB isoform. Changes in energy balance leads to protein expression changes in both transcripts, to the opposite direction and not in a proportional way. Negative energy balance was not in favor of the presence of any particular isoform, while both protein expression levels were not significantly different (P > 0.05). In contrast, at the transition point from negative to positive and on the positive energy balance, there is a significant increase of oG6PDA compared with oG6PDB protein expression (P < 0.001). Both oG6PDH protein isoforms changed significantly toward the positive energy balance. oG6PDA is escalating, while oG6PDB is falling, under the same stimulus (positive energy balance alteration). This change is also positively associated with increasing levels in enzyme activity, 4 weeks post-weaning in ewes’ adipose tissue. Furthermore, regression analysis clearly demonstrated the linear correlation of both proteins in response to the WPW, while energy balance, enzyme activity, and oG6PDA relative protein expression follow the same escalating trend; in contrast, oG6PDB relative protein expression falls in time, similar to both transcripts accumulation pattern, as reported previously.2
Keywords: :
Introduction
Studying the biology of adipose tissue in farm animals and acquiring the knowledge of animal production and health, we may comprehend better the significant resemblances in lipid physiology with emphasis in lipogenesis. There are species-based biases that occur, not only in adipose tissue features but also in the metabolism, as a whole. In all species studied so far, g6pdh is considered as an X-linked gene displaying a “housekeeping” profile, while several variants exist.Citation1,Citation2 Having mentioned these biases in the metabolic pathways, lipogenesis unfolds the hidden selective pressure that nature reveals by the adaptive mechanisms that led ruminants to divert from other herbivorous mammals in order to survive, share mutually natural resources and interact with the other species, as well.
Focusing on the aforementioned metabolic pathway and specifically the biosynthesis of fatty acids starting from acetyl-CoA, there are certainly considerable amounts of reducing agents, like NADPH, which are required for the reductive pathway and leading to fatty acid synthesis. Major important enzymes linked to NADPH production are: cytosolic isocitrate dehydrogenase (IDH), EC 1.1.1.42, cytosolic NADP malic enzyme, EC1.1.1.40, enzyme of pentose phosphate shunt, glucose-6-phosphate dehydrogenase (G6PDH), EC 1.1.1.49, and 6-phosphogluconate dehydrogenase, (6PGDH), EC1.1.1.44. The last two enzymes are the main NADPH suppliers in ruminants, providing 50−80% of the total required NADPH for the fatty acid synthesis.Citation3
G6PDH is a cytosolic enzyme encoded by a housekeeping X-linked gene whose main function is to produce NADPH, a key electron donor in the defense against oxidizing agents and in reductive biosynthetic reactions. It is conventionally referred to, as the rate limiting enzyme of the pentose phosphate pathway in all cells,Citation4 and it is therefore regarded as important in the biosynthesis of the sugar moiety of nucleic acids. In addition, G6PDH apart from its role as NADPH “generator”, the control of key enzymes modulating tissue response, such as 11beta-hydroxysteroid dehydrogenase-1 (11β-HSD-1) that is highly expressed in adipose tissue, liver, and brain, is an additional role.Citation5 In the aforementioned tissue, G6PDH acts primarily as an oxo-reductase to generate active glucocorticoid placed in the endoplasmic reticulum (ER).Citation5
As far as the lipogenic activity of G6PDH is concerned, it has been demonstrated that hepatic G6PDH is regulated by nutritional signals, including high-carbohydrate diet, polyunsaturated fatty acids, and hormonal signals such as insulin, glucagon, thyroid, and glucocorticoids but is also participating in the reductive biosynthesis of fatty acids and cholesterol.Citation6 Previous research reports that enzymatic activity and expression levels of G6PDH were significantly elevated in, with pleiotropic role, white adipose tissue of obese models, and diet-induced obesity mice.Citation7 It is well known in obesity, that adipose tissue exhibits increased oxidative stress, which is a major contributor to metabolic disorders such as insulin resistance, playing also a central role in nutrient sensing and energy homeostasis.
Concerning ovine g6pdh, several studies have been conducted on enzymatic level showing the reaction of enzyme activity to different stimuli.Citation8-Citation10 At the molecular level, g6pdh has been well studied in many eutherian species. The g6pdh gene, is remarkable for its genetic diversity and there are many variants of g6pdh, mostly produced from missense mutations, that have been described with wide range levels of enzyme activity, while in humans are associated with clinical symptoms.Citation11 Complete characterized sequences have been already published for human, mouse and rat genes,Citation12-Citation15 while many studies have shown the regulation of gene expression in respect to various stimuli.Citation2 G6PDH is an ubiquitously expressed molecule and glucose-6-phosphate dehydrogenase enzyme activity is in accordance with the energy uptake.Citation2
Previous work in our laboratory, has reported the ovine g6pdh gene cloning and characterization,Citation16-Citation18 elucidating the presence of two differentially expressed ovine g6pdh transcripts (og6pda and og6pdb). The og6pdb gene product is the result of a splicing event leading to a transcript with 27 extra nucleotides, in total. This extra segment causes a frameshift in the polypeptide chain resulting in changes around the area of the substrate binding site as an enlarged binding “pocket”. It has been hypothesized that the oG6PDB transcript could probably result in a low G6PDH activity,Citation16 as analogous events have been previously reported in sheep.Citation19,Citation20 Thus, the presence of a novel alternative transcript of ovine g6pdh gene, mirrors a potentially novel role in lipid metabolism. Another published workCitation21 from our lab has studied biochemical parameters for metabolic enzymes involved in lipogenesis in ewes from the same breed and at a similar lactation stage, as in this work. That report has shown the effects of energy balance on a few biochemical parameters like insulin, free fatty acids, and β-hydroxybutyric acid concentrations.Citation21 In detail, lower values for insulin, higher levels of growth hormone (GH) and increased rates for the latter two acids were observed at the negative energy balance in the period of the first 2 weeks of lactation, while this period lasted only 5 days in Karagouniko breed. Citation21 These results share similarities with this work, concerning the negative energy balance “dominant” protein isoform, oG6PDB. As in our work, we had intense interest to investigate the mechanism of og6pdh gene regulation upon energy balance changes. We conducted our research at the translational level, to identify the putative protein oG6PDH isoforms.
