Abstract
Peptide nucleic acid (PNA) is one of the most widely used synthetic DNA analogs. Conjugation of functional molecules to PNA is very effective to further widen its potential applications. For this purpose, here we report the synthesis of several ligand monomers and introduced them to PNA. These ligand-modified PNAs attract cerium ion and are useful for site-selective DNA hydrolysis. It should be noted that these ligands on PNA are also effective even under the conditions of invasion complex.
Introduction
DNA-binding small molecules, which recognize a specific DNA sequence, have been intensively studied for their wide-ranging potential applications. In addition to excellent pioneering works such as triplex-forming oligonucleotidesCitation1-Citation4 and pyrrole-imidazole polyamides,Citation5,Citation6 many DNA analogsCitation7-Citation13 have also been reported in this field. Peptide nucleic acid (PNA),Citation14,Citation15 which was developed by Nielsen’s group, is one of the most successful synthetic DNA analogs. Instead of conventional sugar-phosphate backbone of DNA, PNA has a N-(2-aminoethyl)glycine backbone and no negative charge on its backbone. Consequently it can form an immensely stable duplex with complementary DNA through the Watson-Crick base pairing because there is no electrostatic repulsion between PNA and negatively-charged DNA strands. This non-negative backbone is distinctively different from other DNA analogs, and PNA can even interact with double-stranded DNA, forming a unique invasion complex (e.g., duplex, triplex and double-duplex invasion modes).Citation16,Citation17 Furthermore, this invasion can alter the local structure of DNA at a predetermined site and has been applied to various applications including inhibition of enzymatic reaction,Citation18,Citation19 manipulation of DNA nanostructureCitation20 and construction of artificial DNA cutter (vide infra).Citation21,Citation22 In addition to the high affinity to DNA, PNA also discriminates mismatched sequence with high accuracy.Citation23,Citation24 Its sequence-discrimination ability is superior to corresponding DNA, and PNA has been frequently utilized for the probe to recognize specific DNA sequence.Citation25
In this study, we functionalize PNA by attaching several metal-chelating ligands (especially multiphosphonate ligands) to enhance its attractive features as a DNA-recognizing molecule and widen its potential applications ().Citation26-Citation34 Furthermore, it is confirmed that the ligand moiety effectively binds a metal ion even when PNA is forming invasion complex. Significant features of ligand-modified PNAs have been evidenced by means of site-selective DNA scission with Ce(IV).
Results and Discussion
Synthesis of ligand monomers for the introduction to PNA
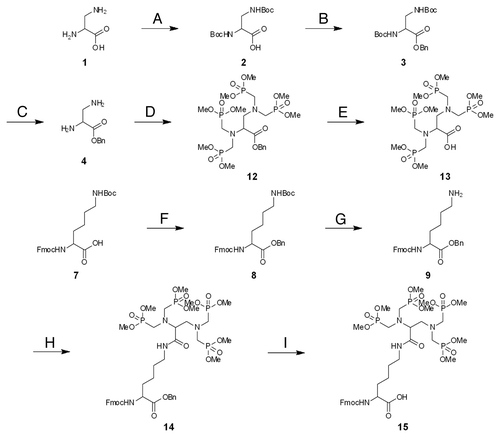
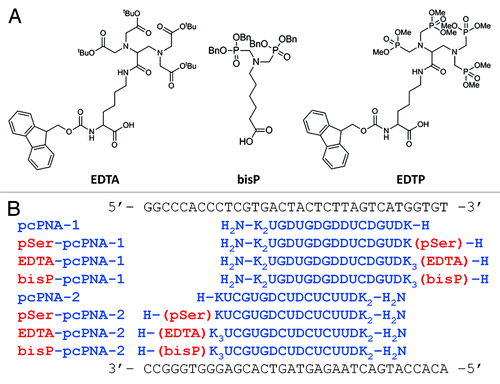
