Abstract
PROPPINs are a family of PtdIns3P and PtdIns(3,5)P2-binding proteins. The crystal structure now unravels the presence of two distinct phosphoinositide-binding sites at the circumference of the seven bladed β-propeller. Mutagenesis analysis of the binding sites shows that both are required for normal membrane association and autophagic activities. We identified a set of evolutionarily conserved basic and polar residues within both binding pockets, which are crucial for phosphoinositide binding. We expect that membrane association of PROPPINs is further stabilized by membrane insertions and interactions with other proteins.
PROPPINs (β-propellers that bind phosphoinositides), a WD40-protein family highly conserved from yeast to human, specifically bind PtdIns3P and PtdIns(3,5)P2 via a FRRG motif. The three homologous Saccharomyces cerevisiae PROPPINs Atg18, Atg21 and Hsv2 differently affect autophagy subtypes. Atg18 is a core autophagy protein, which acts during autophagosome biogenesis as an adaptor molecule, which recognizes the presence of PtdIns3P at autophagosome precursors. Atg18 mediates, in a complex with Atg2 and the integral membrane protein Atg9, the retrieval of Atg9 during autophagosome maturation. Atg21 is restricted to specific autophagic subtypes such as the Cvt pathway. The role of Hsv2 is hardly understood; its absence only partially affects piecemeal microautophagy of the nucleus. The yeast PROPPINs are also recruited in a PtdIns3P-dependent manner to endosomes. Atg18 is further part of a PtdIns(3,5)P2-dependent protein complex at the vacuolar membrane, where it regulates the PtdIns3P 5-kinase Fab1 and vesicular transport back to the Golgi apparatus.
PROPPINs Contain Two Putative Lipid-Binding Sites
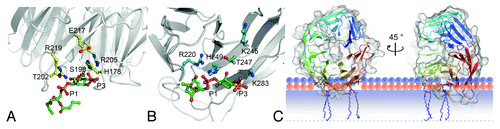
Whereas PtdIns3P-binding domains such as the FYVE- and PX-domains are structurally well characterized, it remained elusive how PROPPINs recognize phosphoinositides. We now solved the 3.0-Å crystal structure of Kluyveromyces lactis Hsv2. It folds into a seven-bladed β-propeller, each blade containing four antiparallel β-strands (A to D, from the inside to the outside). The last blade is completely formed by the carboxy terminus in a rare nonvelcro closure. We found that residues larger than the glycine of the phosphoinositide-binding FRRG-motif would result in steric hindrance with the second arginine of the double-arginine motif, explaining the conservation of this residue. The FRRG-motif localizes to the circumference of the β-propeller. Astonishingly, the two arginines of the motif are located within two distinct basic pockets. In our structure each is occupied by a sulfate ion, which might indicate the positions of phosphate groups. The phosphates of PtdIns3P are 6.5-Å apart, while the sulfates in our structure have a distance of 16.2-Å. The sulfates are further separated by strand D of blade 5 and the loop to strand A of blade 6. We thus speculated that PROPPINs might contain two distinct phosphoinositide binding sites.
Membrane Association and Biological Activity Require both Binding Pockets
Both potential binding pockets are formed by a set of polar and charged residues, which are highly conserved both among the PROPPINs within one organism and among different species. We replaced these residues in S. cerevisae Atg18 (ScAtg18) and ScHsv2 by alanine and analyzed the membrane association of GFP fusion proteins in S. cerevisiae cells. Additionally, we used a liposome flotation assay with PtdIns3P-containing liposomes. This led to the identification of a set of residues in each binding site, which are important for membrane association (). Single mutations within either site 1 or 2 only partially affect macroautophagy and the Cvt pathway, but a combination of one mutation in each site leads to more severe effects. This shows that both binding sites act in concert.
Figure 1. Docking of PtdIns3P into binding site 1 (A) and 2 (B) of K. lactis Hsv2. (C) Docking studies suggest binding of PROPPINs perpendicular to the membrane. Note that parts of the protein are predicted to insert into the membrane. The phosphates of the phospholipids are shown in orange; their polar groups in violet. The shaded bar depicts the hydrophobic part of the membrane. Only the upper membrane leaflet is shown. The figures are adopted from Krick et al. PNAS 109:E2042.
Both Sites Bind PtdIns3P
Similar to PROPPINs, the PH domain of Slm1 contains a double-arginine motif, where each arginine is part of a distinct lipid-binding site. Interestingly, for Slm1 one site binds phosphoinositides, but the other binds phosphorylated sphingolipids. To quantitatively analyze the lipid specificity of both PROPPIN sites we first used reflectometric interference spectroscopy. In this approach, binding of isolated GST-ScHsv2 to a silicon wafer coated with a lipid bilayer was measured spectroscopically. We observe no binding to a lipid bilayer consisting of only phosphatidylcholine, but effective binding (Kd 1.3 ± 0.2 µM) in the presence of 3% PtdIns3P. Single mutations in either of the two sites abolish binding to PtdIns3P-containing bilayers, clearly demonstrating that both sites bind phosphoinositides. In addition, we determined through isothermal titration calorimetry (ITC) that the binding of ScHsv2 to PtdIns3P on liposomes occurs with a molar ratio of 0.50 ± 0.06 and a Kd of 0.67 ± 0.04 µM. This confirms the presence of two individual phosphoinositide binding sites in PROPPINs. We assume that the lower Kd determined in the ITC measurements is caused by the presence of additional lipids such as phosphatidylethanolamine in the liposomes and differences in membrane curvature.
Our data demonstrate that the affinity of a single lipid-binding site is not enough for effective membrane binding; therefore, PROPPINs combine two sites in close proximity. Indeed, most FYVE domains must dimerize for efficient binding to PtdIns3P at endosomes. Our in silico docking studies suggest that perpendicular binding of the propeller to membranes further results in membrane insertion of the loops connecting strands 6C with 6D and 7C with 7D (). This generates additional hydrophobic and electrostatic interactions and strengthens membrane association.
What Differentiates Binding of PROPPINs to PtdIns3P or PtdIns(3,5)P2?
Our protein lipid overlay assays and analysis of PtdIns(3,5)P2-vacuole homeostasis combined with in silico docking suggest that both sites can bind both types of phosphoinositides. We assume that interactions with other proteins are one of the main reasons for the recruitment of PROPPINs to different membranes. Indeed, the function of WD40 β-propellers as protein interaction scaffolds is well established. Accordingly, the PtdIns3P-dependent PAS-localization of Atg18 requires the presence of the peripheral membrane protein Atg2 and the integral membrane protein Atg9. On the other side the PtdIns(3,5)P2-dependent complex at the vacuolar membrane contains, in addition to Atg18, the PtdIns3P 5-kinase Fab1, Fig4, Vac7 and Vac14.
Acknowledgments
This work was funded by the DFG through the SFB860 (to M.T. and K.K.).