In order to explore the above role, our research deal with the effect of dietary energy level in ovine G6PDH protein isoform expression, to elucidate the presence of the two ovine G6PDH protein isoforms, the pattern of expression of both protein isoforms as influenced by the energy balance in the course of time post weaning, and the potential physiological role of og6pda/b gene products. Evidence from other species for G6PDH variants in electrophoretically separated protein extractsCitation22,Citation23 was the aspiration in our research, while the pertinent investigation line was lacking in ruminants since the discovery of the second og6pdb transcript.Citation16 Our findings not only shed light and confirm similarities and differences in both, og6pdh transcript accumulationCitation24 and oG6PDH isoform protein expression, in respect to the energy balance changes, but also emphasize the significance of the alternative splicing g6pdb product in the regulation of g6pdh gene expression and oG6PDH enzyme activity.
Results
Results from our immunoblot analysis shown in , were further analyzed by quantitative densitometry and generated the mean values in . In our graph we summarize the data from oG6PDA and oG6PDB protein expression variations as a function of the energy balance in ewes’ adipose tissue. As reported elsewhere by the authors,Citation24 the average milk production as a yield was not statistically different among the week groups post-weaning. Nevertheless, the energy balance changed at the end of first, second, and fourth WPW, i.e., −0.50 ± 0.24 MJ, −0.19 ± 0.14 MJ, and 4.48 ± 0.26 MJ respectively,Citation24 with significant increase toward the fourth WPW.
Figure 1. Representative results of immunoblot analysis of protein extracts of tail-biopsies of animals fed on (1) negative; (2) transit to positive; and (3) positive energy balance. Twenty-five micrograms of protein were loaded on a 8–16% gradient polyacrylamide gel and dry-blotted on PVDF membrane and further incubated with primary G6PDH-specific and HRP-secondary antibody (see Materials and Methods). Samples numbers correspond to the following samples: (1) first WPW; (2) second WPW; and (3) fourth WPW. Protein expression of oG6PDA and oG6PDB was altered through the course of time and the levels of β-actin protein are shown below in the immunoblot. oG6PDH protein isoform expression was normalized to β-actin protein.
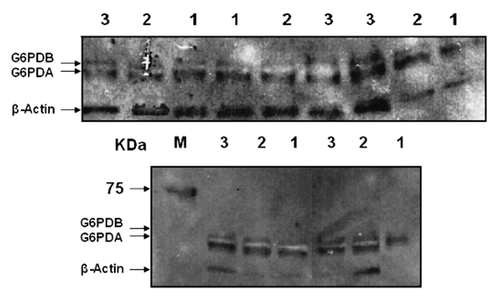
Figure 2. Average relative expression of the oG6PDH protein isoforms in the ovine adipose tissue. (A) Average relative expression of the oG6PDA and oG6PDB protein. Relative amounts of protein were determined in each sample with β-actin protein as the housekeeping gene product and endogenous control. Each sample was analyzed from each biopsy consisted of 7 animals. (B) Average relative expression of oG6PDA and oG6PDB proteins in the ovine adipose tissue. Data were merged in 2 groups according to their negative or positive energy balance. oG6PDA and oG6PDB protein expression vs. negative or positive energy balance. Relative amounts of protein were determined in each sample and β-actin protein as the endogenous control. Values are in arbitrary units (mean ± SEM, n = 6, P > 0.05, ns, not significant).
Our densitometry data in and show the relative protein expression of oG6PDA and oG6PDB isoform, where energy balance switch from the negative (first and second week) to the positive (fourth week) giving rise to a concomitant trend in protein expression of G6PDA. Specifically, oG6PDA protein isoform rises from 1.052 ± 0.071 (first week) and 1.265 ± 0.209 (second week) to 1.596 ± 0.1982 (fourth week) with significant changes between first and fourth week (P < 0.05). On the contrary, oG6PDB relative protein expression shows a more significant decline which is observed with more significant changes (P < 0.01) between the first and second WPW from 1.011 ± 0.096 and 0.548 ± 0.099 respectively, to the fourth week (0.528 ± 0.058), compared with the oG6PDA relative protein expression levels.
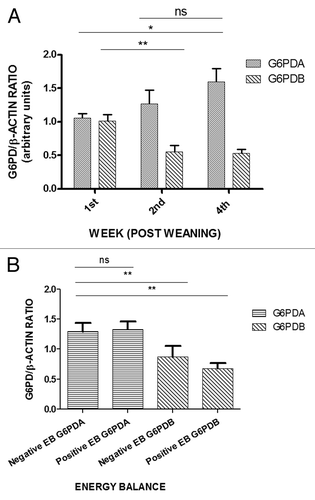
Figure 3. (A) Left-side graphs demonstrate the average relative expression of oG6PDA and oG6PDB proteins in the ovine adipose tissue, compared with the right-side graph of oG6PDH enzyme-specific activity (nkat) vs. WPW. One way ANOVA vs first WPW was performed to evaluate significant changes followed by the Bonferroni multiple comparison test. Values are in arbitrary units (mean ± SEM, *P < 0.05, **P < 0.01, ns = not significant). (B) Average relative expression of the oG6PDH protein isoforms in the ovine adipose tissue vs oG6PDH enzyme activity. x–y graph was plotted from mean ± SEM in every point. Points I, II, and IV indicate first, second WPW, and fourth WPW, respectively. Points II and IV show statistically significant differences between the two isoforms. M symbol is the cross-section of the two curves, where oG6PDA and oG6PDB has equal relative protein expression, and enzyme activity starts at 7.256 ± 3.040 (nkat). The values from the curve are relative expression (arbitrary units/nkat) (mean ± SEM, n = 6, **P < 0.01, ***P < 0.001).
Importantly, the calculated oG6PDA/oG6PDB ratio from in protein expression between these two protein isoforms shows an increase in the course of time (P < 0.01, P < 0.001) from the first week (1.077 ± 0.0610) and second week (2.515 ± 0.467) to the fourth week (2.977 ± 0.218) (data not shown), which is consistent with the similar pattern of the g6pda/g6pdb transcript accumulation ratio, reported elsewhere.Citation24 Hence, there is a significant increase in the oG6PDA compared with the oG6PDB protein levels, and the attributed oG6PDA/oG6PDB ratio (data not shown) in the protein expression level increase is due to the smaller scale changes in G6PDB protein expression, compared with oG6PDA ( and ). Specifically, there are significant changes in both protein isoform expression focused, on one hand in oG6PDA protein isoform expression changes between the distal time points i.e., first and fourth WPW, while on the other hand, statistical analysis concerning oG6PDB protein isoform expression shows significant changes between the first and second as well as between first and fourth WPW. Interestingly, significant changes in both protein isoform expression were not observed between the second and fourth WPW ( and ).
