Abstract
PNAs conjugated to carrier peptides have been employed for the targeting of miRNA precursor, with the aim to develop molecules able to interfere in the pre-miRNA processing. The capability of the molecules to bind pre-miRNA has been tested in vitro by fluorescence assayes on Thiazole Orange labeled molecules and in vivo, in K562 cells, evaluating the amount of miRNA produced after treatment of cells with two amounts of PNAs.
Keywords: :
Introduction
Pre-miRNAs are RNA hairpins, ≈70 bases long, generated in the nucleus starting from primiRNAs after the cleavage operated by the nuclease Drosha. Pre-miRNAs are exported into the cytoplasm by exportin 5, where they are further processed by the nuclease Dicer to give the mature miRNAs.Citation1,Citation2 It has been demonstrated that miRNAs regulate in a post transcriptional fashion several eukaryotic genes, being involved in numerous processes such as cell proliferation, cell differentiation, apoptosis and also in the progress of diseases as cancer, cardiovascular disorders and Alzheimer.Citation3-Citation6Oligonucleotides or analogs perfectly complementary to the target miRNA (the antagomiRs) have been successfully employed to interfere in the miRNA function, allowing for the discovery of the functions of miRNA in cells.Citation7-Citation9 Recently antagomiRs have also been identified as tools to inhibit the function of those miRNAs identified to be implicated in human diseases.Citation10,Citation11 AntagomiRs targeting multiple miRNAs have been proposed for the interference in the miRNA function;Citation12 silencing of miRNA-10b by antagomiRs has been reported to suppress lung metastasis in a tumor model in mice.Citation13 Chemically modified nucleic acid have also been investigated as antagomiRs.Citation14,Citation15 LNACitation16,Citation17 and PNACitation18 have been employed for the inhibition of the miRNA-122, a liver specific miRNA upregulated in HIV-1 infected cells.Citation19,Citation20 PNA based antagomiR efficiently inhibited the function of miRNA 155 in primary B cells.Citation21 Recently, it has been demonstrated that a PNA oligomer conjugated to a polyarginine carrier peptide inhibits the activity of miRNA-210, a miRNA associated with hypoxia, upregulating its mRNA target and ultimately resulting in a decrease of the γ-globin mRNA.Citation22
In this work we propose a new approach to interfere in the miRNA function, based on the use of molecules able to bind the precursor of miRNA, pre-miRNA. In the pre-miRNA bases belonging to the stem are not perfectly complementary; therefore it is reasonable to think that the RNA duplex formed after pairing of mismatched bases has a low stability and may be opened by a high affinity exogenous modified oligonucleotide. Starting from this observation, we hypothesized that the mismatched duplex of the pre-miRNA could be easily opened by PNA oligomers, perfectly complementary to the sense strand of the pre-miRNA. PNAs are in fact the optimal candidates to this aim, as they show high specificity of binding toward complementary RNAs; the ability to open duplex complexes has been widely demonstrated on DNA duplexes.Citation23-Citation25 PNAs designed to bind the pre-miRNA target are expected to modify the pre-miRNA structure and give strand invasion; this will likely result in the inhibition of the recognition and processing by Dicer, which will ultimately prevent the production of the mature miRNA. The double localization of pre-miRNA (nucleus and cytoplasm) provides a double chance for its recruiting, enhancing the probability of the anti-premiR to successfully execute its job. Finally, anti-premiR are not expected to interfere in the RT-PCR experiments which are usually performed to evaluate the amount of miRNA produced.
We designed a PNA anti-premiRs perfectly complementary to the sense strand of pre-miRNA 210, in the region which is complementary to the mature miRNA. We conjugated the PNA to two different carrier peptides, in order to increase its cellular uptake. We choose carrier peptides as the fragment 48–60 of the HIV TAT and the Nuclear Localization Signal, reported able to carry PNA oligomers respectively in the cytoplasm and in the nucleus.Citation26-Citation28 In order to verify the ability of the designed PNA to give strand invasion on the pre-miRNA, we conjugated the PNA to the thiazole orange (TO), a probe which lights-up upon hybridization. Fluorescence experiments performed with the TO-modified PNA revealed that the TAT-PNA oligomer efficiently binds the pre-miRNA in vitro. PNAs were also conjugated to FITC and employed in FACS experiments for a preliminary evaluation of their cellular uptake. Finally the amount of miRNA produced in cells K562 treated with the anti-premiR was evaluated by quantitative RT-PCR.
Results
Synthesis of the PNA oligomers
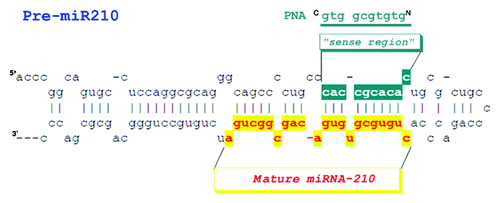
The PNA sequence was designed to bind the pre-miRNA strand region complementary to the mature miRNA (). The region of the pre-miRNA which is targeted by our PNA shows two mismatches. For our studies we choose the precursor of miRNA-210, a miRNA induced by hypoxia and associated to an enhancement in the expression of γ globin genes in differentiating erythroid cells.Citation29,Citation30
Peptides and PNA were obtained by Fmoc chemistry solid phase synthesis following standard procedures.Citation31,Citation32 Thiazole orange -PNA conjugates were synthesized on solid phase employing a synthetic carboxy functionalized TO derivative.Citation33 FITC derivatives were obtained coupling at the N-terminus of the peptide-PNA oligomers, the amino-hexanoic linker first and FITC then. All products were purified by RP-HPLC and characterized by LC-MS. PNA sequences are reported in .
