Abstract
Regulated degradation of cellular components by lysosomes is essential to maintain biological homeostasis. In mammals, three forms of autophagy, macroautophagy, microautophagy and chaperone-mediated autophagy (CMA), have been identified. Here, we showed a novel type of autophagy, in which RNA is taken up directly into lysosomes for degradation. This pathway, which we term “RNautophagy,” is ATP-dependent, and unlike CMA, is independent of HSPA8/Hsc70. LAMP2C, a lysosomal membrane protein, serves as a receptor for this pathway. The cytosolic tail of LAMP2C specifically binds to almost all total RNA derived from mouse brain. The cytosolic sequence of LAMP2C and its affinity for RNA are evolutionarily conserved from nematodes to humans. Our findings shed light on the mechanisms underlying RNA homeostasis in higher eukaryotes.
Degradation of macromolecules by lysosomes is a fundamental event for biological homeostasis. In mammals, three types of autophagy, macroautophagy, microautophagy and chaperone-mediated autophagy (CMA), have been identified.Citation1,Citation2 LAMP2 is a major lysosomal membrane protein with a single transmembrane region.Citation3 Lack of LAMP2 causes Danon disease, which is characterized by cardiomyopathy, myopathy and variable mental retardation.Citation4 LAMP2-deficient mice show increased mortality between 20 and 40 days of age.Citation5 LAMP2A, one of three splice variants of LAMP2, serves as a receptor for CMA through its interaction with substrate proteins on the lysosomal membrane surface.Citation6-Citation8 In CMA, substrate proteins are unfolded by HSPA8/Hsc70, and then directly imported into the lysosomal lumen. Each of three splice variants, LAMP2A, LAMP2B and LAMP2C, possesses an identical lumenal region and different cytosolic sequences.Citation3 The cytosolic sequence of LAMP2C is completely conserved among chicken, mouse and human, while those of LAMP2A and LAMP2B are not.Citation3 The C-terminal sequences of fly and nematode LAMP orthologs show the strongest homology to that of LAMP2C among human LAMP2s (). These observations suggest that LAMP2C is evolutionarily the primary form of LAMP, and that it plays an important role in lysosomes. However, the precise functions of LAMP2C remain unknown.
Figure 1. The cytosolic sequence of LAMP2C interacts with RNA-binding proteins. (A) Identity levels of the cytosolic sequences of fly and nematode LAMPs to that of human LAMP2s. (B) A schematic representation of biotin-conjugated peptides. (C) Proteins that interact with the cytosolic sequences of LAMP2s were analyzed by pull-down assay followed by LC-MS/MS analysis of LAMP2C peptide-interacting proteins. (D) Expression levels of Lamp2c mRNA in mouse tissues. Values are the means ± SE (n = 3 to 4). (E) Pull-down assay and LC-MS/MS analysis of LAMP2C peptide-interacting proteins from mouse brain. The distribution of the identified proteins is shown.
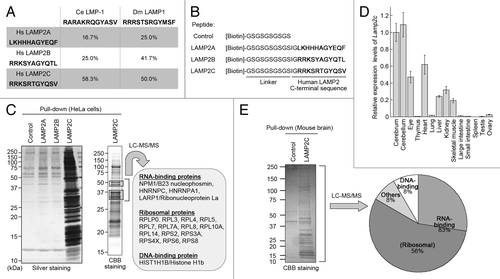
To clarify the specific functions of LAMP2C and LAMP2B, we searched for proteins that interact with the cytosolic sequences of each LAMP2. We have previously shown that a peptide construct of the cytosolic sequence of human LAMP2A is useful as a tool for monitoring proteins interacting with this sequence.Citation9 In the present study, we prepared peptide constructs of LAMP2B and LAMP2C cytosolic sequences (). A pull-down assay using these peptides with HeLa cell lysate showed that numerous proteins bound specifically to the cytosolic sequence of LAMP2C (). Mass spectrometry analysis was applied to identify the interacting proteins in the molecular weight range 30 to 50 kDa, which contained the most intense bands, as a pilot study. Interestingly, all of the identified proteins were nucleic acid-binding proteins, and predominantly RNA-binding proteins (RBPs) such as ribosomal proteins and hnRNPs (; Table S1).