By organizing our data according to the energy balance (absolute negative or positive values) in 2 groups, ewes in negative energy balance and ewes in positive energy balance, and following similar statistical analysis the graph in , was plotted, illustrating relative protein expression values versus the group of either negative or positive energy balance. This “intervention” was useful to understand the “net effect” of energy balance in WPW. It is clearly shown, that negative energy balance oG6PDA protein expression had an almost 2-fold increase (P < 0.01) compared with oG6PDB, i.e., (1.294 ± 0.143) compared with (0.678 ± 0.088) respectively. Equally, in the group of the positive energy balance oG6PDA protein expression had at least 2-fold increase (P < 0.01) compared with oG6PDB, i.e., (1.332 ± 0.129) compared with (0.678 ± 0.088) respectively. Nevertheless, the energy balance change did not affect significantly, either oG6PDA or oG6PDB relative protein expression (P > 0.05) (). Furthermore, the total contribution from both isoforms in oG6PD protein expression from the negative energy balance data group, to the positive, from first WPW to fourth WPW results in a fall at the protein level, nevertheless these changes between the negative and positive energy balance groups were not statistically significant for both protein expression (oG6PDA and oG6PDB), and only affected expression changes between the isoforms, as shown from the graph ().
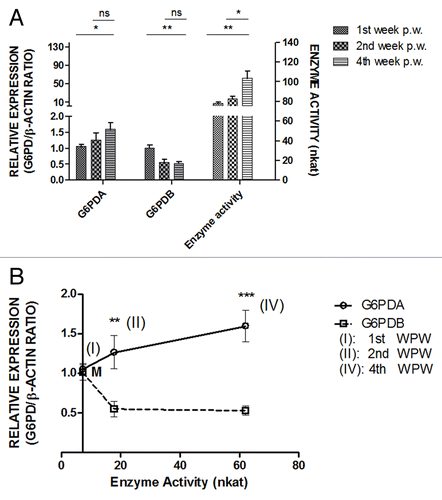
To further analyze the energy balance effect on protein expression levels, regression analysis of the data was performed as shown in . Both oG6PDA and oG6PDB relative protein expression linear regression analysis, demonstrate how far protein expression and energy balance values deviate from a putative linear relationship. The R squared values were estimated, RoG6PDA2 = 0.25 and RoG6PDB2 = 0.35 and post-hoc analysis test and best-fit values estimates for the y intercepts at x = 0.0, were aoG6PDA = 0.886 ± 0.203 and aoG6PDB = 1.021 ± 0.127, (i.e., aoG6PDB ≈aoG6PDA consistent with oG6PDBrelative expression ≈oG6PDArelative expression) while slopes as determined from linear equations were, boG6PDA = 0.178 ± 0.076 and boG6PDB = −0.139 ± 0.047 and P values (PoG6PDA < 0.05, PoG6PDB < 0.01). Interpreting these data, is clearly demonstrated the linear correlation of both proteins in response to the WPW and/or energy balance changes. In , is also shown from the graphs that the cross-section of both lines characterizes no change in protein isoform expression (equilibrium state), during the biopsy week and/or energy balance (MJ), at the protein expression level. Interestingly, from the graphs (), this cross-point is located at the relative protein expression level, ca. 1.0. Under these conditions and at approximate protein expression level, ca. 0.96, this equilibrium occurred before the end of first biopsy week (). The same cross-point also coincides with negative energy balance, where both lines meet at ca. −6.5 MJ ().
Figure 4. (A) Protein expression ratio (relative to β-actin) of oG6PDH isoforms (oG6PDA and oG6PDB) in ovine adipose tissue as a function of WPW. Linear regression analysis was performed to test the linear relationship between protein expression of protein forms of oG6PDH and energy balance. A positive oG6PDA and negative oG6PDB protein expression correlation was found in relation to the energy balance changes. The slope differences between the best-fit curves were significant (***P < 0.001). The insert depicts the equation formulae for both best-fit lines. (B) Protein expression ratio (relative to β-actin) of oG6PDH isoforms (oG6PDA and oG6PDB) in ovine adipose tissue as a function of energy balance (MJ). Linear regression analysis was performed to test the linear relationship between protein expression of protein forms of oG6PDH and energy balance. A positive oG6PDA and negative oG6PDB protein expression correlation was found in relation to the energy balance changes. The slope differences between the best-fit curves were significant (***P < 0.001). The insert depicts the equation formulae for both best-fit lines.
As far as the oG6PDH enzyme activity is concerned, shows statistically significant values increase toward the fourth WPW, where this activity reaches maximum values on the fourth WPW, as shown by the enzyme activity, nanokatal (nkat) at the right y-axis, in . also shows a comparison with both protein isoform expression levels to demonstrate the concomitant trend of oG6PDA and the contrary trend of oG6PDB relative protein expression, simultaneously with the escalating G6PDH enzyme activity in the samples.
In order to evaluate our data further and rule out issues, such as differences in increased enzyme content vs. activity, we plotted on a graph relative expression as a function of enzyme activity (). As shown in , enzyme activity starts from a minimum (ca. 6.93 nkat, first WPW), at equilibrium point where both isoforms are equally expressed (cross-point M in ), reaching a maximum in the enzyme activity, which has been detected at, ca. 62.06 nkat, fourth WPW) for both protein isoforms. At this point, where the maximum of values in oG6PDA relative protein expression is 1.596 ± 0.198, compared with the lowest values in oG6PDB relative protein expression which is 0.528 ± 0.058, in the fourth WPW, the relative protein expression per enzyme activity is 0.026 ± 0.023 and 0.008 ± 0.003 (). A positive association is shown (), for the oG6PDH enzyme activity changes (nkat) (x-axis), with enzyme activity starting from 7.256 ± 3.040, 17.82 ± 5.027, with maximum at 62.06 ± 14.985 nkat (P < 0.01), at time points I (first WPW), II (second WPW), and IV (fourth WPW), respectively (), and oG6PDA relative protein expression starting from 1.052 ± 0.071, 1.265 ± 0.210 with maximum at 1.596 ± 0.198 arbitrary units, at the same time points (). This is in contrast with oG6PDB relative protein expression (opposite trend) starting from 1.011 ± 0.096, 0.549 ± 0.099 with minimum at 0.528 ± 0.058, from the negative to the positive energy balance (first, second, and the fourth WPW, respectively) (). In both cases, the linear trend for G6PDA expression and enzyme correlation is met with good correlation coefficient (r2 = 0.954) in the range of enzyme activity values, whereas correlation coefficient for oG6PDB expression and enzyme activity is lower (r2 = 0.459). In detail, oG6PDA protein isoform expression continues to rise in a linear fashion with the enzyme activity (r2 = 0.954), while oG6PDB protein isoform expression continues to fall, reaching an almost plateau (value at ca. 0.008 ± 0.003 expression/nkat) (). Interestingly, further statistical analysis has shown, that the former curve fits well to a classic linear curve while the latter fits better to one-phase decay curve compared to the linear form estimated earlier (i.e., goodness of fit, rnonlinear2 = 0.57 > rlinear2 = 0.459) ().