Table 1. Sequences of the PNA, peptides and DNA
Strand invasion experiments
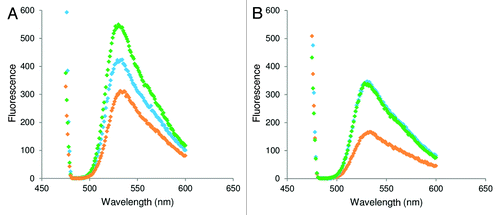
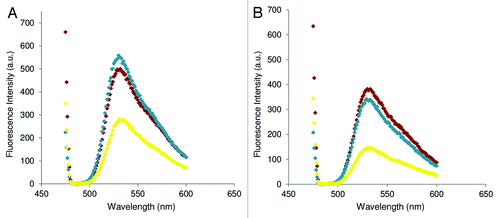
Fluorescence experiments were performed on TO-PNA-TAT to assess the ability of our molecules to bind the pre-miRNA. The TO-PNA-TAT was incubated with the DNA duplex (DNA/DNAc) (see for sequences) at 25°C and 37°C in a 1:1 molar ratio and the variation in the fluorescence emission, (excitation set at 470 nm) with respect to the single strand TO-PNA-TAT was monitored (). An increase in the fluorescence signal was observed, consistent with the hybridization of PNA to the DNA duplex at both temperatures. As a control, measurements of fluorescence were performed also on the preformed TO-PNA -TAT /DNAc duplex, to detect the fluorescence intensity displayed when the PNA fully hybridizes the DNA. Spectra for the preformed duplex and invasion complex were found superimposable at 37°C, while at 25°C the fluorescence intensity for the strand invasion complex was found lower as compared with that of the pre-formed duplex, suggesting that the efficiency of invasion is temperature dependent. A lower intensity in the emission of the probe was observed at 37°C. To investigate the effect of the peptide and temperature on the efficiency of strand invasion, a different peptide (K4) was conjugated to the PNA. The TO-PNA-K4 construct was employed in strand invasion experiments: at 25° and 37°C the fluorescence intensity of the strand invasion complex and the pre-formed TO-PNA-K4/DNAc duplex was very similar ().
miRNA downregulation
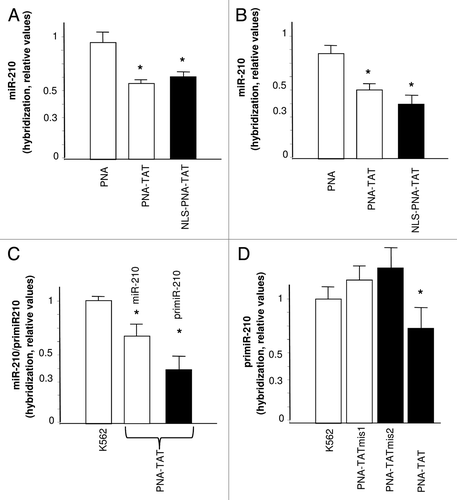
The first step necessary for obtaining anti-premiR activity in cells is their effective cellular uptake.Citation10,Citation22,Citation34 Unfortunately, it is firmly established that the delivery of unmodified PNAs to target cells is not efficientCitation35-Citation38 Therefore, we first determined whether the PNA-TAT and NLS-PNA-TAT bind to K562 cells. shows the FACS analysis performed using FITC-PNA-TAT, and FITC-NLS-PNA-TAT after 48 h incubation of K562 cells in the absence (-), or in the presence of 2 μM concentrations of the fluorescein-labeled molecules. We previously demonstrated that K562 cell system is very useful for studies of PNA uptake, when these molecule are administered at 2 μM for 48 h.Citation22 shows that when the PNA is vehiculated by TAT and NLS peptides, it is readily incorporated into K562 cells. While the FACS analysis are expected to represent true internalization of PNAs assayed by fluorescence microscopy, studies based on confocal analyses are required to determine the intracellular distribution of these PNAs. The K562 cell line has proved to be an excellent experimental system for evaluating the activity of molecules against miRNA-210, since the expression of this microRNA is increased following mithramycin (MTH) induced erythroid differentiation.Citation30 shows the quantification by real time PCR of miRNA-210 in MTH-induced K562 cells treated for 48 h with 1 μM and 2 μM PNA-TAT, and NLS-PNA-TAT. Both PNA-TAT, and NLS-PNA-TAT inhibit the miRNA-210 specific hybridization signal (); this result is compatible with inhibition of accumulation of miRNA-210 in MTH-treated K562 cells treated with these TAT and TAT/NLS conjugated anti-premiR PNAs. Furthermore, shows that treatment with PNA-TAT leads to inhibition of miR-210 as well as primiR-210 hybridization signals. Despite the fact that unrelated PNAs do not display non-specific effect on RT-PCR (data not shown), since PNA oligomers can be potent and specific inhibitors of PCR, we used mutant PNA-TAT oligomers. Interestingly, no effect was found using the mutated PNA-TAT molecules PNA-TAT mis1 and PNA-TAT mis2 ().
