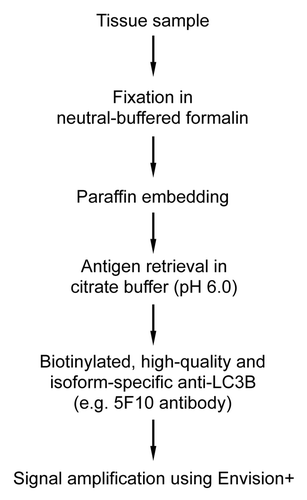
Abstract
Transmission electron microscopy (TEM) is an indispensable standard method to monitor macroautophagy in tissue samples. Because TEM is time consuming and not suitable for daily routine, many groups try to identify macroautophagy in tissue by conventional immunohistochemistry. The aim of the present study was to evaluate whether immunohistochemical assessment of macroautophagy-related marker proteins such as LC3, ATG5, CTSD/cathepsin D, BECN1/Beclin 1 or SQSTM1/p62 is feasible and autophagy-specific. For this purpose, livers from starved mice were used as a model because hepatocytes are highly sensitive to autophagy induction. ATG7-deficient mouse livers served as negative control. Our findings indicate that unambiguous immunodetection of LC3 in paraffin-embedded tissue specimens was hampered due to low in situ levels of this protein. Maximum sensitivity could only be obtained using high-quality, isoform-specific antibodies, such as antibody 5F10, in combination with Envision+ signal amplification. Moreover, LC3 stains were optimal in neutral-buffered formalin-fixed tissue, immersed in citrate buffer during antigen retrieval. However, even when using this methodology, LC3 monitoring required overexpression of the protein, e.g., in GFP-LC3 transgenic mice. This was not only the case for the liver but also for other organs including heart, skeletal muscle, kidney and gut. Immunohistochemical detection of the autophagy-related proteins ATG5, CTSD or BECN1 is not recommendable for monitoring autophagy, due to lack of differential gene expression or doubtful specificity. SQSTM1 accumulated in autophagy-deficient liver, thus it is not a useful marker for tissue with autophagic activity. We conclude that TEM remains an indispensable technique for in situ evaluation of macroautophagy, particularly in clinical samples for which genetic manipulation or other in vitro techniques are not feasible.
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved subcellular process for bulk digestion and elimination through lysosomal enzymes of the cell’s own proteins and entire organelles; for example, when they are obsolete or irreparably injured and need to be replaced.Citation1,Citation2 Intracellular material that needs to be autophagocytized is wrapped up by a flattened membranous structure, known as the phagophore or isolation membrane, whose free margins fuse with one another to form an autophagosome. Autophagosomes eventually fuse with lysosomes to become autolysosomes, so that their contents can be digested by lysosomal enzymes. The process generally acts as a housekeeping mechanism, and is crucial to the maintenance of normal cellular homeostasis. However, when stimulated by cellular stress conditions, autophagy may also function as a self-cannibalization pathway that promotes cell survival under unfavorable conditions.
Different methodologies have been developed to detect autophagy both in vitro and in vivo.Citation3-Citation18 Detection guidelines have also been established for the correct use and interpretation of such methods.Citation19-Citation21 Still demonstration of autophagy in tissue has not received extensive evaluation, particularly in clinical specimens for which genetic techniques (e.g., GFP-LC3 overexpression), autophagic flux measurements and injection of specific dyes are inconceivable. As a consequence, improved methods for detection of autophagy in human tissue relative to the techniques possible with cells in culture still need to be elaborated. Because the presence of autophagic vacuoles is by far the most important morphological feature of cells with autophagic activity, demonstration of these structures by transmission electron microscopy (TEM) remains the gold standard to assess autophagy in tissue.Citation3-Citation6,Citation21 It offers high-resolving power (0.1–0.4 nm) and hence, provides much more detailed information about the cell’s morphology as compared with conventional light microscopy. However, interpreting TEM images is rather subjective and it can be difficult to distinguish autophagosomes from lysosomes, endosomes or other structures in the cell.Citation20 Detection of autophagy in inflamed tissue by TEM is even more problematic. Some inflammatory cells such as monocytes and macrophages have a strong phagocytic potential, rendering it difficult to determine via TEM whether the vacuoles in their cytoplasm result from autophagy or heterophagy. Immunoelectron microscopy with antibodies against autophagosomal marker proteins would allow specific detection of autophagic structures,Citation5 but this method remains labor-intensive and requires well-trained personnel.
Given the many pitfalls in correctly identifying autophagic vacuoles by TEM, many groups have tried to identify autophagy in tissue by conventional immunohistochemistry.Citation9-Citation18 Advantages of immunohistochemistry include a fast turnaround time (typically 2 d vs 5–7 d for TEM), low cost, ease of performance and widespread familiarity. However, most immunohistochemical staining protocols for autophagy-associated proteins have not been validated (e.g., by using autophagy-deficient tissue as negative control) or optimized to obtain high sensitivity. Using wild-type and ATG7-deficient liver tissue as a model for immunohistochemical detection of autophagy, the aim of the present study was to evaluate the feasibility and specificity of immunohistochemical stainings for autophagy-related proteins.
Results
Generation of liver-specific Atg7 knockout mice
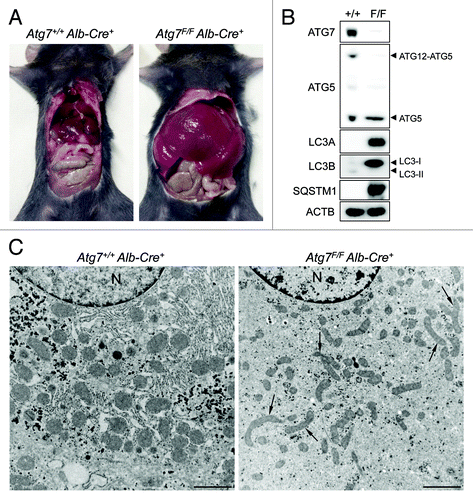
The essential autophagy gene Atg7 was deleted in liver by cross-breeding mice homozygous for the Atg7Flox allele (further referred to as Atg7F/F) with a transgenic mouse strain that expresses the Cre recombinase under control of the mouse albumin enhancer/promoter. Gross inspection of Atg7F/F Alb-Cre+ mice revealed severe enlargement of the liver as compared with Atg7+/+ Alb-Cre+ control mice (). Western blots of Atg7F/F Alb-Cre+ liver samples confirmed lack of ATG7 expression (). ATG7 deficiency was associated with defective autophagy as evidenced by the accumulation of SQSTM1/p62, increased LC3-I/LC3-II ratio’s for both LC3A and LC3B and decreased levels of ATG12-ATG5. In contrast to control mice, Atg7F/F Alb-Cre+ mice showed an abnormal ultrastructure of the liver as characterized by the accumulation of elongated and often deformed mitochondria (). Other differences in Atg7F/F Alb-Cre+ vs. wild-type mice included a decrease in glycogen granules and endoplasmic reticulum ().
Figure 1. ATG7 deficiency in mouse liver causes hepatomegaly and reveals signs of defective autophagy. (A) Gross anatomical view of a representative Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mouse (5 mo old). (B and C) western blot (B) and ultrastructural analysis (C) of liver samples from Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice. Ultrastructural inspection of Atg7F/F Alb-Cre+ livers reveals accumulation of elongated and deformed mitochondria (black arrows) as well as a decrease in glycogen granules and endoplasmic reticulum. Livers from Atg7+/+ Alb-Cre+ mice show normal cell morphology. Scale bar, 2 μm. n, nucleus.
Nutrient deprivation induces autophagy in liver
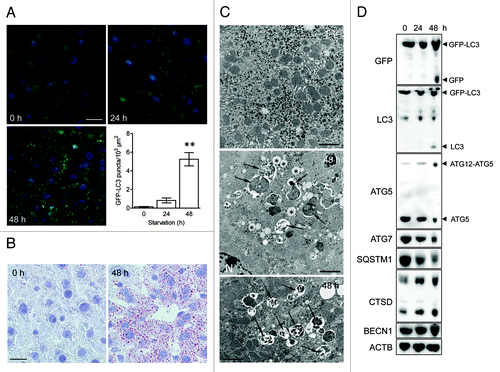
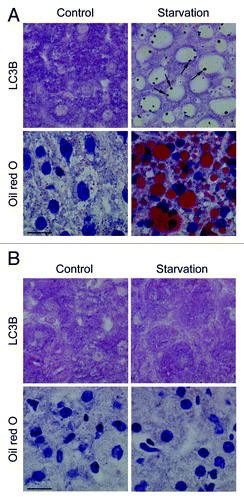
To validate immunocytochemical methods for the detection of autophagy, nutrient deprivation was used as a trigger. GFP-LC3B transgenic mice underwent starvation for 24 or 48 h. In contrast to liver from fed mice showing diffuse GFP-LC3 expression in the cytoplasm, livers from starved mice contained many intense puncta/dot-like GFP-LC3B structures (). A maximum number of GFP-LC3B dots was found after 48 h starvation. A striking morphological event during starvation was the “fatty change” of the liver (). Lipid droplets accumulated in hepatocytes of starved liver, especially around blood vessels. Furthermore, TEM revealed autophagic vacuoles in starved hepatocytes, but not in control liver (). Western blots showed cleavage of GFP-LC3B after 48 h starvation (). GFP-LC3B cleavage was associated with enhanced levels of the ATG12-ATG5 conjugate and CTSD, as well as with decreased levels of ATG7 and SQSTM1 ().