PNA is synthesized by standard solid phase peptide synthesis.Citation35 The terminal amino group of PNA backbone is the most convenient target group for the introduction of ligands. Accordingly, the monomers for ligand introduction should have one carboxyl group for the conjugation with PNA and appropriately protected functional groups for the coordination of metal ions. As a metal-chelating ligand, we chose three ligands, ethylenediamine-N,N,N′,N′-tetraacetic acid (EDTA), iminodi(methylphosphonic acid) (bisP) and ethylenediamine-N,N,N′,N′-tetrakis(methylenephosphonic acid) (EDTP), which have high affinity to various metal ions, especially to lanthanide ions ().Citation36-Citation40 Lanthanide ions have various unique and attractive features, and their spectroscopic and magnetic properties, as well as their catalytic functions (vide infra), have been utilized for various purposes. Synthetic routes of these ligand monomers are shown in Schemes 1–3. EDTA monomer was synthesized through repetitive protections and deprotections of functional groups according to conventional method (Scheme 1). Ethylenediamine-N,N,N′,N′-tetraacetic acid (EDTA) skeleton was synthesized from 2,3-diaminopropanoic acid (1) as a starting material and its carboxyl group was converted to benzyl ester to give compound 4. Then ethylenediamine moiety was treated with t-butyl bromoacetate and protected EDTA with additional carboxyl group was obtained. This compound 6 was further conjugated with N2-Fmoc-2,6-diaminohexanoic acid benzyl ester and Fmoc-EDTA monomer 11 was prepared. For the preparation of the EDTP monomer (Scheme 2), 2,3-diaminopropanoic acid benzyl ester (4) was treated with paraformaldehyde and dimethylphosphite in THF and phosphonomethyl group was introduced to its ethylenediamine moiety, giving ethylenediamine-N,N,N′,N′-tetrakis(methylenephosphonic acid) (EDTP) skeleton (Scheme 2). After that, Fmoc-EDTP monomer 15 was prepared by conjugating with N2-Fmoc-2,6-diaminohexanoic acid benzyl ester as described above for the EDTA monomer synthesis. As for the bisP monomer, 6-aminohexanoic acid (16) was used as a starting material and phosphonomethyl group was introduced in a similar way (Scheme 3). This bisP monomer 19 was introduced directly to pcPNA and benzyl group was applied as a protection group of phosphonate because it is less bulky than other EDTA or EDTP monomers. As mentioned above, EDTA and EDTP monomers were conjugated with N2-Fmoc-2,6-diaminohexanoic acid to make the reaction point (carboxyl group) more accessible and increase the coupling yield.
Scheme 1. Synthesis of EDTA monomer for the introduction to PNA. (A) (Boc)2O, K2CO3, H2O/1,4-dioxane = 1/1, 98%; (B) benzyl bromide, Et3N, THF, 89%; (C) TFA, CH2Cl2, 95%; (D) tbutyl bromoacetate, DIEA, DMF, 74%; (E) H2, 10% Pd/C, MeOH, 90%; (F) Z-Cl, DIEA, DMAP, CH2Cl2, quant; (G) TFA, CH2Cl2, quant; (H) 6, DIEA, HOBt, HBTU, DMF, 48% (I) H2, 10% Pd/C, MeOH, 82%.
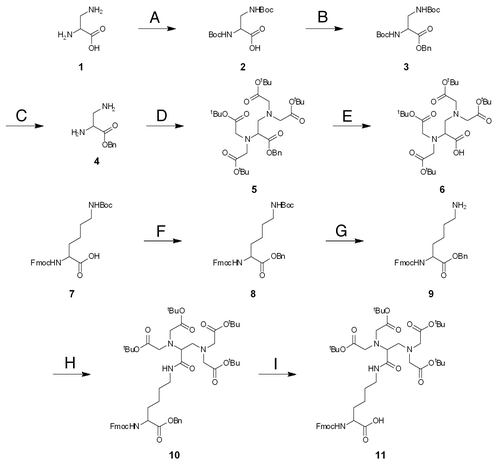
Scheme 3. Benzyl-protected bisP monomer for the introduction to PNA. (A) allyl alcohol, TsOH, benzene, 13%; (B) dibenzyl phosphite, 37% formaldehyde aq, MeOH, 53%; (C) Pd(PPh3)4, pyrrolidine, CHCl3, 87%.
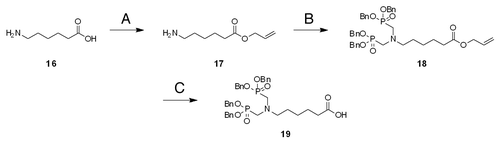
Scheme 2. Fmoc-EDTP monomer for the introduction to PNA. (A) (Boc)2O, K2CO3, H2O/1,4-dioxane = 1/1, 98%; (B) benzyl bromide, Et3N, THF, 89%; (C) TFA, CH2Cl2, 95%; (D) dimethyl phosphite, paraformaldehyde, THF, 43%; (E)H2, 10% Pd/C, MeOH, 90%; (F) Z-Cl, DIEA, DMAP, CH2Cl2, quant; (G) TFA, CH2Cl2, quant; (H) 13, DIEA, HOBt, HBTU, DMF, 48% (I) H2, 10% Pd/C, MeOH, 82%.