Figure 4. FACS analysis showing the uptake of PNA-TAT and NLS-PNA-TAT after 48 h incubation of K562 cells in the absence (-), or in the presence of 2 μM concentrations of the fluorescein-labeled FITC-PNA-TAT [(A) pink line], and FITC-NLS-PNA-TAT [(A) blue line] molecules. In (B) the quantitative determinations are shown, relative to replicate experiments (n = 4).
![Figure 4. FACS analysis showing the uptake of PNA-TAT and NLS-PNA-TAT after 48 h incubation of K562 cells in the absence (-), or in the presence of 2 μM concentrations of the fluorescein-labeled FITC-PNA-TAT [(A) pink line], and FITC-NLS-PNA-TAT [(A) blue line] molecules. In (B) the quantitative determinations are shown, relative to replicate experiments (n = 4).](/cms/asset/eeaec263-4ac7-4091-80ca-a149e4cbc7a2/kdpx_a_10920911_f0004.gif)
Figure 5. (A and B) Quantification by real time PCR of miRNA-210 in Mithramycin-induced K562 cells. Effects of treatment with anti-premiR PNA, PNA-TAT, and NLS-PNA-TAT, 1 μM (A) and 2 μM (B) on miRNA-210 content in cells cultured for 48 h in the presence of MTH. C. Effects on miR-210 and primiR-210 of 72 h treatment of K562 cells with 2 μM PNA-TAT. D. Effects of 24 h treatment with 1 μM PNA-TAT-mis1, PNA-TATmis2 and PNA-TAT on primiR-210. The data represent the average ± SD (n = 3) (* p < 0.05).
Discussion
The discovery of miRNAs significatively widened the comprehension of the mechanisms at the base of gene regulation, offering to scientist new potential targets to interfere in such process. Several studies have been focused on the development of molecules able to bind the mature miRNA, inhibiting the successive binding of the miRNA to its mRNA target. In a very recent paper the precursor of miRNA-205 was targeted by morpholino oligonucleotides in zebrafish embrios. It was demonstrated that morpholino targeting either the 5' and the 3' end of the pri-miRNA, respectively the Drosha and Dicer cleavage site, completely knocked down miRNA-205.Citation39 Antagomirzymes based on DNA and LNA have recently been proposed for the inhibition of miRNA function, and have been demonstrated to silence miRNA-371 and miRNA-373 in vitro and in vivo.Citation40 Recently, it has been reported that targeting mature miRNA-210 by PNAs results in a decrease in miRNA-210 levels as measured by RT-PCR and deep alteration of the miRNA-210-regulated erythroid functions.Citation22 We here propose a new tool for the inhibition of the miRNA function based on PNAs perfectly complementary to a small region of the pre-miRNA, which we will refer to as anti-premiR. The anti-premiRs are expected to interfere with the recognition of the pre-miRNA by Dicer. The mechanism by which Dicer cuts the pre-miRNA is very complex and involves the recognition of the loop by the helicase domain, followed by an inspection of the distance between the 3′ overhang and the terminal loop by the PAZ domain and finally by the processing operated by the RNase domains.Citation41 Binding of the anti-premiR to the pre-miRNA should open locally the duplex and result in either the spreading of the pre-miRNA duplex or in the formation of a P loop containing the PNA/RNA duplex. In both cases the pre-miRNA structure will be modified and the processing should not occur. Such anti-premiRNA may also be useful in investigations on the pre-miRNA processing, as it has been recently demonstrated that pre-miRNA are subject to several regulatory processes involving 3' and 5' cleavage and polyuridylation.Citation42 PNAs were chosen due their high affinity toward nucleic acids, and their stability to degradation by nucleases and proteases, which renders them very useful tools for in vivo applications. The PNA sequence was conjugated to different carrier peptides, chosen to deliver the PNA in the cytoplasm and in the nucleus. All molecules were entirely obtained by solid phase synthesis, purified by HPLC and characterized by electrospray mass analysis. In order to assess the viability of our approach, we had first to demonstrate in vitro that the PNA was able to bind the duplex corresponding to the pre-miRNA. We employed fluorescence experiments on PNA-peptide conjugated to the light up probe thiazole orange (TO) TO-PNA-TAT. We tested this conjugate as we assume that the peptide sequence does not grossly affect the efficiency of the invasion. The TAT peptide bears 9 positive charges and the TAT-NLS 13, therefore it is reasonable to think that they will show similar behavior in strand invasion experiments. In fact, it is reported in the literature that rates of strand invasion by PNA are increased after conjugation of the PNA to a positively charged peptide and increase at 22 and 37°C when the charge of the peptide is doubled.Citation23 A DNA duplex, in lieu of a RNA duplex, was employed for the measurements, as it is more stable to degradation and therefore easy to work with. The DNA strand mimicking the pre-miRNA sense strand was longer as compared with that of pre-miRNA 210 to make sure the TO dye will light up. It has been reported, in fact, that TO dye switches on when the hybridization occurs between complementary sequences at least 28 bases long.Citation43,Citation44 According to data reported in the literature, the fluorescence emission of free and hybridized probe is different.Citation45 In order to assess the stability of the DNA duplex in the experimental conditions chosen for the strand invasion experiments, the melting temperature of the hybrid was measured and was found to be 53.2°C. The fluorescence emission of the TO-PNA-TAT conjugate after incubation with the DNA duplex increases with respect to the emission of the TO-PNA-TAT alone both at 25 and 37°C, and this strongly supports the binding of the PNA to the duplex. The comparison of the spectra recorded for the duplex formed between the TO-PNA-TAT and the DNAc, and that obtained upon incubation of the DNA duplex with the TO-PNA-TAT conjugate further confirms the result. The difference between the fluorescence intensity of the strand invasion complex at 25 and 37°C argues for a difference in the efficiency of invasion at different temperatures, which is independent by the DNA duplex stability. A decrease in the intensity of all TO conjugates signals was observed when the spectra were recorded at 37°C. To further assess the dependence of the binding efficiency on the temperature and on the carrier peptide we performed experiments on a control PNA-peptide conjugate, having 4 positive charges, K4. Interestingly in this case we observe very similar intensities for the strand invasion complex and the duplex, at 25 and 37°C. These results suggest that the efficiency of the strand invasion does not depend only on the temperature; the nature of the peptide, affects the process at 25°C, while at 37°C all PNA-peptide conjugates behave in a similar fashion.