Figure 2. Nutrient deprivation induces autophagy in liver from GFP-LC3 transgenic mice. (A) Examination of GFP fluorescence in liver of GFP-LC3 transgenic mice before (0 h) and after starvation (24 or 48 h). Scale bar, 20 μm. Formation of GFP-LC3 dots during starvation was quantified. **p < 0.01 vs. 0 h (one-way ANOVA, followed by Dunnett test, n = 9). (B) Fatty change of liver tissue as demonstrated by oil red O staining before (0 h) and after starvation (48 h). Scale bar, 20 μm. (C) Ultrastructural characterization of liver tissue from GFP-LC3 mice before (0 h) and after starvation (48 h). The micrographs of starved liver show accumulation of lipid droplets (asterisks) and autophagic vacuoles (arrows). Scale bar, 2 μm. N indicates the nucleus. (D) Western blot analysis of liver from GFP-LC3 mice before and after starvation.
Immunohistochemical detection of LC3 in paraffin-embedded tissue depends on expression level, autophagy induction and sensitivity of the detection method
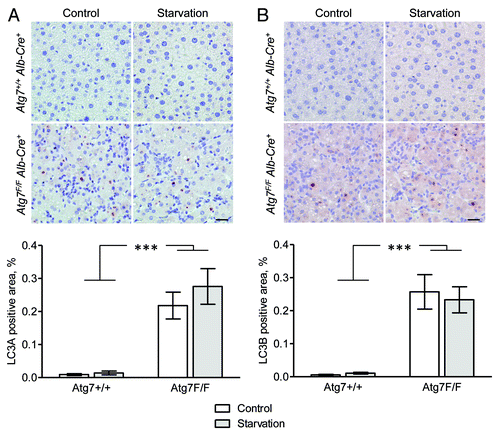
Because mammalian LC3 is one of the most frequently used biomarkers for autophagy both in vitroCitation3,Citation4,Citation19,Citation21 and in situ,Citation11-Citation16 starved and nonstarved livers isolated from Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice were stained for LC3A and LC3B, two LC3 isoforms known to be associated with autophagic membranes.Citation22 For this purpose, we used a highly sensitive immunohistochemical detection method (Vectastain ABC), which is based on the formation of large macromolecular complexes containing avidin and biotinylated horseradish peroxidase. Western blot experiments revealed that rabbit polyclonal LC3A raised against the C-terminal cleavage site of LC3A as well as mouse monoclonal LC3B antibody (clone 5F10) and rabbit monoclonal LC3B antibody (clone D11) raised against the N-terminus of LC3B, were highly specific for recombinant LC3A and LC3B protein, respectively (). However, all tested rabbit polyclonal LC3B antibodies showed crossreactivity with LC3A protein (), and were excluded for detection of LC3B in mouse tissue. Despite the sensitivity of the Vectastain ABC method, liver from Atg7+/+ Alb-Cre+ control mice stained negative for LC3A and LC3B, even after 48 h starvation (). Still, large globular structures of different size (diameter 1–7 μm) stained positive for LC3A and LC3B in liver from autophagy-incompetent Atg7F/F Alb-Cre+ mice (). This staining was independent of the starvation status of the tissue.
Figure 3. Rabbit polyclonal LC3B antibodies show crossreactivity with LC3A protein. Western blots containing recombinant LC3A and LC3B (5–20 ng) protein were probed with the following antibodies: mouse monoclonal antiLC3B (Nanotools), rabbit monoclonal antiLC3B (Cell Signaling), rabbit polyclonal antiLC3A (Abgent) and rabbit polyclonal antiLC3B from Novus Biologicals and Santa Cruz Biotechnology.
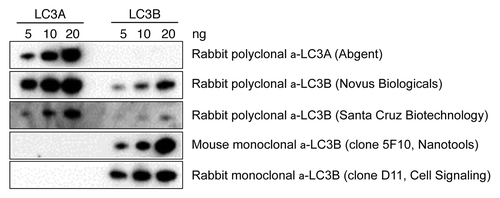
Figure 4. Liver from autophagy-deficient Atg7F/F Alb-Cre+ mice but not from autophagy-competent Atg7+/+ Alb-Cre+ shows immunohistochemical staining for LC3A and LC3B. Liver samples were isolated from fed mice (control) or from mice that underwent starvation for 48 h. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for LC3A (A) and LC3B (B) using rabbit polyclonal anti-LC3A (Abgent, 1:100) and biotinylated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:100) in combination with Vectastain ABC. Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm. The LC3A and LC3B positive area was quantified. ***p < 0.001 vs. Atg7+/+ Alb-Cre+ mice (two-way ANOVA, n = 10).
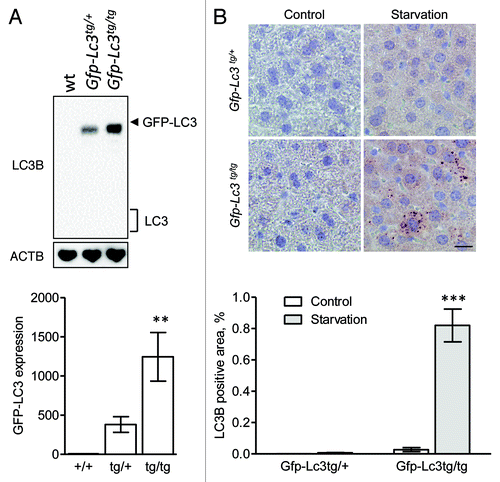
A western blot analysis of different mouse tissue lysates showed that the expression of LC3A and LC3B varies considerably, with the highest expression level found in the brain (). Particularly LC3A was highly expressed in brain tissue. Purkinje cells in the cerebellum showed strong staining for LC3A (), but also neurons in some parts of the cerebrum stained positive for LC3A (), but not for LC3B, suggesting that immunohistochemical detection of LC3 isoforms is feasible provided that the expression level of the target protein is sufficiently high. To further evaluate this hypothesis, livers from GFP-LC3B transgenic mice were examined. Western blots showed that GFP-LC3B was moderately expressed in liver from mice containing one copy of the Gfp-Lc3b transgene (Gfp-Lc3tg/+), but higher levels of GFP-LC3B were found in homozygous Gfp-Lc3tg/tg mice containing two gene copies. Using Vectastain ABC, GFP-LC3B was not detectable in liver from Gfp-Lc3tg/+ mice, neither in nonstarved control conditions nor after 48 h starvation (). Livers from unstarved Gfp-Lc3tg/tg mice did not stain for LC3B either. In contrast, LC3B-puncta indicative of autophagosomes could be detected in liver from starved Gfp-Lc3tg/tg mice ().
Figure 5. The highest expression of LC3A and LC3B in nonstarved control mice was found in brain tissue. (A) Western blot analysis of LC3A and LC3B in different tissue lysates. GAPDH served as a loading control. (B) Immunohistochemical detection of LC3A and LC3B in mouse brain using the Vectastain ABC system. Tissue samples were isolated from fed control mice. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for LC3A (A) and LC3B (B) using rabbit polyclonal anti-LC3A (Abgent, 1:3000) and biotinylated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:100). Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm.
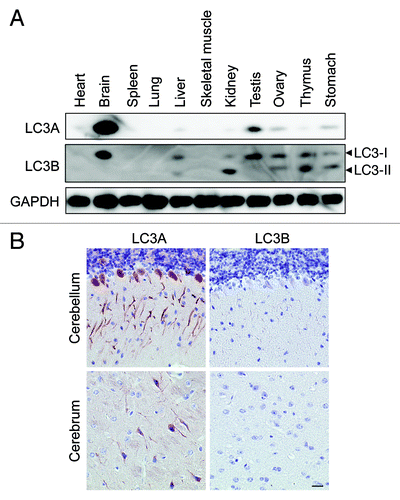
Figure 6. Immunohistochemical detection of LC3B is feasible in liver from starved Gfp-Lc3tg/tg mice using Vectastain ABC. (A) Western blot analysis of LC3B in liver from wild-type mice, heterozygous Gfp-Lc3tg/+ or homozygous Gfp-Lc3tg/tg transgenic mice. The GFP-LC3 expression in each group was quantified. **p < 0.01 vs. wild-type mice (one-way ANOVA, followed by Dunnett test, n = 4). (B) Immunohistochemical detection of LC3B in liver from Gfp-Lc3tg/+ or Gfp-Lc3tg/tg mice using the Vectastain ABC system. Liver samples were isolated from fed mice (control) or from mice that underwent starvation for 48 h. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for LC3B using biotinylated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:100). Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm. The LC3B positive area was quantified. ***p < 0.001 vs. control (two way ANOVA, followed by Bonferroni post test, n = 10).