PNAs with or without these ligands were synthesized by employing both Boc- and/or Fmoc-based solid phase peptide synthesis.Citation35 Here, two lysine residues were additionally introduced to the ligand-modified PNAs because the negative charges of carboxyl or phosphonate groups may decrease the invasion efficiency of PNA by the electrostatic repulsion to negatively-charged DNA. As shown in , the target sequence of PNA is the chromophore-coding region of blue fluorescent protein (BFP), which emits blue fluorescence and is one of the reporter proteins like EGFP.Citation41-Citation43 These PNAs were detached from the resin, purified by HPLC, and characterized by MALDI-TOF MS. Results of MS measurements are shown in Figure S1.
Electrophoretic mobility shift assay (gel shift assay) for the confirmation of invasion complex formation
For the evaluation of the invasion ability of the ligand-modified PNA, electrophoretic mobility shift assay (gel shift assay) was performed by using several PNAs under two concentration conditions. As shown in , shifted bands were clearly observed and it was experimentally confirmed that these ligand-modified pcPNAs can form invasion complexes. Furthermore, their efficiencies of invasion-complex formation were sufficient from a practical point of view, compared with the control lane of phosphoserine (pSer)-modified pcPNAs (pSer-pcPNA-1 and pSer-pcPNA-2), which have been investigated in our previous studies.Citation30 This result indicated that functions of modified pcPNAs can be enhanced by using various functional metal ions without any disadvantage.
Evaluation of metal-chelating ability of ligand-modified PNA by the cerium-induced site-selective DNA scission
To further confirm the utility of ligand-modified pcPNAs, we used cerium ion as a coordination metal and applied these pcPNAs to a site-selective hydrolysis of double-stranded DNA. Ce(IV) ion has an extremely-high activity for the hydrolysis of phosphodiester in DNA and has been used as Ce(IV)/EDTA for many DNA-cleavage experiments.Citation44 Furthermore, artificial restriction DNA cutter (ARCUT) has been developed by combining pcPNAs with Ce(IV)/EDTA, and applied to various biological applications (the details of this DNA cutter are described in Fig. S2).Citation21,Citation44 In this ARCUT system, a proximally-positioned cerium ion at target site is very important for the efficient DNA hydrolysis. Therefore the coordination ability of ligand on pcPNA should be evaluated by the promotion of scission efficiency.
Recently, Ce(IV) prepared by the oxidation of Ce(III) on the EDTP ligand was utilized for the site-selective DNA scission.Citation39 Thus, we decided to combine Ce(III) and ligand-modified pcPNAs for the confirmation of their metal coordination behavior. Here the coordination of Ce(III) to ligands is a key for the effective DNA hydrolysis because Ce(IV) itself is prone to form hydroxide gel around pH 7.0 and the oxidation of Ce(III) should occur on the ligand to prevent its precipitation. For the site-selective scission, linearized BFP plasmid (4,733 bp) was used. If the desired scission occurred, 1.7- and 3.0-kbp bands should be formed. First, pcPNAs and DNA were mixed and incubated at 50°C for 1 h to form invasion complex. Then, the reaction was started by adding NaCl and Ce(NO3)3 solutions to reaction mixtures. The agarose gel electrophoresis patterns are shown in . In lanes 1 and 2 where EDTA-pcPNA-1 and EDTA-pcPNA-2 were used, the desired site-selective scission was detected. On the other hand, there is no clear scission band in control lanes 3 and 4 using unmodified pcPNAs and this result indicates that the EDTA ligands on pcPNAs coordinate Ce(III) ion and the Ce(III) is readily oxidized to catalytically-active Ce(IV). In addition, effects of introduced EDTA and bisP on the site-selective DNA scission by Ce(IV)/EDTA was also evaluated (). In lane 4 using bisP-pcPNA-1 and bisP-pcPNA-2, the desired scission bands were clearly observed. Compared with EDTA-pcPNA, bisP-pcPNA signify a slight increase in the efficiency of the site-selective DNA scission (yields of DNA cleavage using EDTA-pcPNA and bisP-pcPNA are 6 and 12%, respectively). The scission manner is proved to be hydrolytic (not oxidative) by the experiment with scavengers (DMSO, NaN3); see Figure S3. This excludes the possibility that any contaminated metal ion other than cerium is responsible for the present DNA cleavage. The coordination of cerium ion on the ligands is crucial for the efficient DNA hydrolysis because no scission band was observed in control lanes using unmodified PNA. Accordingly, the ligands on pcPNAs work as a metal-chelating unit and bisP is superior to EDTA as a ligand to Ce(III) and/or Ce(IV).