It is worth noting that the PNA sequence employed is not a polypirimidine sequence, but is nonetheless able to bind to the duplex mimicking the pre-miRNA; in consideration of its sequence, the PNA will not be able to form a triplex, but in principle it should form a duplex with the pre-miRNA strand sense, displacing the complementary strand. These results are in line with those reported in the literature, showing strand invasion on DNA duplexes also by mixed sequence PNA. Having demonstrated the binding of anti-premiR to the pre-miRNA mimicking duplex, we next explored the ability of our molecules to be taken up by cells and to downregulate miRNA expression in vivo. Evaluation of the PNA uptake was performed by FACS using PNAs conjugated to fluorescein. PNAs conjugated to carrier peptides as TAT and TAT-NLS display efficient uptake by K562 cells. This result is in line to what reported in the literature.Citation34 Quantitative analysis of the miRNA produced after treatment of the cells with PNA-peptide anti-premiR reveals a reduction of the amount of miRNA 210 in treated cells, consistent with the inhibition of pre-miRNA processing mediated by the anti-premiR PNAs. These data were supported by the finding that PNA-TAT inhibits also primiR-210 and this effect was found fairly specific, since the two mutant PNA-TAT molecules (PNA-TATmis1 and PNA-TATmis2) were not effective. Effects observed of PNA on pri-miRNA lead to the hypothesis that the synthetic PNA is able to bind primary miRNA and therefore likely reaches the cell nucleus. While analysis of other PNA mutants are required to draw conclusions about sequence specificity, these results encourage further investigations aimed at the molecular characterization of the effect of molecules targeting miRNA precursors, also in comparison to the effects of molecules targeting mature miRNAs.
The use of molecules targeting pre-miRNAs represents a new approach for the interference in miRNA function. This has important implications from a theoretical point of view, helping in elucidating miRNA functions. In addition, in consideration of the involvement of microRNAs in human pathologies, our findings are of interest also with respect to biomedical applications aimed at the design of more effective anti-miRNA molecules to be employed in the so called “miRNA therapeutics.” In this paper we firmly demonstrate that PNA oligomers conjugated to carrier peptides targeting the sense strand of the pre-miRNA 210 are able to strand invade the miRNA precursor duplex in vitro and in vivo. The biological effect of these anti-premiR molecules has been analyzed on K562 cells, demonstrating that the anti-premiR PNAs were able to reduce the amount of miRNA-210. Further investigations will be performed to validate the effect(s) of such molecules on other pre-miRNAs and to determine the effect of PNA based anti-premiR on all the other biological functions controlled by miRNA-210.
Material and Methods
Materials and general procedures
Protected N-Fmoc-amino acid derivatives, acetic anhydride and coupling reagents have been purchased from Novabiochem. Activators HBTU, HOBT and HATU were purchased at Inbios (Italy). Fmoc-PNA-cytosine(Bhoc)-OH, Fmoc-PNA-thymine-OH, Fmoc-PNA-guanine(Bhoc)-OH, Fmoc-PNA-adenine(Bhoc)-OH were obtained by Link Technologies. Acetonitrile for LC-MS, N,N-dimethylformamide for solid phase synthesis, DIPEA, Dichloromethane were from Romil Pure Chemistry, N-methylmorpholine from Fluka and piperidine from Biosolve. Fmoc-PAL-PEG-PS (0.18 mmol/g) resin was from Applied Biosystems. All other chemicals were supplied by Sigma-Aldrich and were used without other purification. Preparative purification was performed on a Shimadzu LC-8A, equipped with a SPD-M10 AV diode array detector. Preparative HPLC was performed on a Phenomenex Jupiter 10μm Proteo (90Ǻ 250 × 10.00 mm) column with a flow rate of 5 mLmin−1. 1H NMR spectra were recorded on a Varian 400 MHz spectrometer. Proton chemical shifts are reported in ppm (δ) relative to the solvent reference (d6-DMSO, d 2.50). Data are reported as follows: chemical shift {multiplicity [singlet (s), doublet (d), triplet (t), quartet (q) and multiplet (m)], integration}. Carbon NMR spectra were recorded on a Varian 500 (125 MHz) spectrometer with complete proton decoupling. Carbon chemical shifts were reported in ppm (δ) relative to TMS with the respective solvent resonance as the internal standard (DMSO, 39.5). LC-MS analyses were performed on a LC-MS Thermo Finnigan with an electrospray source (MSQ) on a Phenomenex Jupiter 5 μ C18 (300 Å, 150 × 460 mm) column with a flow rate of 0.8 mLmin−1 at room temperature. Fluorescence spectroscopy was performed on a Varian Spectrophotometer.