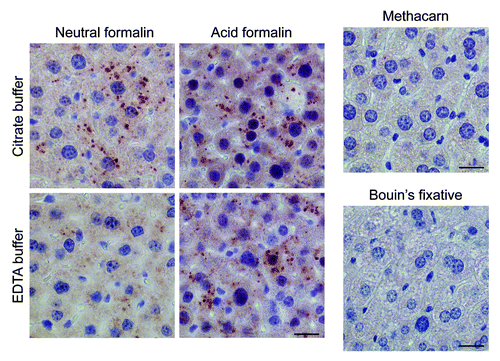
Using different fixatives and buffer solutions, we next tried to optimize the conditions for LC3B staining. Optimal staining was obtained with starved liver fixed in neutral buffered formalin and immersed in citrate buffer (pH 6.0) during antigen retrieval (). Immersion of neutral formalin-fixed tissue in EDTA buffer (pH 8.0) inhibited staining (), whereas fixation in acid formalin yielded more background, although LC3B puncta were clearly detectable also after treatment with EDTA buffer (). Tissues fixed in the alcohol-based fixative methacarn or in Bouin’s fixative did not yield positive staining ().
Figure 7. Optimal immunohistochemical detection of LC3B requires processing of tissue samples in a suitable fixative. Liver samples were isolated from Gfp-Lc3tg/tg mice that underwent starvation for 48 h. After fixation in different fixatives for 24 h, tissues were paraffin-embedded and stained for LC3B using biotinylated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:100) and Vectastain ABC. For formalin-fixed samples, heat-mediated antigen retrieval was performed either in citrate buffer (pH 6.0) or in EDTA buffer (pH 8.0). Scale bar, 20 μm.
Surprisingly, frozen, acetone-fixed liver samples from starved wild-type mice showed strong LC3B staining compared with nonstarved control samples (), albeit puncta were not observed. Frozen liver sections from autophagy-deficient Atg7F/F Alb-Cre+ mice also stained positive for LC3B, though in a starvation-independent manner (). Staining of frozen livers isolated from Gfp-Lc3b transgenic mice required strong dilution (1:30,000) of the LC3B antibody to allow a difference in staining between starved and nonstarved samples (). However, even with strongly diluted LC3B antibodies, differences in staining intensity between starved and nonstarved livers from Gfp-Lc3btg/tg mice were negligible. Because a standard fixation with acetone might be too rigorous for the structure of frozen sections, we also tried LC3B staining without fixation and after short fixation (5 min) with 4% paraformaldehyde or methacarn. Stainings of unfixed or paraformaldehyde-fixed frozen samples were similar as compared with acetone-fixed sections (results not shown). Methacarn fixation of frozen sections did not allow LC3B staining (not shown). Furthermore, it is important to note that all LC3B stainings were obtained using horseradish peroxidase, which requires pretreatment of sections with hydrogen peroxidase in methanol to inactivate endogenous peroxidase. Paraformaldehyde-fixed frozen sections of wild-type liver that were stained using alkaline phosphatase showed LC3B-positive dots around lipid droplets (). These droplets were abundantly present after starvation (). However, if frozen sections were fixed with acetone, LC3B dots could not be detected (not shown). Moreover, starved liver from Atg7F/F Alb-Cre+ mice showed neither lipid droplet formation nor LC3B staining (). Other tissues from starved wild-type mice (heart, skeletal muscle, gut) were tested for LC3B-positive dots in paraformaldehyde-fixed frozen sections, but did not accumulate lipid droplets and stained negative for LC3B when using alkaline phosphatase. One exception was starved kidney that revealed both lipid droplets and LC3B-positive dots on their surface, similar to starved liver (data not shown).
Figure 8. Immunohistochemical staining of LC3B is enhanced in frozen liver sections. Liver samples were isolated from Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice (A) or from transgenic Gfp-Lc3tg/+ or Gfp-Lc3tg/tg mice (B). Some animals were fed normal chow (control), others underwent starvation for 48 h. After fixation in acetone, frozen sections were stained for LC3B using biotinylated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:1,000 [Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ samples] or 1:30,000 [Gfp-Lc3tg/+ and Gfp-Lc3tg/tg samples]) and Vectastain ABC. Scale bar, 40 μm.
![Figure 8. Immunohistochemical staining of LC3B is enhanced in frozen liver sections. Liver samples were isolated from Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice (A) or from transgenic Gfp-Lc3tg/+ or Gfp-Lc3tg/tg mice (B). Some animals were fed normal chow (control), others underwent starvation for 48 h. After fixation in acetone, frozen sections were stained for LC3B using biotinylated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:1,000 [Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ samples] or 1:30,000 [Gfp-Lc3tg/+ and Gfp-Lc3tg/tg samples]) and Vectastain ABC. Scale bar, 40 μm.](/cms/asset/e048f347-0a4e-4bc3-9163-8da4ce5e1834/kaup_a_10922968_f0008.gif)
Figure 9. LC3B dots are detectable in frozen liver sections using a staining procedure with alkaline phosphatase and are localized on the surface of lipid droplets. Liver samples were isolated from Atg7+/+ Alb-Cre+ (A) or Atg7F/F Alb-Cre+ mice (B) that were fed normal chow (control) or underwent starvation for 48 h. After fixation in 4% paraformaldehyde, frozen sections were stained for LC3B using biotinylated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:1,000) and Vectastain ABC-AP kit, containing biotinylated alkaline phosphatase instead of biotinylated horseradish peroxidase. LC3B-positive dots (arrows) were detected on the surface of lipid droplets. These structures could be stained using oil red O. Scale bar, 20 μm.
Besides Vectastain ABC, the Dako Envision+ system is an ultrasensitive immunohistochemical staining technique that has recently been used to detect LC3 in autophagic vacuoles of paraffin-embedded tissue.Citation17 However, in analogy with Vectastain, Envision+ in combination with an isoform-specific rabbit monoclonal anti-LC3B () failed to detect LC3B in liver samples isolated from fed or starved wild-type mice (). Similar results were obtained with rabbit polyclonal LC3A (not shown). Nonetheless, LC3-positive puncta could be detected in livers from starved Gfp-Lc3tg/+ mice, even though the LC3B positive area was highly limited (0.3 ± 0.1%, ). Large LC3B-positive areas (3.2 ± 0.3%, ) that were much larger than those obtained with Vectastain ABC (0.8 ± 0.1%, ), were found after staining of LC3B in liver from Gfp-Lc3tg/tg mice, suggesting that Envision+ is the preferred method to obtain maximum staining sensitivity.
Figure 10. Envision+ enhances the immunohistochemical detection of LC3B in GFP-LC3 transgenic mice. Liver samples were isolated from fed wild-type, Gfp-Lc3tg/+ or Gfp-Lc3tg/tg mice (control) and from mice that underwent starvation for 48 h. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for LC3B using rabbit monoclonal anti-LC3B (clone D11, Cell Signaling, 1:100 [wild-type]; 1:300 [Gfp-Lc3tg/+] or 1:1,000 [Gfp-Lc3tg/tg]) and Envision+. Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm. The LC3B positive area was quantified. ***p < 0.001 vs. control (two-way ANOVA, followed by Bonferroni post test, n = 10).
![Figure 10. Envision+ enhances the immunohistochemical detection of LC3B in GFP-LC3 transgenic mice. Liver samples were isolated from fed wild-type, Gfp-Lc3tg/+ or Gfp-Lc3tg/tg mice (control) and from mice that underwent starvation for 48 h. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for LC3B using rabbit monoclonal anti-LC3B (clone D11, Cell Signaling, 1:100 [wild-type]; 1:300 [Gfp-Lc3tg/+] or 1:1,000 [Gfp-Lc3tg/tg]) and Envision+. Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm. The LC3B positive area was quantified. ***p < 0.001 vs. control (two-way ANOVA, followed by Bonferroni post test, n = 10).](/cms/asset/a3b90d44-830f-4aa5-9e68-2a7883096a65/kaup_a_10922968_f0010.gif)
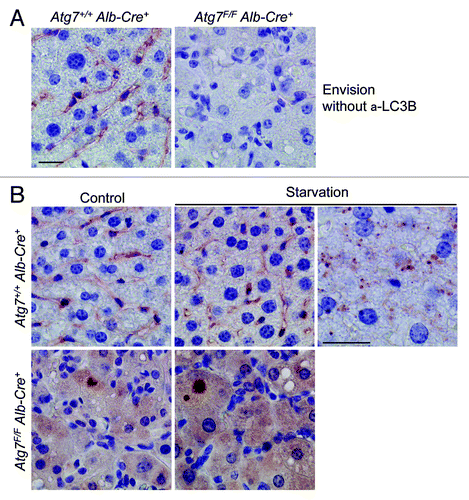
Because Envision+ combined with mouse monoclonal LC3B (clone 5F10) may lead to detection of LC3B-positive puncta,Citation17 we also tried this condition on mouse liver. Apparently, anti-mouse Envision+ reagent caused high background staining in wild-type liver, but not in autophagy-deficient liver (). Neither control liver nor starved liver from wild-type mice showed LC3B-specific staining (). However, tiny LC3B-positive dots were detected in hepatocytes around blood vessels of starved liver (), but not in other regions of the same liver. The immunoreactive area of these dots was negligible (estimated area < 0.01%) and not quantifiable due to background staining.
Figure 11. Livers from starved wild-type mice show background staining with anti-mouse Envision+. (A) Anti-mouse Envision+ without primary antibody results in background staining of Atg7+/+ Alb-Cre+ liver, but not of Atg7F/F Alb-Cre+ liver. (B) Liver samples were isolated from fed Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice (control) or from mice that underwent starvation for 48 h. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for LC3B using unconjugated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:100), in combination with Envision+. Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm. One panel of starved Atg7+/+ Alb-Cre+ hepatocytes (around blood vessel) was taken at higher magnification and illustrates tiny LC3B positive puncta.