To examine the protein interactions under more physiological conditions, we then performed a pull-down assay using mouse brain lysate because, among various tissue types, Lamp2c mRNA showed the highest expression levels in the brain (). Lamp2c was expressed in neurons while it was hardly detected in glial cells (Fig. S1). Mass spectrometry analysis of all protein bands detected in the pull-down assay revealed that LAMP2C peptide predominantly interacted with a wide range of RBPs (; Table S1).
We next examined whether the interactions of RBPs with the cytosolic sequence of LAMP2C were mediated by RNA. The protein interactions of LAMP2C peptide were almost completely abolished by pretreatment of the lysate with protease-free RNase A in a pull-down assay (). Substitutions of phenylalanines at the RNA-binding sites of HNRNPA1, which has a typical RNA-binding motif,Citation10,Citation11 completely abolished its interaction with LAMP2C peptide (). These data suggest that the interactions of RBPs with the cytosolic tail of LAMP2C are indirect associations mediated by RNA.
Figure 2. The cytosolic sequence of LAMP2C directly interacts with RNA. (A) Protein interactions of LAMP2C peptide were analyzed by pull-down assay using brain lysates preincubated with or without RNase A. (B) Pull-down assay using lysates of HeLa cells transfected with the indicated constructs. (C) A pull-down assay was performed using brain lysate, and RNA was detected with EtBr. The intense signal in the input lane (*) is presumably degraded RNA in the brain lysate. (D) Interactions of purified total RNA with cytosolic sequence of LAMP2C. Amounts of RNA remaining in the flow-through fraction were quantified by measuring OD260 (n = 4). (E) Interactions of purified total RNA with cytosolic sequences of nematode and fly LAMPs.
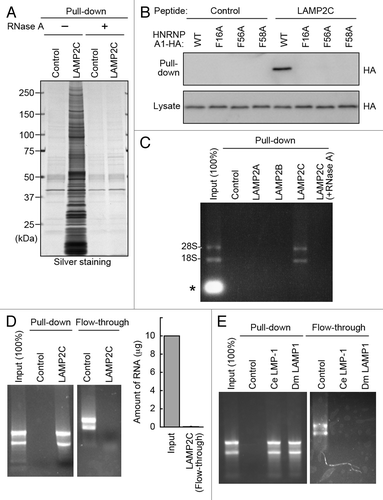
We then investigated the interaction between RNAs and the cytosolic sequence of LAMP2C. A pull-down assay in the same experimental condition using mouse brain lysate showed an interaction of RNA specifically with the LAMP2C peptide among LAMP2 constructs (). Using purified total RNA derived from mouse brain, the cytosolic sequence of LAMP2C was shown to bind directly to a wide range of RNAs (). The RNA was hardly detected in the flow-through after being pulled down with LAMP2C peptide (), indicating that LAMP2C peptide bound to almost all RNAs. An endogenous interaction between LAMP2C and RNA was also observed (Fig. S2). In a pull-down assay using purified RNA, RNA partly interacted with the cytosolic sequence of LAMP2B, but not with that of LAMP2A (Fig. S3). Considering that LAMP2B peptide did not interact with RNA in mouse brain lysate, this interaction may be nonphysiological. Intriguingly, the C-terminal sequences of both fly and nematode LAMP orthologs exhibited high affinity for RNA, similar to that of LAMP2C ().
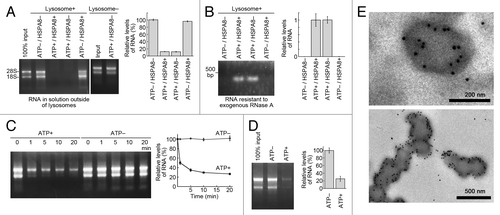
Considering the relationship between LAMP2A and CMA, we hypothesized a novel autophagic pathway that directly imports RNA into lysosomes via LAMP2C. The standard method for monitoring direct uptake of molecules into lysosomes is a cell-free system using isolated lysosomes.Citation12-Citation14 Freshly isolated lysosomes were prepared from mouse brain (Fig. S4). We confirmed that the isolated lysosomes were intact and that CMA was working properly in a cell-free system (Fig. S5B). For monitoring uptake of RNA, the lysosomes were incubated with total RNA in the presence or absence of ATP and/or HSPA8. Subsequently, the lysosomes were precipitated by centrifugation, and the levels of RNA remaining in the supernatant were analyzed. In parallel, to assess the presence of RNA taken up inside the lysosomes, we degraded RNA outside of lysosomes by RNase A treatment after the incubation, and then analyzed the levels of RNase A-resistant RNA. As a result, the amount of RNA outside of lysosomes was dramatically reduced only in the presence of ATP (). Levels of RNA were reduced over time in the presence of ATP, but remained unchanged in the absence of ATP (). RNA resistant to exogenous RNase A was detected only in lysosomes incubated in the presence of ATP (). These results indicate that RNA is taken up by lysosomes in a process that is dependent on ATP. Immunoelectron microscopy of the lysosomes incubated with ATP and RNA, or with ATP only, also showed the presence of RNA taken up in lysosomes (; Fig. S6). Unlike CMA, HSPA8 had no effect on the uptake of RNA into isolated lysosomes ().