Peptide-PNA conjugates synthesis
All peptides were synthesized by solid phase peptide synthesis as C-terminally amidated derivatives following standard Fmoc chemistry protocol on a Fmoc-PAL-PEG-PS resin (0.18 mmol/g).Citation31 PNAs were grown on the peptides anchored to the solid phase following procedures reported in the literature.Citation32 The conjugates were cleaved off the resin and deprotected by treatment with a solution of 78% TFA, m-cresol 20%, 2% TIS for 3 h at room temperature.
All conjugates were purified by RP-HPLC using a gradient of acetonitrile (0.1% TFA) in water (0.1% TFA) from 5 to 50% in 30 min and characterized by LC-MS, using a gradient of acetonitrile (0.05% TFA) in water (0.05% TFA) from 5% to 50% applied over 30 min.
PNA (Da): Calculated: 2814.6; [M+2H]2+: 1408.3; [M+3H]3+: 939.2; [M+4H]4+: 704.6. Found: 2815.0; [M+2H]2+: 1408.5; [M+3H]3+: 939.3; [M+4H]4+: 704.7.
PNA -TAT (Da): calculated: [M+3H]3+: 1549.4; [M+4H]4+: 1162.2; [M+5H]5+: 929.9; [M+6H]6+: 775.1; found: [M+3H]3+: 1551.5; [M+4H]4+: 1163.8; [M+5H]5+: 931.3; [M+6H]6+: 776.3.
NLS-PNA-TAT (Da): calculated: [M+3H]3+: 1837.9; [M+4H]4+: 1378.7; [M+5H]5+: 1103.2; [M+6H]6+: 919.1; [M+7H]7+: 788.2; [M+8H]8+: 689.8; found: [M+3H]3+: 1839.2; [M+4H]4+: 1379.6; [M+5H]5+: 1103.9; [M+6H]6+: 920.1; [M+7H]7+: 788.8; [M+8H]8+: 690.3.
PNA-TAT mis1 (Da): calculated: [M+3H]3+: 1540.5; [M+4H]4+: 1155.6; [M+5H]5+: 924.7; found: [M+3H]3+: 1540.5; [M+4H]4+: 1155.6; [M+5H]5+: 931.3; [M+6H]6+: 924.7.
PNA-TAT mis2 (Da): calculated: [M+3H]3+: 1540.5; [M+4H]4+: 1155.6; [M+5H]5+: 924.7; found: [M+3H]3+: 1540.5; [M+4H]4+: 1155.6; [M+5H]5+: 931.3; [M+6H]6+: 924.7.
TO-PNA-Peptide conjugates synthesis
The TO dye was prepared according to procedures described in the literature. The dye was coupled to the PNA-peptide conjugates anchored to the resin. 2.5 equivalents of TO, 2.49 equivalents of HOBT/HBTU (0.45 M solution in DMF) and 3.5 equivalents of NMM were incubated with the resin for 1 h. Cleavage and deprotection of the conjugates were performed as described earlier. Purification and characterization was performed following the protocols described for peptide-PNA conjugates.
TO: UV-Vis: λmax (H2O) 502 nm (ε = 66400 M−1cm−1); 1H NMR (400 MHz, DMSO) d 1.38 (m, 2H, CH2), 1.56 (m, 2H, CH2), 1.86 (m, 2H, CH2), 2.21 (t, J = 7.2 Hz, 2H, CH2), 4.03 (s, 3H, NCH3), 4.61 (t, J = 7.2 Hz, 2H, NCH2), 6.94 (s, 1H, CH), 7.40 (m, 2H, Ar), 7.62 (m, 1H, Ar), 7.77 (m, 2H, Ar), 8.00 (m, 1H, Ar), 8.07 (d, J = 4 7.69 Hz, 1H, Ar), 8.15 (d, J = 9.16 Hz, 1H, Ar), 8.65 (d, J = 6.96 Hz, 1H, Ar), 8.81 (d, J = 8.62 Hz, 1H, Ar); 13C NMR (125 MHz, DMSO): d 175.6, 161.2, 149.7, 145.6, 141.7, 138.2, 134.5, 129.4, 128.0, 127.0, 126.0, 125.4, 125.1, 124.1, 119.3, 114.2, 109.0, 89.3, 55.2, 35.1, 34.9, 29.8, 26.7, 25.3
TO-PNA-TAT (Da): calculated: [M+3H]3+: 1678.3; [M+4H]4+: 1259.1; [M+5H]5+: 1007.6; [M+6H]6+: 839.6; [M+7H]7+: 719.8; [M+8H]8+: 630.1; [M+9H]9+: 560.1; found: [M+3H]3+: 1676.2; [M+4H]4+: 1257.4; [M+5H]5+: 1006.2; [M+6H]6+: 838.6; [M+7H]7+: 718.9; [M+8H]8+: 629.6; [M+9H]9+: 597.0.
TO-PNA-K4 (Da): calculated: [M+3H]3+: 1239.1; [M+4H]4+: 929.6; [M+5H]5+: 743.9; [M+6H]6+: 620.1; found: [M+3H]3+: 1239.0; [M+4H]4+: 929.2; [M+5H]5+: 743.2; [M+6H]6+: 620.0.