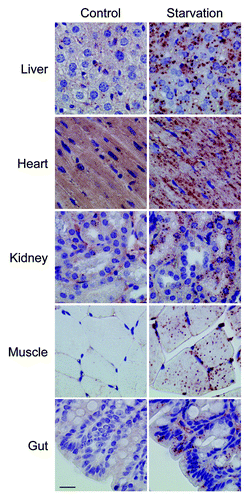
To extrapolate our results to other tissues, samples of heart, skeletal muscle, lung, kidney and gut were isolated from starved wild-type or Gfp-Lc3tg/tg mice. Tissue samples were fixed in neutral buffered formalin and embedded in paraffin. In all conditions tested, Envison+ did not allow LC3B staining of tissue from starved wild-type mice (not shown). In addition, monoclonal anti-LC3B (clone D11) with Envision+ stained none of the tested organs isolated from starved Gfp-Lc3tg/tg mice (not shown). Only monoclonal anti-LC3B (5F10) with Envision+ led to LC3B-positive puncta in the different Gfp-Lc3tg/tg tissue samples (). Lung tissue showed strong background staining with Envison+ reagent and was excluded from further analysis. Tissues from heart, kidney, striated muscle and gut in addition to livers from starved Gfp-Lc3tg/tg mice serving here as a positive control, revealed abundant LC3B staining ().
Figure 12. Monoclonal 5F10 LC3B antibody combined with Envision+ allows the immunohistochemical detection of LC3B in different organs of starved Gfp-Lc3tg/tg mice. Gfp-Lc3tg/tg mice were fed regular chow (control) or underwent starvation for 48 h. Tissue samples from different organs were collected and fixed in neutral buffered formalin for 24 h. Thereafter, tissues were paraffin-embedded and stained for LC3B using unconjugated mouse monoclonal anti-LC3B (clone 5F10, Nanotools, 1:1000) and Envision+. Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm.
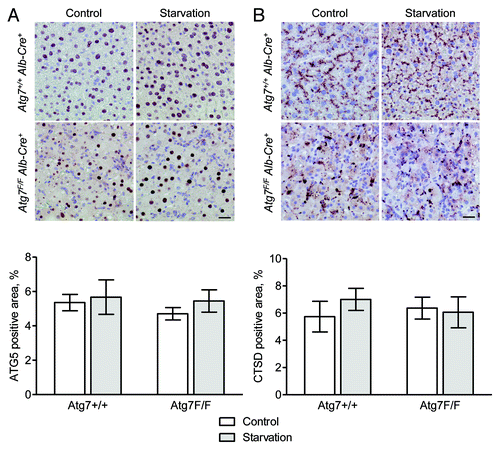
ATG5 and CTSD are not good marker proteins for immunohistochemistry-based autophagy detection
Liver from both Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice showed nuclear ATG5 staining (). After starvation, we did not observe any change in the immunohistochemical staining pattern as compared with nonstarved controls. Because cytoplasmic and not nuclear staining was expected, nuclear ATG5 staining was confirmed by a second primary antibody (Lifespan, data not shown). Additional stainings for ATG5 were performed on other tissues, yielding strong nuclear and cytoplasmic ATG5 staining in kidney tissue (data not shown). Staining for CTSD, just like ATG5, was not different between starved and nonstarved controls (). Livers from Atg7+/+ Alb-Cre+ mice showed a typical granular staining in the cytoplasm, which was more irregular in size and intensity in Atg7F/F Alb-Cre+ samples.
Figure 13. ATG5 and CTSD are not suitable targets for the immunohistochemical detection of autophagy in liver. Liver samples were isolated from fed Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice (control) or from mice that underwent starvation for 48 h. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for ATG5 (A) or CTSD (B) using rabbit polyclonal anti-ATG5 (Abcam, 1:100) and rabbit monoclonal anti-CTSD (clone EPR3057Y, Abcam, 1:1000), respectively, in combination with Vectastain ABC. Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm. The ATG5- and CTSD-positive area was quantified. Neither the effect of starvation nor the results between Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice were statistically significant.
BECN1 shows modest upregulation after starvation
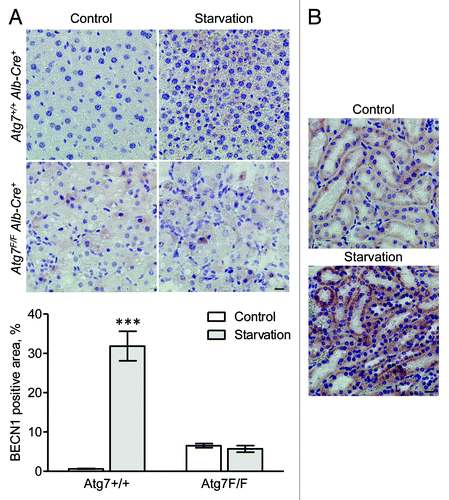
BECN1 was not detectable in liver from fed Atg7+/+ Alb-Cre+ mice, but showed staining after starvation (). Livers from Atg7F/F Alb-Cre+ mice showed diffuse background staining for BECN1, both in starved and nonstarved conditions (). In contrast to liver, BECN1 expression was clearly detectable in the convoluted tubules of the kidney cortex (). Staining in the tubules was more pronounced after starvation.
Figure 14. BECN1 is upregulated in mouse tissue after starvation. (A) Liver samples were isolated from fed Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice (control) or from mice that underwent starvation for 48 h. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for BECN1 using rabbit polyclonal anti-BECN1 (Lifespan, 1:1,000) and Vectastain ABC. Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar 20 μm. The BECN1 positive area was quantified. ***p < 0.001 vs. control (two-way ANOVA with Bonferroni post test, n, 10). (B) BECN1 immunohistocehmical staining in kidney from Atg7+/+ Alb-Cre+ mice before and after starvation (48 h). Scale bar, 20 μm.
Autophagy-deficient livers contain large SQSTM1-positive inclusions in the cytoplasm
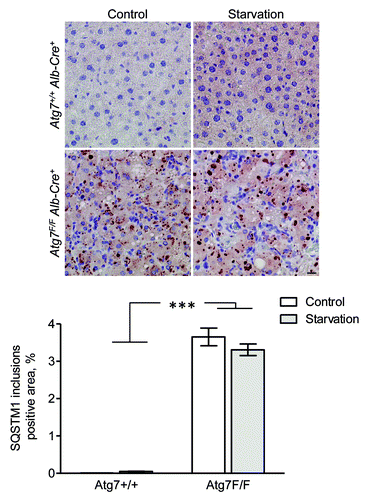
SQSTM1 was undetectable in liver from fed fed Atg7+/+ Alb-Cre+ mice, but showed diffuse cytoplasmic staining after starvation (). Livers from Atg7F/F Alb-Cre+ mice revealed many SQSTM1-positive inclusions in the cytoplasm, both in starved and nonstarved conditions ().
Figure 15. SQSTM1 accumulates in cytoplasmic inclusions of liver from autophagy-deficient mice. Liver samples were isolated from fed Atg7+/+ Alb-Cre+ and Atg7F/F Alb-Cre+ mice (control) or from mice that underwent starvation for 48 h. After fixation in neutral buffered formalin for 24 h, tissues were paraffin-embedded and stained for SQSTM1 using rabbit polyclonal anti-SQSTM1 (Sigma, 1:5,000) and Vectastain ABC. Heat-mediated antigen retrieval was performed in citrate buffer (pH 6.0). Scale bar, 20 μm. The positive area of SQSTM1 inclusions was quantified. ***p < 0.001 vs. Atg7+/+ Alb-Cre+ mice (two-way ANOVA, n, 10).
Discussion
In recent years, autophagy has been associated with a plethora of different pathological conditions including heart disease, cancer, neurodegeneration and infectious diseases.Citation23-Citation25 Considering the potential therapeutic implications of the pharmacological modulation of autophagy in many of these pathological conditions, the development of a fast and simple detection method for autophagy in clinical tissue samples has become a major priority. Increasing evidence suggests that immunohistochemical staining of LC3 could be a valuable technique for evaluation of autophagy in situ, particularly in the field of cancer. Indeed, immunohistochemical assessment of LC3 shows that expression of LC3 is intensified in cancer cells compared with normal tissues,Citation13,Citation18,Citation26 which incites researchers to assume increased autophagic activity in such tumors. Unfortunately, since the immunoreactivity for LC3 in cancer tissue is most often granular without clear visualization of LC3-positive puncta, it remains unclear whether it truly reflects autophagosome formation or merely LC3 protein expression and/or aggregation. On the other hand, large LC3A-positive amorphous material, typically enclosed within LC3-positive cytoplasmic vacuoles, has recently been identified by immunohistochemistry in different tumor types.Citation16,Citation27,Citation28 The presence of these ‘stone-like structures’ may reflect accumulation of autophagic debris that cannot be degraded by the available lysosomal pool, rather than an active autophagic process. The main objective of the present study was to determine whether autophagosome formation could be easily detected in (normal) tissue using standard immunohistochemical techniques. An important criterion was that the immunohistochemical stainings should give a clear, unambiguous pattern of dot-like structures (puncta) reflecting autophagic vacuoles, analogous with detection of autophagosomes in cultured cells. To this end, we used liver from starved mice as a model because hepatocytes are highly sensitive to autophagic conditions and rapidly turn on their autophagic-lysosomal degradation pathway upon starvation in an attempt to supply the organism with glucose and amino acids.Citation29 Indeed, it has been calculated that autophagic protein degradation in liver, which proceeds at a rate of approximately 1.5% of total liver protein/h under nutrient-rich conditions, is enhanced 2- to 3-fold during starvation,Citation30 and results in a nearly 40% overall loss of liver protein after 48 h of starvation.Citation31 In the present study, livers from GFP-LC3 mice undergoing starvation for 48 h revealed a large number of autophagosomes, as determined by confocal microscopy and TEM. Moreover, cleaved GFP-LC3 as well as reduced levels of SQSTM1, which is degraded by autophagy, could be found, suggesting initiation of an active autophagic process.