Figure 3. Uptake and degradation of RNA by isolated lysosomes. (A and B) Uptake of RNA by isolated lysosomes. Isolated lysosomes were incubated with purified total RNA (5 μg) in the presence or absence of ATP (energy regeneration system) and/or HSPA8. Levels of RNA remaining in solution outside of lysosomes (A) and levels of RNA resistant to exogenous RNase A (B) were analyzed (n = 3). (C) Lysosomes were incubated with 10 μg of RNA in the presence or absence of ATP. At the indicated times, the levels of RNA remaining outside of lysosomes were analyzed (n = 3). (D) Degradation of RNA by isolated lysosomes. Lysosomes and RNA were incubated with or without ATP. Total levels of RNA in the incubated samples were analyzed (n = 3). (E) Immunogold labeling of RNA in isolated lysosomes incubated with ATP and RNA. Immunogold labeling was performed using an anti-rRNA antibody followed by anti-mouse IgG coupled with 10 nm of gold particles. Gold particles were observed in the lysosomes.
We further investigated the degradation of RNA in lysosomes. Isolated lysosomes and RNA were incubated with or without ATP, and the total levels of RNA in the samples were analyzed. The total levels of RNA were reduced in a manner that was dependent on ATP (; Fig. S7), indicating ATP-dependent degradation of RNA by isolated lysosomes. Taken together, our results revealed an autophagic pathway that directly imports RNA into lysosomes for degradation. Hereinafter, we refer to this pathway as RNautophagy.
Finally, we assessed whether LAMP2C mediates RNA degradation by RNautophagy. Cells overexpressing LAMP2C exhibited a significant enhancement in the overall levels of RNA degradation, which was not observed in those overexpressing LAMP2A or LAMP2B (; Fig. S8). Lysosomes isolated from cells overexpressing LAMP2C also showed elevated levels of RNautophagy (). Additionally, the levels of RNautophagy were significantly reduced in lysosomes derived from LAMP2-deficient mouse brains (). Moreover, a significant accumulation of total RNA was observed in the brains of LAMP2 knockout mice (). Collectively, these results indicate that LAMP2C functions as a receptor for RNautophagy. In the brains of the knockout mice, RBPs such as ribosomal protein S6 (RPS6), RPL8 and HNRNPA1 were upregulated (). RBPs may be upregulated as a response to the accumulation of RNA, since RPL8 was not directly taken up by lysosomes (Fig. S5). Also, the upregulation of RBPs did not seem to be a result of CMA impairment, because representative substrates for CMA, GAPDH and SNCA/α-synuclein,Citation15,Citation16 were not accumulated in the brains of the knockout mice (). Consistent with this notion, RPL8, RPS6 and HNRNPA1 do not contain a KFERQ motif, a sequence required for targeting to CMA.Citation17,Citation18 The constancy of CMA substrates in the knockout brains may be due to compensation by other degradation systems.Citation13
Figure 4. LAMP2C mediates lysosomal degradation of RNA. (A) RNA turnover in HeLa cells transfected with each LAMP2 isoform or empty vector (n = 3). (B and C) Uptake of RNA into lysosomes isolated from HeLa cells transfected with LAMP2C or empty vector (n = 3) (B). Uptake of RNA into lysosomes isolated from the brains of wild-type (WT) and LAMP2 knockout (KO) mice (n = 3) (C). Levels of RNA uptake were measured by subtracting the levels of RNA remaining in solution outside of lysosomes from the levels of input RNA. (D) Relative levels of total RNA in the brains of WT and LAMP2 KO mice (KO: n = 3, WT: n = 4). (E) Levels of RBPs and CMA substrate proteins in the brains of WT and LAMP2 KO mice were analyzed by immunoblotting (n = 3/group). (F) Model for the possible mechanism of RNautophagy. LAMP2C on the lysosomal membrane recognizes RNA, and the RNA is then imported into the lysosomal lumen in an ATP-dependent manner, followed by its degradation. The requirement for ATP in RNautophagy suggests an involvement of ATPases such as RNA helicases in the process on the lysosomal membrane.