Figure 3. (A) Site-selective scission of linearized BFP plasmid DNA by pcPNAs with or without EDTA for the confirmation of its metal-coordination behavior. Lane M, 1,000 bp ladder; Lanes 1 and 2, EDTA-pcPNA-1/EDTA-pcPNA-2; Lanes 3 and 4, pcPNA-1/pcPNA-2. Reaction conditions: [linearized BFP plamid DNA] = 4 nM, [each of pcPNAs] = 100 nM, [Hepes (pH 7.0)] = 5 mM, [NaCl] = 100 mM and [Ce(NO3)3] = 20 μM (Lanes 1 and 3) or 30 μM (Lanes 2 and 4) at 50°C for 17 h under air. (B) Comparison of the effect of ligands introduced to the terminal of pcPNAs on the site-selective DNA scission by using Ce(III) salt. Lanes 1, DNA only; Lane 2, DNA + Ce(III); Lanes 3, EDTA-pcPNA-1/EDTA-pcPNA-2; Lane 4, bisP-pcPNA-1/ bisP-pcPNA-2. Reaction conditions: [linearized BFP plamid DNA] = 4 nM, [each of pcPNAs] = 100 nM, [Hepes (pH 7.0)] = 5 mM, [NaCl] = 100 mM and [Ce(NO3)3] = 30 μM at 50°C for 17 h under air.
![Figure 3. (A) Site-selective scission of linearized BFP plasmid DNA by pcPNAs with or without EDTA for the confirmation of its metal-coordination behavior. Lane M, 1,000 bp ladder; Lanes 1 and 2, EDTA-pcPNA-1/EDTA-pcPNA-2; Lanes 3 and 4, pcPNA-1/pcPNA-2. Reaction conditions: [linearized BFP plamid DNA] = 4 nM, [each of pcPNAs] = 100 nM, [Hepes (pH 7.0)] = 5 mM, [NaCl] = 100 mM and [Ce(NO3)3] = 20 μM (Lanes 1 and 3) or 30 μM (Lanes 2 and 4) at 50°C for 17 h under air. (B) Comparison of the effect of ligands introduced to the terminal of pcPNAs on the site-selective DNA scission by using Ce(III) salt. Lanes 1, DNA only; Lane 2, DNA + Ce(III); Lanes 3, EDTA-pcPNA-1/EDTA-pcPNA-2; Lane 4, bisP-pcPNA-1/ bisP-pcPNA-2. Reaction conditions: [linearized BFP plamid DNA] = 4 nM, [each of pcPNAs] = 100 nM, [Hepes (pH 7.0)] = 5 mM, [NaCl] = 100 mM and [Ce(NO3)3] = 30 μM at 50°C for 17 h under air.](/cms/asset/3883261f-3e6b-42ba-9eab-e460499183d8/kdpx_a_10920727_f0006.gif)
Conclusion
In this study, we made an attempt to widen potential applications of PNA by introducing metal-chelating ligands. The synthesized ligand monomers were successfully introduced to PNA strands. The pcPNAs with ligand moiety (EDTA and bisP) can invade double-stranded DNA with comparable efficiency as usual pcPNAs. Furthermore, these ligands can attract metal ion even under the conditions of invasion complex and functionalization of pcPNA was experimentally confirmed by site-selective DNA hydrolysis using Ce(III). Judging from the scission yields, it was found that bisP is superior to EDTA as a ligand to cerium ion. Here, only cerium ion was conjugated with PNA, but the present method is also applicable to the conjugation of variety of metal ions. For example, gadolinium for contrast agent of MRI (magnetic resonance imaging), gallium, samarium, technetium and yttrium for PET (positron-emission tomography) or radiotherapy are good candidates. Furthermore, these ligand-modified PNAs have two independent functions (DNA-recognition and metal-coordination domains), and they could perform as attractive functional molecules by merging these two aspects. For example, an effective drug delivery system (DDS) may be constructed by combining site-specific targeting ability based on complementarity of PNA strand and radiotherapeutic agent composed of metal-ligand complex. Consequently, various prospective applications can be expected by utilizing appropriate present technologies, and then future growth potential of these modified pcPNAs is assumedly high.
Materials and Methods
Target DNA and PNA additives
For the preparation of ligand-modified pcPNAs, Boc-protected 2-thiouracil and 2,6-diaminopurine monomers were used with commercially available Boc-protected PNA monomers (from ASM), N-Fmoc-O-benzyl-L-phosphoserine (from Novabiochem) for control PNAs and (4-methylbenzhydryl)amine resin (from ABI) (please consult ref. Citation35 for detailed procedures of the preparation 2-thiouracil and 2,6-diaminopurine monomers and pcPNA synthesis). After the final removal of Boc or Fmoc group on pcPNA, the pcPNA strand was removed from the resin by “low-high TFMSA method.” All these modified and unmodified pcPNAs were purified by reversed-phase HPLC and characterized by MALDI-TOFMS (Bruker, AutoFLEX).