Given that LC3 is expressed as different isoforms in mammalian cellsCitation32 and that many commercialized LC3-antibodies are not isoform specific,Citation33 we first tested whether the LC3 antibodies used in this study could bind to recombinant LC3A and/or LC3B. Both proteins are currently the best-studied in situ markers for autophagy. Our results confirm that crossreactivity with other isoforms may exist. However, when using appropriate (isoform-specific) LC3 antibodies in combination with a highly sensitive immunohistochemical detection technique (Vectastain ABC), we detected neither LC3A nor LC3B in both normal and starved mouse liver. Still, large globular structures of different size stained positive for LC3A and LC3B in liver from Atg7F/F Alb-Cre+ mice, which are characterized by defective autophagy in hepatocytes due to deletion of the essential autophagy gene Atg7. This finding was not surprising since autophagy-deficient cells could accumulate protein aggregates or cellular structures, in which LC3 can be incorporated in an autophagy-independent manner.Citation34,Citation35 Therefore, immunohistochemical stainings for LC3 should be interpreted with caution, particularly if defective autophagy is suspected.
Our findings are consistent with previous reports from our group showing that LC3 can be detected via immunohistochemistry in liver tissue, but only when this protein is overexpressed.Citation11,Citation12 A western blot analysis of different mouse tissue lysates revealed that the expression of LC3A and LC3B may vary considerably. Expression of LC3A and LC3B is relatively weak in liver as well as in many other organs such as heart, spleen and lung. However, LC3A was highly expressed in the brain, allowing immunohistochemical detection, particularly in the Purkinje cells of the cerebellum. In this light, it is also noteworthy that protein levels of both LC3A and LC3B were strongly increased in liver from Atg7F/F Alb-Cre+ mice, a finding that may explain the aberrant LC3 immunoreactivity in such tissue as mentioned above. Therefore, to further validate LC3 as a potential biomarker for autophagosome formation in situ, we decided to examine liver sections from transgenic mice that systemically overexpress GFP-LC3B. LC3B-positive puncta indicative of autophagosomes could be detected in liver from starved GFP-LC3 mice that were homozygous for the transgene. Livers from heterozygous GFP-LC3B mice, showing less abundant LC3 expression, or homozygous GFP-LC3B mice that did not undergo starvation, were negative for LC3 staining, indicating that both initiation of autophagy and sufficiently high expression levels of LC3 are required for immunohistochemical detection.
To ensure an optimal detection of LC3 in tissue, we tested several fixatives, buffer solutions, antibody concentrations and embedding media. Even though there is no ideal universal fixative for the demonstration of an antigen in situ, most antigens can be successfully detected in formalin-fixed paraffin-embedded tissue sections. Given that formalin is a cross-linking fixative, the discovery and development of antigen retrieval techniques further stimulated the use of formalin as a routine fixative for immunohistochemistry in many research laboratories. However, for certain cell antigens precipitant fixatives (e.g., methacarn) or combination fixatives such as Bouin’s fixative (combining formalin with the precipitant picric acid) are recommended. In the present study, optimal LC3 staining was achieved with neutral-buffered formalin-fixed tissue specimens, immersed in citrate buffer (pH 6.0) during antigen retrieval. Liver samples fixed in the precipitant fixative methacarn or Bouin’s fixative did not stain for LC3. Possibly, sufficient cross-linking is essential for fixing small proteins such as LC3, which may not be large enough to be made insoluble by precipitant fixatives. Bouin’s fixative contains formalin, but also picric acid, resulting in a highly acidic solution (pH 1.5–1.9) that is therefore not always suitable for antigen detection. Another important issue is that some antigens do not survive paraffin embedding so that frozen sections are needed. For example, a disadvantage of paraffin-embedded tissue is that all lipids are extracted. LC3 is lipidated before associating with autophagosomes. In this regard, it could be assumed that the LC3 protein is partially lost in the paraffin-embedding process. Frozen liver samples from starved wild-type mice showed upregulation of LC3B staining vs. nonstarved liver, which probably reflects transcriptional induction of LC3 as previously described.Citation21 Furthermore, LC3 could be detected in frozen sections at very low antibody concentrations, indicating that frozen sections offer a much more sensitive detection platform than paraffin-embedded tissue. However, because the morphology and resolution of frozen sections are poor, the typical LC3 puncta as observed in paraffin-embedded tissue could not be observed. In our opinion, this finding makes frozen sections unsuitable for the detection of autophagosomal structures.
Importantly, stainings of frozen (or paraffin-embedded) tissue were performed using horseradish peroxidase. This staining procedure requires inactivation of the endogenous peroxidase with destructive agents such as hydrogen peroxidase and methanol. To avoid the use of chemicals that could dissolve small membranous structures, we tried to stain frozen sections using alkaline phosphatase. Interestingly, paraformaldehyde-fixed, frozen sections of starved liver showed many LC3B dots around lipid droplets (LDs), which is consistent with previous reports showing that LC3B is localized on the surface of LDs and critically involved in LD formation.Citation35,Citation36 Autophagy-deficient frozen liver did not accumulate LDs and stained negative for LC3B. The dotted staining pattern in wild-type liver was lost if sections were fixed in acetone, indicating that lipid extraction with organic solvents (or during paraffin embedding) may cause a significant loss of LC3 protein. However, an analysis of different frozen tissue specimens including heart, kidney, skeletal muscle and gut that were stained for LC3B using alkaline phosphatase revealed that this approach fails to detect autophagic vacuoles. Besides liver, only kidney showed LC3B dots because of its ability to accumulate LDs after starvation.
Recently, an optimized method for the detection of LC3 by immunohistochemistry in autophagic vacuoles of paraffin-embedded tissue was reported.Citation17 Staining was based on Envision+ immunodetection, which is an extremely sensitive system that allows signal amplification via a hydrophilic polymer (dextran) conjugated to secondary antibodies and multiple (up to 100) horseradish peroxidase molecules. The polymer does not contain avidin or biotin so that nonspecific staining resulting from endogenous avidin-biotin activity is eliminated or significantly reduced. Although this method of signal amplification, like any indirect immunofluorescence, is not quantitative and can only be qualitative, Envision+ offered superior detection as compared with avidin-biotin based signal amplification systems such as Vectastain ABC, at least for LC3. Nonetheless, important limitations in LC3 detection should be taken into account. First, anti-LC3 antibodies combined with Envision+ signal amplification failed to stain LC3 in most tissues from starved wild-type mice. In liver, however, few LC3B puncta could be detected, but they were only locally present (in hepatocytes around blood vessels), not throughout the entire liver, and were small in size and thus hard to find. Second, an important prerequisite for the detection of puncta in the liver with Envision+ was the use of the LC3B mouse monoclonal antibody 5F10 which clearly offered the highest sensitivity over other related antibodies. Based on these findings, we assume that Envision+ immunohistochemical stainings with antibody 5F10 in starved wild-type liver were performed at the detection limit. Much better results were obtained in GFP-LC3 transgenic mice, also with other organs such as skeletal muscle, heart and kidney. A disadvantage of mouse monoclonal antibody 5F10 is that LC3 detection on mouse tissue is complicated by high levels of background staining, at least when indirect immunohistochemical detection methods are used such as Envision+. Background is attributed to binding of secondary anti-mouse antibody to endogenous mouse IgGs in the tissue being stained, and to Fc receptors on inflammatory cells. Background can be inhibited by blocking endogenous IgGs with Fab fragments of anti-mouse IgGs, preferably in combination with the use of a biotin-conjugated Fab fragment anti-mouse IgG (instead of whole IgG secondary antibody). This approach requires short incubation steps and is therefore much less sensitive than the protocol used in the present study, thus not recommendable for LC3 detection.