So conclusively, we observed changes with the same trend, as has been shown previously for g6pda relative transcript accumulationCitation24 and oG6PDH protein expression vs. enzyme activity, as well. , is also emphasizing the fact described earlier in and that significant changes in relative protein expression were observed at the time points (WPW) where the detected enzyme activity is in the second or fourth WPW. The significant changes between the two oG6PDA and oG6PDB isoform protein expression, were observed between the second and fourth WPW, and not at point I (first WPW) where enzyme activity at its lowest value (7.256 ± 3.040) (). Interestingly, the distance of these expression curves after the second WPW, tend to a parallel direction reaching their maximum difference in relative protein isoform expression with the same enzyme activity (ca. 62 nkat), i.e. maximum for oG6PDA and minimum for oG6PDB (1.596 ± 0.198 vs 0.528 ± 0.0585 respectively).
Discussion
Previous studies have reported the cloning and molecular characterization of the ovine g6pdh gene as well as its promoter region.Citation16-Citation18 These studies have analyzed and confirmed the presence of two og6pdh transcripts at the cDNA level. These alternative spliced og6pdh gene products, i.e., the expressed protein isoforms as shown in our data, accumulate evidence on the disparate physiological role of these transcripts. By studying the influence of the energy balance alterations on the og6pdh transcript accumulationCitation24 and protein levels, these patterns of expression could shed light on the translational control of the oG6PD protein product(s) and further elucidate the potential physiological role of the recently identified second (g6pdb) alternative transcript.Citation16,Citation24
As, shown from our protein analysis and immunodetection results, on both transcript products i.e., oG6PDA and oG6PDB, there is a concomitant rise in the accumulation of these proteins on the first, and a falling pattern on the second protein respectively, following the transition from negative to positive energy balance, in the adipose tissue of the ewes. This change is explained from the fact, that animals fed with high carbohydrate diet are led to higher energy balance, i.e., high energy excess and positive energy balance, higher enzyme activities in the lipogenesis, such as fatty acid synthase, acetyl CoA carboxylase, malic enzyme, and oG6PDH expression levels, compared with the previous lower energy state. All the aforementioned factors are coordinated depending on the energy demands at a certain time, resulting to the enhancement or attenuation of the final g6pdh gene product and finally its protein synthesis. Previous work regarding the aforementioned changes at the g6pdh mRNA and protein level has been extensively reported.Citation2,Citation25
Published work has shown that G6PDH-deficient mice had increased oxidative stress, decreased NADPH levels, decreased glutathione reduced form (GSH) levels, as well as increased markers of lipid peroxidation.Citation26 In the above oxidative reactions glutathione contribution is rate-limiting depending on cysteine, which is the rate-limiting factor in cellular glutathione synthesis.Citation27 G6PDH is also essential for embryo development, as the complete knockout of g6pdh gene was embryonically lethal.Citation28,Citation29
In our data, the observed values i.e., the rising trend in protein expression of oG6PDA from the negative to the positive energy balance, is concomitant with the other estimated parameters. Hence, is directly related to oG6PDA transcription level,Citation24 protein expression and total enzyme activity ( and ). In addition to that, og6pdb alternative transcript accumulation,Citation24 as well as protein expression, show similar but opposite to og6pda transcript trend ( and ); however the oG6PDA protein preponderance over oG6PDB protein in the course of the WPW is apparent.
Previous work in our lab has reported another study for the g6pda/b transcripts,Citation24 which in comparison with our data at the protein level, suggest that og6pdh gene transcript accumulation is more abundant and stable, in terms of producing mRNA, than total oG6PDH protein, while this is also more intense for oG6PDB isoform.Citation24 This difference with the previous report is also apparent from our and , where the max x-fold levels of protein expressed were below 2x-fold for oG6PDA and almost 0.5x-fold for oG6PDB protein expression during the fourth WPW, compared with 3x-fold and 1.5x-fold expression in relative transcript levels for og6pdh gene respectively.Citation24 Therefore, estimating from the above observations, the relative protein expression to the transcript accumulation ratio is around ~66% for oG6PDA and ~33% for oG6PDB, suggesting that the level of expressed proteins are marginally correlated with the level of mRNA transcripts. This is not unusual, as changes in gene expression level are frequently not reflected at the protein level. The current gene regulatory events are not known and require further investigation, nevertheless both transcripts/proteins share equal importance. This intensive transcription yielding lower oG6PDH isoform protein levels could be investigated further with more promising scrutiny, by microarray technology with studies of functional regulation that could, in principle, be extended to detect not only the changes in the overall expression of a gene, but also changes in its splicing pattern between different tissues. Interestingly, the results from the translational events in these two transcripts reveal a differential regulation, as responses and roles between them in adipose cells apparently differ. In fact, alternative splicing events consist of a control mechanism in gene regulation, enzyme activity, and function, which generally adapt the functional enzyme production according to the metabolic conditions, in time. It is very likely, that this is applied in oG6PDH protein expression and enzyme activity where the functional oG6PDH protein is reflected on the ratio of these isoforms, as mentioned earlier.