The 119-bp double-stranded DNA having the target sequence of synthesized pcPNAs was prepared by PCR and purified by QIAquick PCR Purification Kit (from Qiagen).
For the site-selective DNA scission, the target BFP plasmid DNA (4,733 bp) was prepared from pEGFP-N1 plasmid DNA (Clontech), which is coding closely-related green fluorescent protein (EGFP), by introducing mutations to give the corresponding BFP gene. The plasmid DNA was linearized by StuI and then used as a substrate for the site-selective DNA scission.
N2-/Fmoc/-N6-EDTP-2,6-diaminohexanoic acid (11)
The benzyl ester 10 (185.9 mg) was hydrogenated under an atmosphere of hydrogen over 10% Pd on carbon (61.9 mg) in methanol (15 ml) at room temperature for 2 h. The mixture was filtered, concentrated under reduced pressure to give the desired product as white powder, yield 151.0 mg (89%). 1H NMR (CDCl3, δ in ppm) 7.90 (s, 1 H, –CONHCH2–), 7.76 (m, 2 H, –ArH Fmoc), 7.62 (m, 2 H, –ArH Fmoc), 7.41–7.30 (m, 4 H, –ArH Fmoc), 5.73 (d, 1 H, J = 5.70 Hz, –NHCH–), 4.36–4.24 (m, 4 H, –NHCH– and –CH2CHAr Fmoc, overlapped), 4.00–3.92 (m, 1 H, –NCHCO–), 3.78–3.73 (m, 24 H, –OCH3), 3.41–3.16 (m, 12 H, –CONHCH2–, –CHCH2N– and –NCH2P–, overlapped), 1.99–1.32 (m, 6 H, –CHCH2CH2CH2–).
N2-Fmoc-N6-EDTA-2,6-diaminohexanoic acid (15)
The benzyl ester 14 (438.6 mg) was hydrogenated under an atmosphere of hydrogen over 10% Pd on carbon (72.6 mg) in methanol (10 ml) at room temperature for 3 h. The mixture was filtered, concentrated under reduced pressure to give the desired product as white powder, yield 325.4 mg (82%). 1H NMR (CDCl3, δ in ppm) 8.38 (m, 1 H, –CONHCH2–), 7.75 (m, 2 H, –ArH Fmoc), 7.60 (m, 2 H, –ArH Fmoc), 7.41–7.29 (m, 4 H, –ArH Fmoc), 5.65 (m, 1 H, –NHCH–), 4.36–4.22 (m, 4 H, –NHCH– and –CH2CHAr Fmoc, overlapped), 3.59–3.45 (m, 8 H, –NCH2P–), 3.40 (m, 2 H, –NCH2CH–), 3.33 (br s, 2 H, –NHCH2–), 2.95 (br s, 1 H, –NCHCO–), 1.89–1.32 [m, 42 H, –CHCH2CH2CH2– and –O(CH3)3, overlapped].
N,N-bis(methylenephosphonic acid tetrabenzyl ester) aminohexanoic acid (19)
The allyl ester 3 (508.0 mg) and tetrakis(triphenylphosphine)palladium(0) (83.4 mg) were dissolved in dichloromethane (10 ml) and stirred at 0°C. To the solution was added pyrrolidine (70 μl) and stirred at 0°C for 4 h. The reaction mixture was diluted with dichloromethane (20 ml) and washed with 1 N HCl aq (25 ml). The organic phase was dried over sodium sulfate, concentrated under reduced pressure and purified by silica column chromatography (eluting with 5% methanol in dichloromethane) to give the desired product as colorless oil, yield 426.9 mg (89%). 1H NMR (CDCl3, δ in ppm) 7.36–7.29 (m, 20 H, –ArH), 5.03–4.93 (m, 8 H, ArCH2-), 3.13 (d, 4 H, J = 8.4 Hz, –NCH2P–), 2.74 (t, 2 H, J = 6.95 Hz, εCH2), 2.24 (t, 2 H, J = 7.45 Hz, αCH2), 1.53 (tt, 2 H, both of J = 7.4 Hz, βCH2), 1.37 (m, 2 H, δCH2), 1.20 (m, 2 H, γCH2).