Finally, it is noteworthy that many autophagy-related proteins, other than LC3, have been used as an immunohistochemical marker for autophagy.Citation9,Citation10,Citation12,Citation14 However, the results of these studies need to be interpreted with caution since some of these proteins can accumulate in conditions of decreased autophagic activity. For example, granular cytoplasmic ubiquitin inclusions constituted an attractive marker for cellular degeneration of cardiomyocytes via autophagy in the early 2000s,Citation37-Citation39 but more recent evidence showed that these inclusions may also result from a malfunction in the autophagic pathwayCitation40 or from structural changes in the protein substrates, halting their degradation.Citation41 In the present study, we examined ATG5, CTSD, BECN1 and SQSTM1 as alternative candidate markers for in situ detection of autophagy. Staining for ATG5 and CTSD was not different between starved samples and nonstarved (or autophagy-deficient) controls. Moreover, ATG5 was found in the nucleus, which does not seem to be autophagy-related. In contrast, BECN1 showed upregulation after starvation and might as such be useful for the detection of autophagy. However, expression of BECN1 has also been found to be upregulated during tumorigenesis and has an important prognostic value in various tumor types.Citation42-Citation46 It is presently unclear whether increased BECN1 expression in cancer can be attributed to autophagy induction. Furthermore, the overexpression in starved liver, as shown in the present study, is relatively poor. In other organs, such as the kidney, we found strong basal levels of BECN1, suggesting that BECN1 is not the most ideal marker to monitor autophagy induction. SQSTM1 showed diffuse cytoplasmic staining in liver from starved wild-type mice, which is again not ideal to monitor autophagy. Interestingly, SQSTM1 could be found in large cytoplasmic inclusions of autophagy-deficient liver. These inclusions result from a marked accumulation of SQSTM1, followed by the formation of large aggregates containing SQSTM1 and ubiquitinated proteins.Citation47 Similar protein aggregates have been identified in various human disorders including neurodegenerative diseases, liver disorders and cancer.Citation48,Citation49
In conclusion, the unambiguous detection of LC3 as a marker protein for autophagy via routinely used immunohistochemical techniques is hampered due to low in situ levels of this protein in paraffin-embedded tissue. Therefore, the immunohistochemical detection of LC3 in paraffin-embedded tissue depends on a highly sensitive detection method (). Optimal LC3 immunostains could only be obtained when using neutral-buffered formalin-fixed tissue specimens, immersed in citrate buffer (pH 6.0) during antigen retrieval. Another critical step during LC3 staining was the use of a high-quality, isoform-specific antibody, such as antibody 5F10, in combination with the Envision+ signal amplification system to obtain maximum sensitivity. However, even this sensitive detection method required overexpression of LC3 (e.g., by using GFP-LC3 mice) to achieve staining. The immunohistochemical detection of other autophagy-related proteins such as ATG5, CTSD or BECN1 is not recommended to monitor autophagy, due to lack of differential gene expression or doubtful specificity.
Materials and Methods
Antibodies
The following mouse monoclonal antibodies were used: anti-ACTB/β-actin (clone AC-15, Sigma-Aldrich, A5441) and anti-LC3B (clone 5F10, Nanotools, 0231-100/LC3-5F10 or 0231-100BIOTIN/LC3-5F10). Rabbit polyclonal antibodies were anti-LC3B (Novus Biologicals, NB100-2331), anti-MAPLC3 (H50, Santa Cruz, sc-28266), anti-ATG5 (Abcam, ab78073; Lifespan, LS-B1843), anti-ATG7 (Sigma-Aldrich, A2856), anti-LC3A (Abgent, AP1805a), anti-SQSTM1 (Sigma, P0067), anti-GFP (Abcam, ab6556) and anti-BECN1 (Lifespan, LS-B3202). Anti-CTSD (clone EPR3057Y, Abcam, ab75852) and anti-LC3B (clone D11, Cell Signaling, 3868) were rabbit monoclonal antibodies. Horseradish peroxidase-conjugated swine anti-rabbit immunoglobulins (P0399) and rabbit anti-mouse immunoglobulins (P0260) were purchased from Dako. Crossreactivity of the LC3 antibodies with different LC3 isoforms was tested using human recombinant LC3A (Abnova, H00084557-P01) and LC3B (Abnova, H00081631-P01) proteins.
Animals and tissue processing
Mice homozygous for a vector that disrupts Atg7 in exon 14 by Cre-loxP technology (further referred to as Atg7F/F mice)Citation40 were housed in a temperature-controlled environment with 12 h light/dark cycles, and received food and water ad libitum. To obtain liver-specific Atg7 knockouts, Atg7F/F mice were crossbred with Alb-Cre+ transgenic mice (Jackson, stock number 003574) containing Cre recombinase under control of a mouse albumin enhancer/promoter. Genotyping of offspring was performed by PCR using standard procedures. Parameters for PCR amplification were 94°C for 5 min and 35 cycles of 98°C for 20 sec, 68°C (60°C in case of wild-type Atg7) for 30 sec and 72°C for 90 sec. The following primers were used: 5′-TTTGCCTGCATTACCGGTCGATGCAAC-3′ (forward) and 5′-TGCCCCTGTTTCACTATCCAGGTTACGGA-3′ (reverse) for Cre recombinase; 5′-TGGCTGCTACTTCTGCAATGATGT-3′ (forward), 5′-CAGGACAGAG-ACCATCAGCTCCAC-3′ (reverse) and 5′-GATCTTCATAAGGTGCTAGAACATGC-AGG-3′ (reverse for wild-type Atg7 allele) for Atg7. GFP-LC3 transgenic mice (RIKEN BioResource Center, strain GFP-LC3#53)Citation50 containing a rat GFP-LC3 fusion under control of the chicken ACTB promoter, were genotyped as previously described.Citation51 For starvation studies, mice (12 weeks of age) were deprived of food for 24 or 48 h, but had free access to drinking water. After sacrifice of starved and nonstarved animals, tissues were harvested and fixed in either 4% neutral-buffered formalin (Merck, 1.03999) or acid formalin (Fisher Scientific, F/1501/PB15), Bouin’s fixative (71.4% saturated picric acid [Merck, 623], ~9% formalin [Fisher Scientific, F/1501/PB15], 4.8% glacial acetic acid [Merck, 1.00063]) or methacarn (60% methanol [Merck, 1.06009], 30% 1,1,1-trichloroethane [Merck, 8749], 10% glacial acetic acid) for 24 h prior to paraffin embedding. Alternatively, non-fixed tissue specimens were embedded in Richard-Allan Scientific Neg-50 Frozen Section Medium (Thermo Scientific, 6502) using liquid nitrogen, and stored at −80°C. All experiments were approved by the Ethical Committee of the University of Antwerp.
Western blotting
Freshly isolated tissue samples were homogenized in an appropriate volume of Laemmli sample buffer (Bio-Rad, 161-0737). Samples were then heat-denatured for 5 min in boiling water and loaded on NuPAGE 12% (Novex, NP0343BOX) or 4–12% Bis-Tris polyacrylamide gels (Novex, NP0323BOX). In some experiments, ready-to-use INSTA-Blot mouse tissue lysates (Imgenex, from 40101–40112) were applied. After electrophoresis in NuPAGE MOPS SDS Running Buffer (Novex, NP0001), proteins were transferred to an Immobilon-P Transfer Membrane (Millipore, IPVH304F0) according to standard procedures. Membranes were blocked in Tris-buffered saline [20 mM TRIS-HCl, pH 7.6 (Acros, 424575000), 137 mM NaCl (BDH Prolabo, 27810.295)] (TBS) containing 0.05% Tween-20 (Sigma-Aldrich, P1379) (TBS-T) and 5% non-fat dry milk (Bio-Rad, 170-6404) for 1 h. Membranes were then probed overnight at 4°C with primary antibodies in antibody dilution buffer (TBS-T containing 1% nonfat dry milk), followed by 1 h incubation with horseradish peroxidase-conjugated secondary antibody at room temperature. Antibody detection was accomplished with SuperSignal West Pico (Thermo Scientific, 34080) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific, 34096) using a Lumi-Imager (Roche, Mannheim, Germany).
Immunohistochemical staining of paraffin-embedded tissue
Sections (5 μm thick) of formalin-fixed tissue were dewaxed in toluene (BDH Prolabo, 28701.364, 2 × 5 min), immersed in isopropanol (BDH Prolabo, 20922.364, 2 × 5 min) and rehydrated in distilled water (5 min). Endogenous peroxidase activity was quenched by incubation in 3% hydrogen peroxide (Merck, 1.07210) in water for 5 min. Slides were then washed in water and incubated for 10 min at 37°C with trypsin buffer [10 mM TRIS-HCl, pH 7.8, 0.1% CaCl2 (Merck, 2382), 0.1% NaCl] containing 0.01% trypsin (Fluka, 93613). After rinsing in water, heat-mediated antigen retrieval was performed in boiling citrate [1.6 mM citric acid (Fluka, 27488), 6.9 mM sodium citrate-5,5-hydrate (Merck, 6431), pH 6.0] or EDTA (UCB, 87046079; 10 mM, pH 8.0) buffer for 10 min using a microwave. Slides were cooled down for 30 min at room temperature and then rinsed twice with distilled water and once with staining buffer [10 mM TRIS-HCl, pH 7.6, 0.9% NaCl, 0.1% Triton X-100 (Sigma-Aldrich, T8787), 0.004% thimerosal (Sigma-Aldrich, T5125)]. Nonspecific protein binding was blocked for 20 min by normal goat serum (Vectastain ABC Kit, Vector Laboratories, PK-4001) or normal horse serum (Vectastain ABC Kit, Vector Laboratories, PK-4002), depending on the animal in which the secondary antibody was raised. Subsequently, the primary antibody was applied overnight in a damp box, followed by 2 × 5 min immersion in staining buffer. Next, the slides were incubated for 30 min with biotinylated anti-rabbit or anti-mouse immunoglobulins (Vectastain ABC Kit, Vector Laboratories), 1:200 diluted in staining buffer. In case the primary antibody was already biotinylated, blocking with normal serum and incubation with biotinylated secondary antibodies were omitted. Meanwhile, Vectastain ABC Reagent was prepared by diluting avidin (reagent A, Vectastain ABC Kit) and biotinylated horseradish peroxidase (reagent B, Vectastain ABC Kit) 1:200 and 1:100, respectively, in staining buffer. In some staining reactions, the anti-mouse or anti-rabbit Envision+ system (Dako, K4000 or K4002) was used rather than Vectastain ABC reagent. Envision+ solutions were ready-to-use and did not require biotinylated primary antibodies. After washing (2 × 5 min in staining buffer), slides were incubated for 1 h with Vectastain ABC reagent (or undiluted Envision+), rinsed (2 × 5 min) in staining buffer and distilled water, prior to incubation with peroxidase substrate solution [20 mg 3-amino-9-ethylcarbazole (Sigma-Aldrich, A5754) dissolved in 24 ml DMSO (Fisher Scientific, D/4121/PB15), then mixed with 200 ml sodium acetate, pH 5.2 (Merck, 6268), and supplemented with 4 ml 1:100 diluted hydrogen peroxide in water] until the desired stain intensity developed. Finally, sections were rinsed in distilled water, counterstained for 1 min using Carazzi’s hematoxylin, rinsed in water and mounted under glass coverslips.