As shown from the graphs ( and ), the 2 og6pdh gene transcripts and its products are functioning in two opposite directions to supply the cell with the required oG6PDH protein isoform levels and enzyme activity. Moreover, the ratio obtained by calculating the relative protein expression of the two isoforms, i.e., oG6PDA/oG6PDB ratio (data not shown), show an upward trend from the negative to the positive energy balance, due to the greater protein level changes in favor of oG6PDA. These data become more apparent as presented in . By this graph, we interested to show what the correlation is between protein expression for these two protein isoforms, as detected by the immunoblots, and the enzyme activity, in order to rule out differences in increased enzyme content vs. activity. As shown from , it is possible to have no change in enzyme content (plateau area in these curves), but measurable changes in activity, as the number of protein molecules remain invariable, but enzyme activity reaches a maximum, modulated possibly by negative feedback of the isoforms themselves (). Importantly, oG6PDB isoform expression is the lowest in terms of protein expression for the maximum measured enzyme activity in return. It seems that at the positive energy balance until the fourth WPW, less oG6PDB protein is required in order oG6PDH enzyme activity remains unaffected fulfilling its role in the adipose cell, simultaneously with oG6PDA protein isoform (), We cannot estimate activation/inhibitory sites on these oG6PDH enzyme activities and the possible mechanism at this stage, or the enzyme localization in terms of physiological significance. Although, we may assume that the changes in enzyme activity (possibly due to, either increase in the number of activation sites in oG6PDA isoform or blend with molecules of oG6PDB as this could result in lower oG6PDH activityCitation16,Citation19,Citation20) could be also due to posttranslational modifications, and the dominant protein isoform (oG6PDA) at the maximum level, is “rendered” by these modifications more active and remain possibly the major oG6PDH activated enzyme at the positive energy balance. Interestingly, the estimated intercepts of both protein expression (boG6PDA and boG6PDB) show, that expressing oG6PDA protein there is a ca. 1.30x-fold more direct response, as an effect of the energy balance changes, than oG6PDB protein isoform (). This point will be further elucidated by experimenting on the individual enzyme affinities of the two isoforms for their substrates, or ideally by producing their recombinants and further scrutinize their kinetic parameters.
oG6PDH enzyme regulation and activity along with the energy balance, is functionally linked to the protein components and expression levels of the total oG6PDH in terms of protein and enzyme activity. Although, we do not know the mechanism in this activity and the link with these protein components, nevertheless this is reminiscent of a balanced protein regulation from a more or less “stable” pool of the oG6PDH protein from various deposited resources, reserved to be channeled to produce the enzyme and its activity on demand of energy requirement.Citation30 Thus, at low glucose, or energy, the flux of G6PDH for ATP generation to the cytosol, with cycling, is adjusted by the protein levels and the isoform in favor, in order to maintain a threshold redox state of NADPH. When higher levels of energy is provided through the diet, there is an increased flux through cytosolic oG6PDH enzyme, while adipose tissue provides sufficient G6PDH for both ATP synthesis rates and cytosolic NADPH generation, which is allosterically stimulated by NAPD+ to increased rates of ATP and NADPH synthesis.Citation30 As shown in , the equilibrium point (cross-point), where both oG6PDA and oG6PDB were equally expressed in the first WPW (ca. third day post-weaning, the relative expression of the two isoforms appeared at the negative values of the curve and energy balance (tick point −6.69 MJ), which is interpreted as less energy balance input and consumption than what is required. Therefore, under these conditions and taking into account the almost linear relationship (), these results imply a starvation regime for the organism, where beyond the cross-point and toward the negative energy balance values, an active role is undertaken by oG6PDB. We cannot predict at this energy stage, if oG6PDB is a potential “regulator” of the other protein isoform (oG6PDA) or if both are regulated by the same control mechanism. Under this mechanism, a negative control feature emerges in oG6PDH activity to protect homeostasis, restoring the necessary enzyme activity and keep an equilibrium between lipogenetic and lipolytic processes, as dictated by the energy balance. The energy balance dependence, also reveals a fine tuning feature with a more sophisticated and complex functional control of G6PDH enzyme(s).
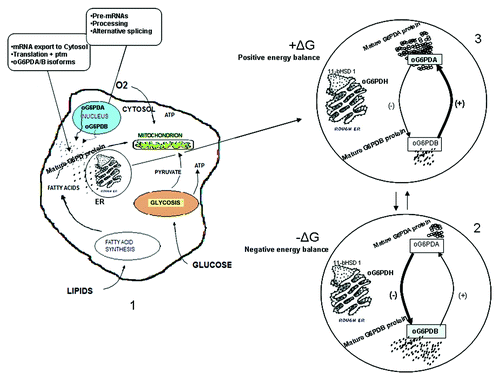
In the model that shaped our thoughts, we show graphically this interaction in detail, as depicted in , for the role of oG6PDH transcripts in energy metabolism. Low energy balance diminishes og6pda transcript production in favor of og6pdb. The same trend is followed for the respective protein isoforms (oG6PDA and oG6PDB). There is a contrary effect exerted by the positive energy balance on the transcript accumulation and protein isoforms, i.e., this favors og6pda gene product. Briefly, upon energy balance changes, more molecules are recruited from the favored protein isoform to supply the enzyme demands, at one time (see circles 2 and 3 in ). Furthermore, the og6pdh gene products seem to have more than one roles to serve, briefly, on one hand in providing NADPH for cytosolic synthesis of fatty acids etc., and on the other hand to regulate key enzymes modulating responses such as 11beta-hydroxysteroid dehydrogenase 1 (11β-HSD1) in adipose tissue, localized in the endoplasmic reticulum (ER) besides oG6PDH, as shown in , revealing a multi-functional profile for this metabolic enzyme.
Figure 5. Theoretical model of the role of oG6PDH transcripts in energy metabolism. The picture depicts the influence of energy balance on ovine G6PDH mRNA and protein isoforms. Negative energy balance has inversely proportional effect on oG6PDA mRNA accumulation and protein isoform. In contrast, positive energy balance has a positive effect on oG6PDA mRNA and negative effect on oG6PDB mRNA accumulation and protein isoform. (1) State of the cell in tissues producing and secreting both transcripts into the cytoplasm. The mRNA molecules produced after processing and alternative splicing, export to the cytosol and translation follows. Each isoform production, depends on the (−) or (+) energy balance. (2 and 3) Negative and positive energy balance favors either oG6PDB or oG6PDA mRNA and protein isoform. The molecules of oG6PDH (° or ”) as depicted in the circles (2 and 3), are for simplicity only (see Discussion). Thick lines and signs show the favored transcript/protein isoform production upon the energy balance state; ptm, posttranslational modifications; ± ΔG, ± energy balance.