Gel shift assay to confirm the formation of invasion complex
Invasion complex composed of the 119-bp double-stranded DNA and pcPNAs with or without ligand was confirmed by gel-shift assay and its efficiency was evaluated. This 119-bp DNA was prepared from BFP plasmid DNA by PCR and the target sequence of pcPNAs is located in its middle. First, pcPNAs and target DNA were incubated in Hepes buffer (pH 7.0) at 50°C for 1 h. After the formation of invasion complex, loading buffer (0.05% bromophenol blue and 30% glycerol in 0.5 × TBE buffer) was added to the solution and then the mixture was subjected to 10% non-denaturing PAGE (PAGE). The bands were detected by staining with GelStar (from FMC). Invasion conditions are as follows; [double-stranded DNA (119 bp)] = 50 nM, [each of pcPNAs] = 200 or 400 nM and [Hepes (pH 7.0)] = 5 mM at 50°C for 1 h.
Evaluation of metal-chelating ability of ligand-modified PNA by the cerium-induced site-selective scission of double-stranded DNA
For the formation of invasion complex, linearized BFP plasmid DNA was incubated with pcPNAs in Hepes buffer (pH 7.0) at 50°C for 1 h. Then NaCl solution was added to the mixture and the site-selective DNA cleavage was started by adding Ce(NO3)3 solution to a final concentration. After a predetermined time, the scission reaction was stopped by adding an ethylenediamine-N,N,N′,N′-tetrakis(methylenephosphonic acid) (EDTP) 0.5 × TBE solution (pH 7.0) and the mixture was further incubated at 50°C for 1 h. After that, the scission products were subjected to 0.8% agarose gel electrophoresis and scission bands were detected by staining with GelStar. Typical cleavage conditions: [DNA] = 4 nM, [each of pcPNAs] = 100 nM, [Hepes (pH 7.0)] = 5mM, [NaCl] = 100mM and [Ce(NO3)3] = 30 μM at 50°C for 17 h under air.
| Abbreviations: | ||
| EDTA | = | ethylenediamine-N,N,N′,N′-tetraacetic acid |
| EDTP | = | ethylenediamine-N,N,N′,N′-tetrakis(methylenephosphonic acid) |
Additional material
Download Zip (1.1 MB)Acknowledgments
This work was partially supported by Grants-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (18001001 and 22000007), and by the Global COE Program for Chemistry Innovation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Thuong NT, Hélène C. Sequence-Specific Recognition and Modification of Double-Helical DNA by Oligonucleotides. Angew Chem Int Ed Engl 1993; 32:666 - 90; http://dx.doi.org/10.1002/anie.199306661
- Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science 1991; 251:1360 - 3; http://dx.doi.org/10.1126/science.2003222; PMID: 2003222
- Maher LJ 3rd, Wold B, Dervan PB. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science 1989; 245:725 - 30; http://dx.doi.org/10.1126/science.2549631; PMID: 2549631
- Fox KR. Targeting DNA with triplexes. Curr Med Chem 2000; 7:17 - 37; PMID: 10637355
- Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr Opin Struct Biol 2003; 13:284 - 99; http://dx.doi.org/10.1016/S0959-440X(03)00081-2; PMID: 12831879
- White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature 1998; 391:468 - 71; http://dx.doi.org/10.1038/36116; PMID: 9461213
- Wojciechowski F, Leumann CJ. Alternative DNA base-pairs: from efforts to expand the genetic code to potential material applications. Chem Soc Rev 2011; 40:5669 - 79; http://dx.doi.org/10.1039/c1cs15027h; PMID: 21431120
- Oka N, Wada T. Stereocontrolled synthesis of oligonucleotide analogs containing chiral internucleotidic phosphorus atoms. Chem Soc Rev 2011; 40:5829 - 43; http://dx.doi.org/10.1039/c1cs15102a; PMID: 21720637
- Bandy TJ, Brewer A, Burns JR, Marth G, Nguyen T, Stulz E. DNA as supramolecular scaffold for functional molecules: progress in DNA nanotechnology. Chem Soc Rev 2011; 40:138 - 48; http://dx.doi.org/10.1039/b820255a; PMID: 20694258
- Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol 2003; 21:74 - 81; http://dx.doi.org/10.1016/S0167-7799(02)00038-0; PMID: 12573856