Immunohistochemical staining of tissue fixed with Bouin’s fixative or methacarn was comparable with that of formalin-fixed tissue, except that sections were not treated with trypsin and that the heat-mediated antigen retrieval step was omitted. All stained sections were analyzed using a color image analysis system (Image Pro Plus 4.1, Media Cybernetics Inc.).
Immunohistochemical staining of Neg-50-embedded, frozen tissue
Sections (6 μm thick) of frozen, Neg-50 embedded tissue were dried for 1 h at room temperature before fixation (5 min) in acetone, 4% paraformaldehyde or methacarn. After drying, endogenous peroxidase activity was quenched by incubation in methanol/water (1:1) containing 0.3% hydrogen peroxide for 20 min. Slides were then washed in water and incubated in modified staining buffer [10 mM TRIS-HCl, pH 7.6, 1% bovine serum albumin (Sigma-Aldrich, A7906)] for 5 min. Nonspecific protein binding was blocked by normal serum, as described above, prior to incubation with the primary antibody. All further steps were comparable with the staining procedure of paraffin-embedded tissue, except that staining buffer was always replaced with the modified staining buffer. In case the primary antibody was biotinylated, blocking with normal serum and incubation with biotinylated secondary antibodies were omitted.
In some experiments, frozen sections were stained using a Vectastain ABC-AP kit (Vector Laboratories, AK-5000) containing biotinylated alkaline phosphatase instead of biotinylated horseradish peroxidase. The procedure is similar with the staining steps for frozen sections described above, except quenching of endogenous peroxidase is no longer required. After incubation with Vectastain ABC reagent, sections were immersed in Fast Blue BB solution [5 ml Tris-buffered naphtol AS-MX phosphate, 2 ml N,N-dimethylformamide (Sigma-Aldrich, D4254), 20 mg napthol AS-MX phosphate (Sigma-Aldrich, N4875), 98 ml Tris-HCL 100 mM pH 8.5] and 30 μl Fast Blue BB (Sigma-Aldrich, F3378; 5% in N,N-dimethylformamide) supplemented with 5 drops of 1 mM levamisole (Sigma-Aldrich, L9756) to inactivate endogenous alkaline phosphatase. When the desired stain intensity developed (usually after 5–20 min), sections were rinsed in distilled water, counterstained for 5 min using Nuclear Fast Red (Klinipath, 642425), rinsed in water and mounted under glass coverslips.
Oil red O staining of Neg-50-embedded, frozen tissue
Sections (6 μm thick) of frozen, Neg-50 embedded tissue were dried for 1 h at room temperature before fixation (5 min) in 4% paraformaldehyde. Thereafter, sections were immersed in propylene glycol (Sigma-Aldrich, P4347) for 2 min and stained in oil red O solution (5 mg/ml, Sigma-Aldrich, O0625) for 1 h. Sections were destained in 85% propylene glycol (1 min), rinsed in saline, counterstained for 5 min using Carazzi’s hematoxylin, rinsed again in saline and mounted under glass coverslips.
Confocal microscopy
For examination of GFP fluorescence in tissue of GFP-LC3 transgenic mice, tissue samples were fixed for 24 h in 4% neutral-buffered formalin prior to being embedded in Neg-50 Frozen Section Medium and stored at −80°C. After mounting of sections (6 μm thick) using Vectashield medium with DAPI (Vector Laboratories, H-1200), images of single confocal planes were taken with a Zeiss LSM 510 confocal microscope (Carl Zeiss GmbH) equipped with an argon laser (excitation: 488 nm) for the visualization of GFP. Images were captured with a water immersion 63× C-Apochromat (numerical aperture 1.2) objective. In order to reduce photobleaching, excitation of GFP was only at 7% of the maximum laser power output (25 mW). A pinhole of 112 μm, giving optical slices of 1.6 μm thickness, and a 500- to 550-nm bandpass filter were used for GFP fluorescence detection. Images were analyzed with LSM 4.2 software (Carl Zeiss).
Transmission electron microscopy
Transmission electron microscopy was performed as previously described.Citation52 In brief, tissue was fixed in 0.1 M sodium cacodylate (Sigma-Aldrich, C0250)-buffered (pH 7.4) 2.5% glutaraldehyde solution (Merck, 1.04239) for 2 h, then rinsed (3 x 10 min) in 0.1 M sodium cacodylate-buffered (pH 7.4) 7.5% saccharose (Gibco BRL, 15712-011) and postfixed in 1% OsO4 solution (Electron Microscopy Sciences, 19134) for 1 h. After dehydration in an ethanol gradient (70% ethanol for 20 min, 96% ethanol for 20 min, 100% ethanol for 2 × 20 min), samples were embedded in Durcupan ACM [Sigma, single component A (44611), single component B (44612) and single component D (44614)]. Ultrathin sections were stained with uranyl acetate (Merck, K3312073) and lead citrate [made with sodium citrate (Fluka, 71405) and lead nitrate (Sigma, 31137)] and examined with a Tecnai G2 Spirit BioTwin electron microscope (FEI) at 80 kV.
Statistical analysis
Results are expressed as mean ± SEM. All analyses were performed using IBM SPSS Statistics software (version 20). p < 0.05 was considered statistically significant. The applied statistical tests are mentioned in the figure legends.