FITC-PNA-Peptide conjugates synthesis
To the resin bound PNA-peptide conjugates, the Fmoc-ε-Ahx-OH was coupled following standard peptide synthesis protocols. After removal of the N-terminal Fmoc, the resin was incubated with five equivalents of FITC dissolved in DMF in the presence of seven equivalents of NMM for 1 h, two times. Cleavage and deprotection of the conjugates were performed as described earlier. Purification and characterization was performed following the protocols described for peptide-PNA conjugates.
FITC-PNA-TAT (Da): calculated: [M+3H]3+: 1716.1; [M+4H]4+: 1287.3; [M+5H]5+: 1030.0; [M+6H]6+: 858.5; [M+7H]7+: 736.1; [M+8H]8+: 644.1. [M+9]9+: 572.7; [M+10]10+: 515.5; found: [M+3H]3+: 1709.8; [M+4H]4+: 1282,6; [M+5H]5+: 1026.7; [M+6H]6+: 855.6; [M+7H]7+: 733.0; [M+8H]8+: 642.0. [M+9]9+: 570.3; [M+10]10+: 513.7.
FITC-NLS-PNA-TAT (Da): calculated: [M+4H]4+: 1504.6; [M+5H]5+: 1203.4; [M+6H]6+: 1003.1; [M+7H]7+: 859.9; [M+8H]8+: 752.5; [M+9]+9: 669.0; [M+10H]10+: 602.2; [M+11H]11+: 547.6; [M+12H]12+: 502.0; [M+13]13+: 463.4; found: [M+4H]4+: 1498.0; [M+5H]5+: 1198.5; [M+6H]6+: 997.6; [M+7H]7+: 856.4; [M+8H]8+: 749.0; [M+9]+9: 665.7; [M+10H]10+: 599.0; [M+11H]11+: 544.3; [M+12H]12+: 499.7; [M+13]13+: 461.0.
Fluorescence
DNA/DNAc duplex was obtained mixing 1μM of DNA and 1μM of DNAc in Phosphate buffer 10mM, 100 mM NaCl, pH 7.0. After the annealing reaction (90°C 5 min, 4°C overnight), strand invasion experiments were performed incubating the TO-PNA–TAT with the duplex at 25°C or 37°C for 30 min, adding 1μM of probe to 1μM of duplex solution (TO-PNA-TAT+ DNA/DNAc ratio 1:1) in a final volume of 2.5 mL (λex = 470 nm, slitex = 2.5, slitem = 10.0).
CD melting
Thermic denaturation experiments were performed using 3 μM of DNA/DNAc duplex at a 1°C/min scan speed, following the CD signal at 276 nm.
FACS analysis
For determination of fluorescence intensity by FACScan (Becton Dickinson), cells were incubated with the indicated concentrations of PNAs for different length of time, harvested and washed. Then 1 × 105 cells were analyzed by the CellQuestTM version 3.3 software (Becton Dickinson), using the FL1 channel to detect fluorescence. The results were expressed as median fold, i.e., the ratio between the median fluorescence intensity values obtained in the presence and absence of treatment, respectively. A graphic presentation of data was finally obtained by histograms, showing the number of cells vs. the expressed fluorescence intensity.
Human cell lines and culture conditions
Human leukemia K562 cells were cultured in humidified atmosphere of 5% CO2/air in RPMI 1640 medium (SIGMA) supplemented with 10% fetal bovine serum (FBS, Analytical de Mori), 50 units/ml penicillin and 50 μg/ml streptomycin.Citation46 Mithramycin (MTH) was from Sigma. Stock solutions of MTH (100 μM) were stored at -20°C in the dark and diluted immediately before the use.Citation46,Citation47 Treatment of K562 cells with mithramycin (MTH) was performed by adding the appropriate drug concentrations at the beginning of the cultures (30,000 cells/ml were seeded).
RNA extraction
Cells were isolated, washed with PBS, lysed with Tri-reagentTM (Sigma Aldrich), according to manufacturer’s instructions. The isolated RNA was washed once with cold 75% ethanol, dried and dissolved in nuclease free pure water before use.Citation22
Real-time quantitative PCR
For microRNA quantification using real-time RT-PCR reagents, the primers and probes were obtained from Applied Biosystems.Citation22 Reverse transcriptase (RT) reactions were performed using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems); real-time PCR was performed according to the manufacturer’s protocols. 300 ng per sample were used for the assays. All RT reactions, including no-template controls and RT-minus controls, were performed in duplicate using the 7700 Sequence Detection System version 1.7 (Applied Biosystems). The relative expression was calculated using the comparative cycle threshold method and as reference U6 snRNA was used to normalize all RNA samples, since it remains constant in the assayed samples by miRNA-profiling and quantitative RT-PCR analysis, as previously reported.Citation22,Citation30
| Abbreviations: | ||
| PNA | = | Peptide Nucleic Acids |
| miRNA | = | microRNA |
| LNA | = | locked nucleic acid, MTH, mithramycin |
Acknowledgments
This work was partially supported by a grant from MIUR (PRIN2009 Prot 20093N774P). R.G. is granted by TELETHON (GGP10124), by Fondazione Cassa di Risparmio di Padova e Rovigo, by C.I.B. and by Associazione Veneta per la Lotta alla Talassemia.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843 - 54; http://dx.doi.org/10.1016/0092-8674(93)90529-Y; PMID: 8252621