- Leumann CJ. DNA analogues: from supramolecular principles to biological properties. Bioorg Med Chem 2002; 10:841 - 54; http://dx.doi.org/10.1016/S0968-0896(01)00348-0; PMID: 11836090
- Kool ET. Preorganization of DNA: Design principles for improving nucleic acid recognition by synthetic oligonucleotides. Chem Rev 1997; 97:1473 - 88; http://dx.doi.org/10.1021/cr9603791; PMID: 11851456
- Goodchild J. Conjugates of oligonucleotides and modified oligonucleotides: a review of their synthesis and properties. Bioconjug Chem 1990; 1:165 - 87; http://dx.doi.org/10.1021/bc00003a001; PMID: 1965782
- Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 1993; 365:566 - 8; http://dx.doi.org/10.1038/365566a0; PMID: 7692304
- Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991; 254:1497 - 500; http://dx.doi.org/10.1126/science.1962210; PMID: 1962210
- Lohse J, Dahl O, Nielsen PE. Double duplex invasion by peptide nucleic acid: a general principle for sequence-specific targeting of double-stranded DNA. Proc Natl Acad Sci U S A 1999; 96:11804 - 8; http://dx.doi.org/10.1073/pnas.96.21.11804; PMID: 10518531
- Haaima G, Hansen HF, Christensen L, Dahl O, Nielsen PE. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine. Nucleic Acids Res 1997; 25:4639 - 43; http://dx.doi.org/10.1093/nar/25.22.4639; PMID: 9358176
- Izvolsky KI, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. Sequence-specific protection of duplex DNA against restriction and methylation enzymes by pseudocomplementary PNAs. Biochemistry 2000; 39:10908 - 13; http://dx.doi.org/10.1021/bi000675e; PMID: 10978178
- Protozanova E, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. Pseudocomplementary PNAs as selective modifiers of protein activity on duplex DNA: the case of type IIs restriction enzymes. Nucleic Acids Res 2003; 31:3929 - 35; http://dx.doi.org/10.1093/nar/gkg450; PMID: 12853608
- Stadler AL, Sun D, Maye MM, van der Lelie D, Gang O. Site-selective binding of nanoparticles to double-stranded DNA via peptide nucleic acid “invasion”. ACS Nano 2011; 5:2467 - 74; http://dx.doi.org/10.1021/nn101355n; PMID: 21388119
- Komiyama M, Aiba Y, Yamamoto Y, Sumaoka J. Artificial restriction DNA cutter for site-selective scission of double-stranded DNA with tunable scission site and specificity. Nat Protoc 2008; 3:655 - 62; http://dx.doi.org/10.1038/nprot.2008.7; PMID: 18388948
- Katada H, Komiyama M. Artificial restriction DNA cutters to promote homologous recombination in human cells. Curr Gene Ther 2011; 11:38 - 45; http://dx.doi.org/10.2174/156652311794520094; PMID: 21182465
- Ratilainen T, Holmén A, Tuite E, Nielsen PE, Nordén B. Thermodynamics of sequence-specific binding of PNA to DNA. Biochemistry 2000; 39:7781 - 91; http://dx.doi.org/10.1021/bi000039g; PMID: 10869183
- Miyajima Y, Ishizuka T, Yamamoto Y, Sumaoka J, Komiyama M. Origin of high fidelity in target-sequence recognition by PNA-Ce(IV)/EDTA combinations as site-selective DNA cutters. J Am Chem Soc 2009; 131:2657 - 62; http://dx.doi.org/10.1021/ja808290e; PMID: 19199631
- Komiyama M, Ye S, Liang XG, Yamamoto Y, Tomita T, Zhou JM, et al. PNA for one-base differentiating protection of DNA from nuclease and its use for SNPs detection. J Am Chem Soc 2003; 125:3758 - 62; http://dx.doi.org/10.1021/ja0295220; PMID: 12656606
- Zelder FH, Mokhir AA, Krämer R. Sequence selective hydrolysis of linear DNA using conjugates of Zr(IV) complexes and peptide nucleic acids. Inorg Chem 2003; 42:8618 - 20; http://dx.doi.org/10.1021/ic034896v; PMID: 14686836
- Watson RM, Skorik YA, Patra GK, Achim C. Influence of metal coordination on the mismatch tolerance of ligand-modified PNA duplexes. J Am Chem Soc 2005; 127:14628 - 39; http://dx.doi.org/10.1021/ja051336h; PMID: 16231915