| Abbreviations: | ||
| ACTB | = | β-actin |
| Alb | = | albumin |
| Atg | = | autophagy-related |
| BECN1 | = | Beclin 1 |
| CTSD | = | cathepsin D |
| GFP | = | green fluorescent protein |
| LC3 | = | microtubule-associated protein 1 light chain 3 |
| LD | = | lipid droplet |
| SQSTM1 | = | sequestosome 1 |
| TEM | = | transmission electron microscopy |
Acknowledgments
The authors are indebted to Rita Van Den Bossche, Hermine Fret, Anne-Elise Van Hoydonck, Lieve Svensson, Francis Terloo and Dominique De Rijck for excellent technical assistance. The authors also thank Dr. Masaaki Komatsu (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) for providing Atg7F/F mice. This work was supported by the Fund for Scientific Research (FWO)-Flanders (projects G.0431.11N, G.0448.11N, G.0443.12N and G.0074.12N) and the University of Antwerp (BOF). The Tecnai G2 Spirit BioTwin TEM was purchased with support of the Hercules Foundation (Hercules Type 2: AUHA004).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814 - 22; http://dx.doi.org/10.1038/ncb0910-814; PMID: 20811353
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728 - 41; http://dx.doi.org/10.1016/j.cell.2011.10.026; PMID: 22078875
- Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol 2004; 36:2491 - 502; http://dx.doi.org/10.1016/j.biocel.2004.02.005; PMID: 15325587
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy 2007; 3:181 - 206; PMID: 17224625
- Swanlund JM, Kregel KC, Oberley TD. Investigating autophagy: quantitative morphometric analysis using electron microscopy. Autophagy 2010; 6:270 - 7; http://dx.doi.org/10.4161/auto.6.2.10439; PMID: 19923921
- Eskelinen EL, Reggiori F, Baba M, Kovács AL, Seglen PO. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy 2011; 7:935 - 56; http://dx.doi.org/10.4161/auto.7.9.15760; PMID: 21566462
- Larsen KB, Lamark T, Øvervatn A, Harneshaug I, Johansen T, Bjørkøy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy 2010; 6:784 - 93; http://dx.doi.org/10.4161/auto.6.6.12510; PMID: 20574168
- Eng KE, Panas MD, Karlsson Hedestam GB, McInerney GM. A novel quantitative flow cytometry-based assay for autophagy. Autophagy 2010; 6:634 - 41; http://dx.doi.org/10.4161/auto.6.5.12112; PMID: 20458170
- Akazawa H, Komazaki S, Shimomura H, Terasaki F, Zou Y, Takano H, et al. Diphtheria toxin-induced autophagic cardiomyocyte death plays a pathogenic role in mouse model of heart failure. J Biol Chem 2004; 279:41095 - 103; http://dx.doi.org/10.1074/jbc.M313084200; PMID: 15272002
- Zhu H, Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Methods Enzymol 2009; 453:343 - 63; http://dx.doi.org/10.1016/S0076-6879(08)04017-2; PMID: 19216915
- Martinet W, De Meyer GRY, Andries L, Herman AG, Kockx MM. In situ detection of starvation-induced autophagy. J Histochem Cytochem 2006; 54:85 - 96; http://dx.doi.org/10.1369/jhc.5A6743.2005; PMID: 16148314
- Martinet W, De Meyer GRY, Andries L, Herman AG, Kockx MM. Detection of autophagy in tissue by standard immunohistochemistry: possibilities and limitations. Autophagy 2006; 2:55 - 7; PMID: 16874065
- Othman EQ, Kaur G, Mutee AF, Muhammad TS, Tan ML. Immunohistochemical expression of MAP1LC3A and MAP1LC3B protein in breast carcinoma tissues. J Clin Lab Anal 2009; 23:249 - 58; http://dx.doi.org/10.1002/jcla.20309; PMID: 19623642
- Ma JF, Huang Y, Chen SD, Halliday G. Immunohistochemical evidence for macroautophagy in neurones and endothelial cells in Alzheimer’s disease. Neuropathol Appl Neurobiol 2010; 36:312 - 9; http://dx.doi.org/10.1111/j.1365-2990.2010.01067.x; PMID: 20102518
- Holt SV, Wyspianska B, Randall KJ, James D, Foster JR, Wilkinson RW. The development of an immunohistochemical method to detect the autophagy-associated protein LC3-II in human tumor xenografts. Toxicol Pathol 2011; 39:516 - 23; http://dx.doi.org/10.1177/0192623310396903; PMID: 21441228
- Sivridis E, Koukourakis MI, Zois CE, Ledaki I, Ferguson DJ, Harris AL, et al. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Am J Pathol 2010; 176:2477 - 89; http://dx.doi.org/10.2353/ajpath.2010.090049; PMID: 20382705
- Rosenfeldt MT, Nixon C, Liu E, Mah LY, Ryan KM. Analysis of macroautophagy by immunohistochemistry. Autophagy 2012; 8:963 - 9; http://dx.doi.org/10.4161/auto.20186; PMID: 22562096
- Ladoire S, Chaba K, Martins I, Sukkurwala AQ, Adjemian S, Michaud M, et al. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy 2012; 8:1175 - 84; http://dx.doi.org/10.4161/auto.20353; PMID: 22647537
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007; 3:542 - 5; PMID: 17611390
- Eskelinen EL. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy 2008; 4:257 - 60; PMID: 17986849
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445 - 544; http://dx.doi.org/10.4161/auto.19496; PMID: 22966490
- Wu J, Dang Y, Su W, Liu C, Ma H, Shan Y, et al. Molecular cloning and characterization of rat LC3A and LC3B--two novel markers of autophagosome. Biochem Biophys Res Commun 2006; 339:437 - 42; http://dx.doi.org/10.1016/j.bbrc.2005.10.211; PMID: 16300744
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069 - 75; http://dx.doi.org/10.1038/nature06639; PMID: 18305538
- Martinet W, Agostinis P, Vanhoecke B, Dewaele M, De Meyer GRY. Autophagy in disease: a double-edged sword with therapeutic potential. Clin Sci (Lond) 2009; 116:697 - 712; http://dx.doi.org/10.1042/CS20080508; PMID: 19323652
- Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol 2012; 226:255 - 73; http://dx.doi.org/10.1002/path.3025; PMID: 21990109
- Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, et al. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol 2008; 33:461 - 8; PMID: 18695874
- Giatromanolaki A, Koukourakis MI, Harris AL, Polychronidis A, Gatter KC, Sivridis E. Prognostic relevance of light chain 3 (LC3A) autophagy patterns in colorectal adenocarcinomas. J Clin Pathol 2010; 63:867 - 72; http://dx.doi.org/10.1136/jcp.2010.079525; PMID: 20876316
- Sivridis E, Giatromanolaki A, Karpathiou G, Karpouzis A, Kouskoukis C, Koukourakis MI. LC3A-positive “stone-like” structures in cutaneous squamous cell carcinomas. Am J Dermatopathol 2011; 33:285 - 90; http://dx.doi.org/10.1097/DAD.0b013e3181f10de0; PMID: 21430508
- Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 2011; 7:727 - 36; http://dx.doi.org/10.4161/auto.7.7.15371; PMID: 21471734
- Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem 1981; 256:7652 - 8; PMID: 7019210
- Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci U S A 1983; 80:2179 - 83; http://dx.doi.org/10.1073/pnas.80.8.2179; PMID: 6340116
- Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 2011; 12:226; http://dx.doi.org/10.1186/gb-2011-12-7-226; PMID: 21867568
- Zois CE, Giatromanolaki A, Sivridis E, Papaiakovou M, Kainulainen H, Koukourakis MI. “Autophagic flux” in normal mouse tissues: focus on endogenous LC3A processing. Autophagy 2011; 7:1371 - 8; http://dx.doi.org/10.4161/auto.7.11.16664; PMID: 21997374
- Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 2007; 3:323 - 8; PMID: 17387262
- Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, et al. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun 2009; 382:419 - 23; http://dx.doi.org/10.1016/j.bbrc.2009.03.039; PMID: 19285958
- Shibata M, Yoshimura K, Tamura H, Ueno T, Nishimura T, Inoue T, et al. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem Biophys Res Commun 2010; 393:274 - 9; http://dx.doi.org/10.1016/j.bbrc.2010.01.121; PMID: 20132792
- Knaapen MW, Davies MJ, De Bie M, Haven AJ, Martinet W, Kockx MM. Apoptotic versus autophagic cell death in heart failure. Cardiovasc Res 2001; 51:304 - 12; http://dx.doi.org/10.1016/S0008-6363(01)00290-5; PMID: 11470470
- Shimomura H, Terasaki F, Hayashi T, Kitaura Y, Isomura T, Suma H. Autophagic degeneration as a possible mechanism of myocardial cell death in dilated cardiomyopathy. Jpn Circ J 2001; 65:965 - 8; http://dx.doi.org/10.1253/jcj.65.965; PMID: 11716248
- Kostin S, Pool L, Elsässer A, Hein S, Drexler HC, Arnon E, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res 2003; 92:715 - 24; http://dx.doi.org/10.1161/01.RES.0000067471.95890.5C; PMID: 12649263
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005; 169:425 - 34; http://dx.doi.org/10.1083/jcb.200412022; PMID: 15866887
- Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci 1998; 21:516 - 20; http://dx.doi.org/10.1016/S0166-2236(98)01276-4; PMID: 9881849
- Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, et al. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS 2007; 115:1344 - 9; http://dx.doi.org/10.1111/j.1600-0463.2007.00858.x; PMID: 18184403
- Li BX, Li CY, Peng RQ, Wu XJ, Wang HY, Wan DS, et al. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy 2009; 5:303 - 6; http://dx.doi.org/10.4161/auto.5.3.7491; PMID: 19066461
- Wan XB, Fan XJ, Chen MY, Xiang J, Huang PY, Guo L, et al. Elevated Beclin 1 expression is correlated with HIF-1alpha in predicting poor prognosis of nasopharyngeal carcinoma. Autophagy 2010; 6:395 - 404; http://dx.doi.org/10.4161/auto.6.3.11303; PMID: 20150769
- Huang JJ, Li HR, Huang Y, Jiang WQ, Xu RH, Huang HQ, et al. Beclin 1 expression: a predictor of prognosis in patients with extranodal natural killer T-cell lymphoma, nasal type. Autophagy 2010; 6:777 - 83; http://dx.doi.org/10.4161/auto.6.6.12784; PMID: 20639699
- Giatromanolaki A, Koukourakis MI, Koutsopoulos A, Chloropoulou P, Liberis V, Sivridis E. High Beclin 1 expression defines a poor prognosis in endometrial adenocarcinomas. Gynecol Oncol 2011; 123:147 - 51; http://dx.doi.org/10.1016/j.ygyno.2011.06.023; PMID: 21741077
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007; 131:1149 - 63; http://dx.doi.org/10.1016/j.cell.2007.10.035; PMID: 18083104
- Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol 2002; 160:255 - 63; http://dx.doi.org/10.1016/S0002-9440(10)64369-6; PMID: 11786419
- Komatsu M, Kageyama S, Ichimura Y. p62/SQSTM1/A170: Physiology and pathology. Pharmacol Res 2012; 66:457 - 62; http://dx.doi.org/10.1016/j.phrs.2012.07.004; PMID: 22841931
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15:1101 - 11; http://dx.doi.org/10.1091/mbc.E03-09-0704; PMID: 14699058
- Kuma A, Mizushima N. Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy 2008; 4:61 - 2; PMID: 17786029
- Dewaele M, Verfaillie T, Martinet W, Agostinis P. Death and survival signals in photodynamic therapy. Methods Mol Biol 2010; 635:7 - 33; http://dx.doi.org/10.1007/978-1-60761-697-9_2; PMID: 20552337