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell 2004; 16:861 - 5; http://dx.doi.org/10.1016/j.molcel.2004.12.002; PMID: 15610730
- Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, et al. An analysis of human microRNA and disease associations. PLoS One 2008; 3:e3420; http://dx.doi.org/10.1371/journal.pone.0003420; PMID: 18923704
- Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007; 13:613 - 8; http://dx.doi.org/10.1038/nm1582; PMID: 17468766
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005; 132:4653 - 62; http://dx.doi.org/10.1242/dev.02073; PMID: 16224045
- Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res 2007; 101:1225 - 36; http://dx.doi.org/10.1161/CIRCRESAHA.107.163147; PMID: 18063818
- van Solingen C, de Boer HC, Bijkerk R, Monge M, van Oeveren-Rietdijk AM, Seghers L, et al. MicroRNA-126 modulates endothelial SDF-1 expression and mobilization of Sca-1(+)/Lin(-) progenitor cells in ischaemia. Cardiovasc Res 2011; 92:449 - 55; http://dx.doi.org/10.1093/cvr/cvr227; PMID: 21856785
- Goljanek-Whysall K, Sweetman D, Abu-Elmagd M, Chapnik E, Dalmay T, Hornstein E, et al. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc Natl Acad Sci U S A 2011; 108:11936 - 41; http://dx.doi.org/10.1073/pnas.1105362108; PMID: 21730146
- Guglielmelli P, Tozzi L, Bogani C, Iacobucci I, Ponziani V, Martinelli G, et al, AGIMM (AIRC-Gruppo Italiano Malattie Mieloproliferative) Investigators. Overexpression of microRNA-16-2 contributes to the abnormal erythropoiesis in polycythemia vera. Blood 2011; 117:6923 - 7; http://dx.doi.org/10.1182/blood-2010-09-306506; PMID: 21527532
- Gambari R, Fabbri E, Borgatti M, Lampronti I, Finotti A, Brognara E, et al. Targeting microRNAs involved in human diseases: a novel approach for modification of gene expression and drug development. Biochem Pharmacol 2011; 82:1416 - 29; http://dx.doi.org/10.1016/j.bcp.2011.08.007; PMID: 21864506
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005; 438:685 - 9; http://dx.doi.org/10.1038/nature04303; PMID: 16258535
- Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z, et al. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res 2009; 37:e24; http://dx.doi.org/10.1093/nar/gkn1053; PMID: 19136465
- Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 2010; 28:341 - 7; http://dx.doi.org/10.1038/nbt.1618; PMID: 20351690
- Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, et al. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res 2009; 37:70 - 7; http://dx.doi.org/10.1093/nar/gkn904; PMID: 19015151
- Ørom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 2006; 372:137 - 41; http://dx.doi.org/10.1016/j.gene.2005.12.031; PMID: 16503100
- Koshkin AA, Wengel J. Synthesis of Novel 2′,3′-Linked Bicyclic Thymine Ribonucleosides. J Org Chem 1998; 63:2778 - 81; http://dx.doi.org/10.1021/jo972239c; PMID: 11672157
- Obika S, Nanbu D, Hari Y, Morio K, In Y, Ishida T, et al. Synthesis of 2'-O,4'-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C-3,-endo sugar puckering. Tetrahedron Lett 1997; 38:8735 - 8; http://dx.doi.org/10.1016/S0040-4039(97)10322-7
- Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991; 254:1497 - 500; http://dx.doi.org/10.1126/science.1962210; PMID: 1962210
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 2007; 315:1579 - 82; http://dx.doi.org/10.1126/science.1136319; PMID: 17322031
- Fabani MM, Gait MJ. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA 2008; 14:336 - 46; http://dx.doi.org/10.1261/rna.844108; PMID: 18073344
- Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KG, et al. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res 2010; 38:4466 - 75; http://dx.doi.org/10.1093/nar/gkq160; PMID: 20223773
- Fabbri E, Manicardi A, Tedeschi T, Sforza S, Bianchi N, Brognara E, et al. Modulation of the biological activity of microRNA-210 with peptide nucleic acids (PNAs). ChemMedChem 2011; 6:2192 - 202; http://dx.doi.org/10.1002/cmdc.201100270; PMID: 22012891
- Zhang X, Ishihara T, Corey DR. Strand invasion by mixed base PNAs and a PNA-peptide chimera. Nucleic Acids Res 2000; 28:3332 - 8; http://dx.doi.org/10.1093/nar/28.17.3332; PMID: 10954602
- Hu J, Corey DR. Inhibiting gene expression with peptide nucleic acid (PNA)--peptide conjugates that target chromosomal DNA. Biochemistry 2007; 46:7581 - 9; http://dx.doi.org/10.1021/bi700230a; PMID: 17536840
- Pensato S, Saviano M, Bianchi N, Borgatti M, Fabbri E, Gambari R, et al. gamma-Hydroxymethyl PNAs: Synthesis, interaction with DNA and inhibition of protein/DNA interactions. Bioorg Chem 2010; 38:196 - 201; http://dx.doi.org/10.1016/j.bioorg.2010.06.002; PMID: 20643471
- Díaz-Mochón JJ, Bialy L, Watson J, Sánchez-Martín RM, Bradley M. Synthesis and cellular uptake of cell delivering PNA-peptide conjugates. Chem Commun (Camb) 2005; 3316 - 8; http://dx.doi.org/10.1039/b503777h; PMID: 15983659
- Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, et al. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem 2001; 276:5836 - 40; http://dx.doi.org/10.1074/jbc.M007540200; PMID: 11084031
- Tonelli R, Purgato S, Camerin C, Fronza R, Bologna F, Alboresi S, et al. Anti-gene peptide nucleic acid specifically inhibits MYCN expression in human neuroblastoma cells leading to cell growth inhibition and apoptosis. Mol Cancer Ther 2005; 4:779 - 86; http://dx.doi.org/10.1158/1535-7163.MCT-04-0213; PMID: 15897242
- Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of microRNA expression: the hypoxic component. Cell Cycle 2007; 6:1426 - 31; http://dx.doi.org/10.4161/cc.6.12.4410; PMID: 17582223
- Bianchi N, Zuccato C, Lampronti I, Borgatti M, Gambari R. Expression of miR-210 during erythroid differentiation and induction of gamma-globin gene expression. BMB Rep 2009; 42:493 - 9; http://dx.doi.org/10.5483/BMBRep.2009.42.8.493; PMID: 19712585
- Le Chevalier Isaad A, Papini AM, Chorev M, Rovero P. Side chain-to-side chain cyclization by click reaction. J Pept Sci 2009; 15:451 - 4; http://dx.doi.org/10.1002/psc.1141; PMID: 19455541
- Avitabile C, Moggio L, D'Andrea LD, Pedone C, Romanelli A. Development of an efficient and low-cost protocol for the manual PNA synthesis by Fmoc chemistry. Tetrahedron Lett 2010; 51:3716 - 8; http://dx.doi.org/10.1016/j.tetlet.2010.05.026
- Mahon KP Jr., Ortiz-Meoz RF, Prestwich EG, Kelley SO. Photosensitized DNA cleavage promoted by amino acids. Chem Commun (Camb) 2003; 1956 - 7; PMID: 12932051
- Fabbri E, Brognara E, Borgatti M, Lampronti I, Finotti A, Bianchi N, et al. miRNA therapeutics: delivery and biological activity of peptide nucleic acids targeting miRNAs. Epigenomics 2011; 3:733 - 45; http://dx.doi.org/10.2217/epi.11.90; PMID: 22126292
- Berthold PR, Shiraishi T, Nielsen PE. Cellular delivery and antisense effects of peptide nucleic acid conjugated to polyethyleneimine via disulfide linkers. Bioconjug Chem 2010; 21:1933 - 8; http://dx.doi.org/10.1021/bc1003586; PMID: 20873710
- Zhou P, Dragulescu-Andrasi A, Bhattacharya B, O’Keefe H, Vatta P, Hyldig-Nielsen JJ, et al. Synthesis of cell-permeable peptide nucleic acids and characterization of their hybridization and uptake properties. Bioorg Med Chem Lett 2006; 16:4931 - 5; http://dx.doi.org/10.1016/j.bmcl.2006.06.052; PMID: 16809033
- Zhou P, Wang M, Du L, Fisher GW, Waggoner A, Ly DH. Novel binding and efficient cellular uptake of guanidine-based peptide nucleic acids (GPNA). J Am Chem Soc 2003; 125:6878 - 9; http://dx.doi.org/10.1021/ja029665m; PMID: 12783535
- Rasmussen FW, Bendifallah N, Zachar V, Shiraishi T, Fink T, Ebbesen P, et al. Evaluation of transfection protocols for unmodified and modified peptide nucleic acid (PNA) oligomers. Oligonucleotides 2006; 16:43 - 57; http://dx.doi.org/10.1089/oli.2006.16.43; PMID: 16584294
- Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol 2007; 5:e203; http://dx.doi.org/10.1371/journal.pbio.0050203; PMID: 17676975
- Jadhav VM, Scaria V, Maiti S. Antagomirzymes: oligonucleotide enzymes that specifically silence microRNA function. Angew Chem Int Ed Engl 2009; 48:2557 - 60; http://dx.doi.org/10.1002/anie.200805521; PMID: 19229913
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol 2011; 18:1153 - 8; http://dx.doi.org/10.1038/nsmb.2125; PMID: 21926993
- Burroughs AM, Kawano M, Ando Y, Daub CO, Hayashizaki Y. pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5′/3′ arm variation including concomitant cleavage and polyuridylation patterns. Nucleic Acids Res 2012; 40:1424 - 37;; PMID: 22058130
- Svanvik N, Westman G, Wang D, Kubista M. Light-up probes: thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal Biochem 2000; 281:26 - 35; http://dx.doi.org/10.1006/abio.2000.4534; PMID: 10847607
- Tonelli A, Tedeschi T, Germini A, Sforza S, Corradini R, Medici MC, et al. Real time RNA transcription monitoring by Thiazole Orange (TO)-conjugated Peptide Nucleic Acid (PNA) probes: norovirus detection. Mol Biosyst 2011; 7:1684 - 92; http://dx.doi.org/10.1039/c0mb00353k; PMID: 21399831
- Nygren J, Svanvik N, Kubista M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers 1998; 46:39 - 51; http://dx.doi.org/10.1002/(SICI)1097-0282(199807)46:1<39::AID-BIP4>3.0.CO;2-Z; PMID: 9612138
- Gambari R, Fibach E. Medicinal chemistry of fetal hemoglobin inducers for treatment of beta-thalassemia. Curr Med Chem 2007; 14:199 - 212; http://dx.doi.org/10.2174/092986707779313318; PMID: 17266579
- Fibach E, Bianchi N, Borgatti M, Prus E, Gambari R. Mithramycin induces fetal hemoglobin production in normal and thalassemic human erythroid precursor cells. Blood 2003; 102:1276 - 81; http://dx.doi.org/10.1182/blood-2002-10-3096; PMID: 12738678