In the disease front, diabetes research has shown that inhibition of G6PDH can be used to inhibit insulin resistance in obese patients who developed insulin resistance.Citation31 They are several isoforms of human G6PDH, but which variant is targeted/inhibited in the insulin resistance, or other existing chronic diseases including heart failure and pulmonary hypertension, has yet to be elucidated.Citation32 Under oxidative stress conditions (polyol pathway), the regeneration of reduced GSH is compromised.Citation33 G6PDH-deficient mice have increased oxidative stress and decreased GSH levels, as the reduced levels of NADPH are deficient to maintain the level of glutathione in the tissues, that protect them against oxidative damage.Citation34 Thus, G6PDH enzyme is regulated by nutritional and hormonal factors, at posttranslational and posttranscriptional level as well as oxidant stress.Citation34 It would be interesting to know what is the outcome of the inhibitory effect, i.e., by translational blocking or gene “knock-out” technology, on one or the other og6pdh gene product isoforms. Moreover, the question to be addressed at this stage is what the control mechanism is that “senses” the energy balance changes prior to gene transcription/splicing/protein isoform “adaptation”, adjusting the oG6PDH enzyme activity.
In conclusion, that transcript accumulation as reported beforeCitation24 is in agreement with the relative protein expression of both isoforms, nevertheless there is difference in the scale and intensity of changes between the transcript and the protein levels, where in the latter the level of signal is lower. However, all values reproducibly show similar trend in the relative oG6PDH transcript variants as detected by real-time PCR and relative protein isoform expression, by immunoblotting. These energy-dependent variables of og6pdh gene products, involved in lipogenesis/lipolysis, are adaptable but concurrent with the energy balance changes, manifesting similarities at two levels of control—transcription and translation—as well as differences in scale of their response, in the course of time (weeks post weaning), in the adipose tissue of the ewes.
Materials and Methods
General (animals, housing, and feeding)
Seven randomly selected lactating ewes of the Chios breed were employed for the study. The animals were in the middle stage of lactation and housed at the experimental farm of the Agricultural University of Athens. The experimental design has been described in detail, in a previous reportCitation24; briefly: ewes were fed individually twice per day, at about 7:00 and 16:00 h on an alfalfa hay (4.14 MJ/kg) and on a concentrated diet. The concentrated diet consists of a basal (7.1 MJ/kg) and a lactation (7.3 MJ/kg) ration. By the end of the second experimental week the amount of diet was set in such level that a negative energy balance was achieved according to ewe’s requirements (0.4 kg alfalfa hay, 0.5 kg basal ratio, 1 kg lactation ratio). The next two weeks, the amount of diet achieved a positive energy balance (0.4 kg alfalfa hay, 0.5 kg basal ratio, 1.5 kg lactation ratio). Water was offered free ad libitum under all experimental conditions. At the end of the first, second, and fourth experimental weeks, a tail biopsy was operated on all animal groups from the subcutaneous adipose tissue. Prior to biopsy ewes were fasted but had free water ad libitum. At the day of sampling ewes were anesthetized with the use of Tiletamine and Zolazepam (Zoletin 50) in 15−20 mg/kg BW. Samples of adipose tissues were snap frozen in liquid nitrogen and stored at −80 °C for further protein extraction. All experimental procedures were approved by the bioethical committee of the Agricultural University of Athens under the guidelines of “Council Directive 86/609/EEC regarding the protection of animals used for experimental and other scientific purposes”.
Adipose tissue protein extraction and protein content determination prior to electrophoresis
Adipose tissue frozen blocks were grinded in pestle and mortar with liquid nitrogen and the powder was collected and mixed with Radio-Immunoprecipitation Assay buffer (RIPA) buffer (R0278, Sigma-Aldrich) containing 1 mM Phenylmethanesulfonyl fluoride (PMSF) (P7626, Sigma-Aldrich) and protease inhibitor cocktail (Roche Diagnostics, 11836170001). Lysates were concentrated by centrifugation and supernatants transferred in new pre-chilled tubes, snap frozen in liquid nitrogen and stored at −80 °C for further analysis.
Total protein from the adipose tissue samples was determined by Bicinchoninic Acid (BCA) protein assay (23227, Thermo Fisher Scientific) in a 96-well plate according to manufacturer instructions. Bovine serum albumin (BSA) standard curve was constructed from various concentrations of BSA in RIPA buffer. The BCA protein assay reaction mixture added to the unknown protein sample as well as standards incubated at 37 °C for 30 min and immediately measured at 562 nm by using a microplate reader. The protein concentrations were determined by comparison to the standard curve.
Sodium dodecyl sulfate PAGE (SDS-PAGE) and immunoblot analysis
The expression of oG6PDA and oG6PDB was assessed in adipose tissue immunoblot analysis by using goat antibody raised against oG6PDH. Prior to this investigation, in order to identify the presence of these variants, epitope peptide tracking was based on evidence found by in silico study, searching for the antibody immunogen sequence homologs in the protein database. The above study equally predicted the putative oG6PDH protein isoforms, and confirmed the capability of this antibody to capture equally the protein targets. The protein blast ([BLASTP 2.2.28] citation http://blast.ncbi.nlm.nih.gov/Blast.cgi) for the immunogen peptide used for the antibody (Acris Antibodies GmbH), identified among other hits for G6PDH, the variant A (accession number: ABD34655.1), and variant B (accession number: NP_001087249.1) from Ovis aries.
The technique was performed as previously described elsewhere.Citation35 Specifically, as a primary antibody goat polyclonal and affinity purified antibody against G6PDH (diluted to 1:12500, AP16595PU-N, Acris Antibodies GmbH), was used. As a secondary antibody, a horseradish peroxidase (HRP) conjugated anti-goat antibody (diluted to 1:7500, ab6741, Abcam) was used. As a detection system, the enhanced chemiluminescence reagents (GE Healthcare) for immunoblotting, were employed as well. Consequently, PVDF membranes were re-incubated with mouse β-actin antibody (diluted to 1:1000, MAB1501, CHEMICON International, Inc.), ensued by a horseradish peroxidase (HRP) conjugated anti-mouse antibody (diluted to 1:10 000, RPN2108, GE Healthcare). The transferred protein gel bands on polyvinylidene difluoride (PVDF) membrane, were visualized by chemiluminescent detection on a medical X-ray film (Fuji safety film, RX 13 cm × 18 cm, RX1318, FUJI) and quantified by scanning densitometric analysis using the ImageJ analysis software.Citation36 Results are from two independent blots, with averaged values from the densitometry (n = 2), while the densitometric data were normalized to β-actin signal intensity.
oG6PDH enzyme assay
One sample from each adipose tissue was frozen at −20 °C for enzyme assays for further analysis. For enzyme assays, about 500 mg of adipose tissue was homogenized in 0.15 M KCl at 4 °C and centrifuged at 4 °C and 6000 g for 45 min. Duplicate measurements of G6PDH (EC 1.1.1.49) activities were performed photometrically at 37 °C, according to the methods described previously.Citation10
The estimated enzyme activity was expressed in nanokatal (nkat), which is the SI unit (transformation of 1 mol of substrate per second).