- Kornyushyna O, Stemmler AJ, Graybosch DM, Bergenthal I, Burrows CJ. Synthesis of a metallopeptide-PNA conjugate and its oxidative cross-linking to a DNA target. Bioconjug Chem 2005; 16:178 - 83; http://dx.doi.org/10.1021/bc0497564; PMID: 15656589
- Füssl A, Schleifenbaum A, Göritz M, Riddell A, Schultz C, Krämer R. Cellular uptake of PNA--terpyridine conjugates and its enhancement by Zn2+ ions. J Am Chem Soc 2006; 128:5986 - 7; http://dx.doi.org/10.1021/ja0571139; PMID: 16669638
- Yamamoto Y, Mori M, Aiba Y, Tomita T, Chen W, Zhou JM, et al. Chemical modification of Ce(IV)/EDTA-based artificial restriction DNA cutter for versatile manipulation of double-stranded DNA. Nucleic Acids Res 2007; 35:e53; http://dx.doi.org/10.1093/nar/gkm052; PMID: 17376805
- Shiraishi T, Hamzavi R, Nielsen PE. Subnanomolar antisense activity of phosphonate-peptide nucleic acid (PNA) conjugates delivered by cationic lipids to HeLa cells. Nucleic Acids Res 2008; 36:4424 - 32; http://dx.doi.org/10.1093/nar/gkn401; PMID: 18596083
- Ferri E, Donghi D, Panigati M, Prencipe G, D’Alfonso L, Zanoni I, et al. Luminescent conjugates between dinuclear rhenium(I) complexes and peptide nucleic acids (PNA) for cell imaging and DNA targeting. Chem Commun 2010; 46:6255 - 7; http://dx.doi.org/10.1039/c0cc00450b; PMID: 20668778
- Gasser G, Pinto A, Neumann S, Sosniak AM, Seitz M, Merz K, et al. Synthesis, characterisation and bioimaging of a fluorescent rhenium-containing PNA bioconjugate. Dalton Trans 2012; 41:2304 - 13; http://dx.doi.org/10.1039/c2dt12114j; PMID: 22183093
- Gasser G, Sosniak AM, Metzler-Nolte N. Metal-containing peptide nucleic acid conjugates. Dalton Trans 2011; 40:7061 - 76; http://dx.doi.org/10.1039/c0dt01706j; PMID: 21541385
- Komiyama M, Aiba Y, Ishizuka T, Sumaoka J. Solid-phase synthesis of pseudo-complementary peptide nucleic acids. Nat Protoc 2008; 3:646 - 54; http://dx.doi.org/10.1038/nprot.2008.6; PMID: 18388947
- Rana TM, Ban M, Hearst JE. Synthesis of A Metal-Ligating Amino Acid Suitable for Solid Phase Assembly of Peptides. Tetrahedron Lett 1992; 33:4521 - 4; http://dx.doi.org/10.1016/S0040-4039(00)61302-3
- Lönnberg T, Suzuki Y, Komiyama M. Prompt site-selective DNA hydrolysis by Ce(IV)-EDTA using oligonucleotide multiphosphonate conjugates. Org Biomol Chem 2008; 6:3580 - 7; http://dx.doi.org/10.1039/b807789d; PMID: 19082159
- Xu Y, Suzuki Y, Lönnberg T, Komiyama M. Human telomeric DNA sequence-specific cleaving by G-quadruplex formation. J Am Chem Soc 2009; 131:2871 - 4; http://dx.doi.org/10.1021/ja807313x; PMID: 19209856
- Lönnberg T, Aiba Y, Hamano Y, Miyajima Y, Sumaoka J, Komiyama M. Oxidation of an oligonucleotide-bound Ce(III)/multiphosphonate complex for site-selective DNA scission. Chemistry 2010; 16:855 - 9; PMID: 19938010
- Aiba Y, Lönnberg T, Komiyama M. Manipulation of single-stranded DNA by using an artificial site-selective DNA cutter composed of cerium(IV)/EDTA and phosphonate-oligonucleotide conjugates. Chem Asian J 2011; 6:2407 - 11; http://dx.doi.org/10.1002/asia.201100238; PMID: 21755600
- Tsien RY. The green fluorescent protein. Annu Rev Biochem 1998; 67:509 - 44; http://dx.doi.org/10.1146/annurev.biochem.67.1.509; PMID: 9759496
- Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science 2003; 300:87 - 91; http://dx.doi.org/10.1126/science.1082520; PMID: 12677058
- Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev Cell 2003; 4:295 - 305; http://dx.doi.org/10.1016/S1534-5807(03)00060-1; PMID: 12636912
- Aiba Y, Sumaoka J, Komiyama M. Artificial DNA cutters for DNA manipulation and genome engineering. Chem Soc Rev 2011; 40:5657 - 68; http://dx.doi.org/10.1039/c1cs15039a; PMID: 21566825
![Figure 2. Gel shift assay for the confirmation of invasion complex composed of DNA and ligand-modified pcPNAs. Invasion conditions; [double-stranded DNA (119 bp)] = 50 nM, [each of pcPNAs] = 200 or 400 nM and [Hepes (pH 7.0)] = 5 mM at 50°C for 1 h.](/cms/asset/7bae30c4-31c8-4f4d-a8e9-50dfeb093de6/kdpx_a_10920727_f0005.gif)