Data analysis
Data are presented as mean ± SEM. We used one way analysis of variance to compare the data, and with a significant variance, we performed Bonferroni post test. Analysis was also confirmed by unpaired t test or nonparametric test (Kruskal–Wallis) followed by the Dunn multiple comparison test. All hypothesis testing was two-sided and a P value of less than 0.05 (P < 0.05) was accepted as significant. Linear regression analysis for repeated measures was also used to determine protein expression changes with energy balance (MJ). Subsequently, a posthoc analysis was applied for intercept (b) comparisons in the equations of the first grade (). Moreover, in order to investigate further the energy balance effect in oG6PDH protein levels, the protein expression data were divided into two groups according to ewes energy balance: (1) group of ewes with negative energy balance (n = 14); and (2) group of ewes with positive energy balance (n = 7). Analysis was performed using GraphPad Prism version 5.04 for Windows (GraphPad Software).
| Abbreviations: | ||
| G6PDH | = | glucose-6-phosphate dehydrogenase |
| oG6PDA | = | ovine G6PDH protein isoform A |
| oG6PDB | = | ovine G6PDH protein isoform B |
| WPW | = | week post-weaning |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to the staff of the Experimental Station in the Agriculture University of Athens, for their generous assistance during the course of the study. The work has been core-funded by the Animal Breeding and Husbandry, laboratory of general and specific animal husbandry, in the department of Animal Science and Aquaculture of Agricultural University of Athens.
References
- Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, Tian X, Stanton RC. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem 1998; 273:10609 - 17; http://dx.doi.org/10.1074/jbc.273.17.10609; PMID: 9553122
- Kletzien RF, Harris PK, Foellmi LA. Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB J 1994; 8:174 - 81; PMID: 8119488
- Vernon RG. Lipid Metabolism in the Adipose Tissue of Ruminant Animals. In: Christie WW, ed. Lipid Metabolism in Ruminant Animal. Oxford, New York: Pergamon Press, 1981:279−362.
- Stryer L. Berg JM, Tymoczko JL. The Rate of the Pentose Phosphate Pathway Is Controlled by the Level of NADP+. In: Stryer L. Berg JM, Tymoczko JL eds. The Metabolism of Glucose 6-Phosphate by the Pentose Phosphate Pathway Is Coordinated with Glycolysis. Biochemistry San 5th ed. Francisco, CA: WH Freeman and Co, 1989.
- London E, Lala G, Berger R, Panzenbeck A, Kohli AA, Renner M, Jackson A, Raynor T, Loya K, Castonguay TW. Sucrose access differentially modifies 11beta-hydroxysteroid dehydrogenase-1 and hexose-6-phosphate dehydrogenase message in liver and adipose tissue in rats. J Nutr 2007; 137:2616 - 21; PMID: 18029473
- Salati LM, Amir-Ahmady B. Dietary regulation of expression of glucose-6-phosphate dehydrogenase. Annu Rev Nutr 2001; 21:121 - 40; http://dx.doi.org/10.1146/annurev.nutr.21.1.121; PMID: 11375432
- Park J, Rho HK, Kim KH, Choe SS, Lee YS, Kim JB. Overexpression of glucose-6-phosphate dehydrogenase is associated with lipid dysregulation and insulin resistance in obesity. Mol Cell Biol 2005; 25:5146 - 57; http://dx.doi.org/10.1128/MCB.25.12.5146-5157.2005; PMID: 15923630
- Thomson JM, Butterfield RM. Changes in body composition relative to weight and maturity of Australian Dorset Horn rams and wethers. 4. Adipose tissue and number in dissected fat portions. Anim Prod 1988; 46:387 - 93; http://dx.doi.org/10.1017/S0003356100018997
- Panopoulou E, Deligiorgis SG, Papadimitriou T, Rogdakis E. Carcass composition, size of fat cells and NADPH-generating dehydrogenases activity in adipose tissue of the fat tailed Chios and the thin tailed Karagouniko sheep breed. J Anim Breed Genet 1989; 106:51 - 8; http://dx.doi.org/10.1111/j.1439-0388.1989.tb00213.x
- Rogdakis E, Charismiadou M, Orphanos S, Panopoulou E, Bizelis I. Cellularity and enzymatic activity of adipose tissue in the Karagouniko dairy breed of sheep from birth to maturity. J Anim Breed Genet 1997; 114:385 - 96; http://dx.doi.org/10.1111/j.1439-0388.1997.tb00524.x; PMID: 21395833
- Benatti U, Morelli A, Frascio M, Melloni E, Salamino F, Sparatore B, Pontremoli S, De Flora A. Glucose 6-phosphate dehydrogenase activity in membranes of erythrocytes from normal individuals and subjects with Mediterranean G6PD deficiency. Biochem Biophys Res Commun 1978; 85:1318 - 24; http://dx.doi.org/10.1016/0006-291X(78)91147-6; PMID: 743300
- Persico MG, Viglietto G, Martini G, Toniolo D, Paonessa G, Moscatelli C, Dono R, Vulliamy T, Luzzatto L, D’Urso M. Isolation of human glucose-6-phosphate dehydrogenase (G6PD) cDNA clones: primary structure of the protein and unusual 5' non-coding region. Nucleic Acids Res 1986; 14:2511 - 22; http://dx.doi.org/10.1093/nar/14.6.2511; PMID: 3515319
- Chen EY, Cheng A, Lee A, Kuang WJ, Hillier L, Green P, Schlessinger D, Ciccodicola A, D’Urso M. Sequence of human glucose-6-phosphate dehydrogenase cloned in plasmids and a yeast artificial chromosome. Genomics 1991; 10:792 - 800; http://dx.doi.org/10.1016/0888-7543(91)90465-Q; PMID: 1889820
- Zollo MZ, D’Urso M, Schlessinger D, Chen EY. Sequence of mouse glucose-6-phosphate dehydrogenase cDNA. DNA Seq 1993; 3:319 - 22; PMID: 8400362
- Ho YS, Howard AJ, Crapo JD. Cloning and sequence of a cDNA encoding rat glucose-6-phosphate dehydrogenase. Nucleic Acids Res 1988; 16:7746; http://dx.doi.org/10.1093/nar/16.15.7746; PMID: 3412913
- Laliotis GP, Argyrokastritis A, Bizelis I, Rogdakis E. Cloning and characterization of an alternative transcript of ovine glucose 6-phosphate dehydrogenase gene: comparative approach between ruminant and non-ruminant species. Gene 2007; a 388:93 - 101; http://dx.doi.org/10.1016/j.gene.2006.10.016; PMID: 17157446
- Laliotis GP, Bizelis I, Argyrokastritis A, Rogdakis E. Cloning, characterization and computational analysis of the 5‘ regulatory region of ovine glucose 6-phosphate dehydrogenase gene Comparative Biochemistry and Physiology. Part B 2007; b 147:627 - 34
- Laliotis GP, Bizelis I, Argyrokastritis A, Rogdakis E. Cloning and characterization of a novel ovine (Ovis aries) cDNA sequence encoding glucose-6-phosphate dehydrogenase, an enzyme involved in lipogenesis. Anim Sci Rev 2008; 38:75 - 86
- Maronpot RR. Erythrocyte glucose-6-phosphate dehydrogenase and glutathione deficiency in sheep. Can J Comp Med 1972; 36:55 - 60; PMID: 4258546
- Calabrese EJ, Moore GS, Ho SC. Low glucose-6-phosphate dehydrogenase (G-6-PD) activity in red blood cells and susceptibility to copper-induced oxidative damage. Environ Res 1980; 21:366 - 72; http://dx.doi.org/10.1016/0013-9351(80)90039-0; PMID: 7408807
- Bizelis I, Charismiadou M, Rogdakis E. Metabolic changes during the perinatal period in dairy sheep in relation to plane of nutrition and breed. II. Early lactation. J Anim Physiol Anim Nutr (Berl) 2000; 84:73 - 84; http://dx.doi.org/10.1046/j.1439-0396.2000.00283.x
- Martinez di Montemuros F, Dotti C, Tavazzi D, Fiorelli G, Cappellini MD. Molecular heterogeneity of glucose-6-phosphate dehydrogenase (G6PD) variants in Italy. Haematologica 1997; 82:440 - 5; PMID: 9299858
- Dolezelová E, Zurovec M, Böhmová M, Sehnal F. Use of two transcription starts in the G6PD gene of the bark beetle Ips typographus. Insect Mol Biol 2006; 15:25 - 32; http://dx.doi.org/10.1111/j.1365-2583.2006.00604.x; PMID: 16469065
- Laliotis GP, Vitsa A, Bizelis I, Charismiadou MA, Rogdakis E. Molecular study of ovine glucose 6-phosphate dehydrogenase gene expression in respect to different energy intake. Comp Biochem Physiol B. Part D 2010; 5:124 - 9
- Salati LM, Szeszel-Fedorowicz W, Tao H, Gibson MA, Amir-Ahmady B, Stabile LP, Hodge DL. Nutritional regulation of mRNA processing. J Nutr 2004; 134:2437S - 43S; PMID: 15333739
- Xu Y, Zhang Z, Hu J, Stillman IE, Leopold JA, Handy DE, Loscalzo J, Stanton RC. Glucose-6-phosphate dehydrogenase-deficient mice have increased renal oxidative stress and increased albuminuria. FASEB J 2010; 24:609 - 16; http://dx.doi.org/10.1096/fj.09-135731; PMID: 19805580
- White CC, Viernes H, Krejsa CM, Botta D, Kavanagh TJ. Fluorescence-based microtiter plate assay for glutamate-cysteine ligase activity. Anal Biochem 2003; 318:175 - 80; http://dx.doi.org/10.1016/S0003-2697(03)00143-X; PMID: 12814619
- Pandolfi PP, Sonati F, Rivi R, Mason P, Grosveld F, Luzzatto L. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J 1995; 14:5209 - 15; PMID: 7489710
- Longo L, Vanegas OC, Patel M, Rosti V, Li H, Waka J, Merghoub T, Pandolfi PP, Notaro R, Manova K, et al. Maternally transmitted severe glucose 6-phosphate dehydrogenase deficiency is an embryonic lethal. EMBO J 2002; 21:4229 - 39; http://dx.doi.org/10.1093/emboj/cdf426; PMID: 12169625
- Pongratz RL, Kibbey RG, Shulman GI, Cline GW. Cytosolic and mitochondrial malic enzyme isoforms differentially control insulin secretion. J Biol Chem 2007; 282:200 - 7; http://dx.doi.org/10.1074/jbc.M602954200; PMID: 17102138
- Park J, Choe SS, Choi AH, Kim KH, Yoon MJ, Suganami T, Ogawa Y, Kim JB. Increase in glucose-6-phosphate dehydrogenase in adipocytes stimulates oxidative stress and inflammatory signals. Diabetes 2006; 55:2939 - 49; http://dx.doi.org/10.2337/db05-1570; PMID: 17065329
- Gupte SA. Targeting the pentose phosphate pathway in syndrome X-related cardiovascular complications. Drug Dev Res 2010; 71:161 - 7; PMID: 20711518
- Ballatori N, Krance SM, Notenboom S, Shi S, Tieu K, Hammond CL. Glutathione dysregulation and the etiology and progression of human diseases. Biol Chem 2009; 390:191 - 214; http://dx.doi.org/10.1515/BC.2009.033; PMID: 19166318
- Singh S, Anand A, Srivastava PK. Regulation and properties of glucose-6-phosphate dehydrogenase: A review. Int J Plant Physiol Biochem 2012; 4:1 - 19
- Triantaphyllopoulos KA, Hussain F, Pinart M, Zhang M, Li F, Adcock I, Kirkham P, Zhu J, Chung KF. A model of chronic inflammation and pulmonary emphysema after multiple ozone exposures in mice. Am J Physiol Lung Cell Mol Physiol 2011; 300:L691 - 700; http://dx.doi.org/10.1152/ajplung.00252.2010; PMID: 21355040
- Rasband WS, ImageJ, U S National Institutes of Health, Bethesda, Maryland, USA, 1997–2012.