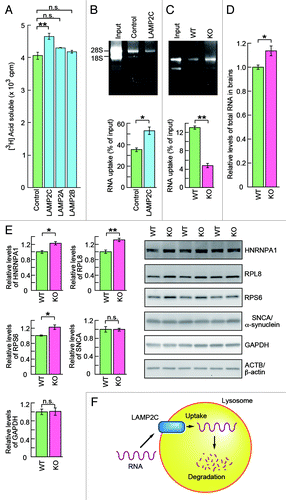
RNA degradation mechanisms in the cytosol independent of lysosomes have long been studied.Citation19 On the other hand, an autophagic pathway selectively targeting RNA has not been elucidated, even though lysosomal RNases and exonucleases have long been recognized.Citation20-Citation22 We have shown here a novel, direct type of autophagy for RNA degradation, which we term RNautophagy, and that LAMP2C acts as a receptor for this pathway. Our data suggest that RNautophagy can contribute to at least 10% to 20% of the total amount of RNA degradation in living cells (). RNautophagy activity was not completely abolished in lysosomes derived from LAMP2-deficient mice (). Thus, we cannot rule out the possibility of LAMP2-independent pathway(s) in RNautophagy.
The RNA-binding capacities of the cytosolic sequences of LAMP family proteins are widely conserved in animals from nematodes to humans (). Together with the fact that a LAMP ortholog has not been found in yeast, RNautophagy likely contributes to RNA metabolism in higher eukaryotes. In the mouse, the expression levels of Lamp2c vary from tissue to tissue, showing notable levels in brain, eye, heart, liver, kidney and skeletal muscle, while the gene was scarcely expressed in other tissues (). The different levels of Lamp2c expression among tissues could be related to requirements for RNA metabolism among tissues. RNautophagy may have an important role in RNA homeostasis in tissues, especially in those expressing LAMP2C at high levels. From this point of view, the high expression level of Lamp2c in neurons is reasonable, because neurons are rich in RNA.Citation23 RNautophagy may play an important role in the maintenance and quality control of neuronal function.
A possible model for the process of RNautophagy is shown in . First, a substrate RNA binds to the cytosolic tail of LAMP2C on the surface of the lysosomal membrane. This process may be regulated by other RNA-interacting proteins in the cytosol, because the cytosolic sequence of LAMP2C itself does not seem to have selectivity in its binding to RNA. Second, RNA is directly imported into the lysosomal lumen. LAMP2C and other proteins may be involved in this translocation process, which is ATP-dependent. RNA-dependent ATPases such as RNA helicases may operate in this process to unfold RNA, similar to the function of HSPA8 in CMA. Third, RNA taken up into lysosomes is degraded by lysosomal enzymes such as acid RNases and exonucleases.
We speculate that DNA could bind to the cytosolic tail of LAMP2C, because LAMP2C peptide interacted with DNA-binding proteins in addition to RBPs (). The interaction between DNA and LAMP2C is currently the subject of ongoing research.
Because a lack of LAMP2 causes Danon disease, RNautophagy dysfunction may contribute to the pathogenesis of that disease. In addition, aberrant expression, function and accumulation of RNA have been considered as key factors in the pathogenesis of various diseases such as macular degeneration, myotonic dystrophies and some neurodegenerative disorders.Citation24 In the immune system, antiviral signaling is thought to be initiated intracellularly, within endosomes/lysosomes containing viral nucleic acid.Citation25 LAMP2 has been used as a macrophage marker.Citation26,Citation27 Synthesis and expression of LAMP2 increase when macrophages are activated.Citation26,Citation27 Given that Lamp2c is highly expressed in a macrophage cell line (Fig. S1), RNautophagy is possibly involved in the immune response. For instance, in certain types of cells, viral RNA could be taken up into lysosomes by RNautophagy and then be involved in antiviral responses via toll-like receptors on the endosomal/lysosomal membrane. The possible involvement of RNautophagy in the immune system and in lysosome- or RNA-related disorders warrants investigation.
Materials and Methods
Uptake and degradation of RNA by isolated lysosomes
Lysosomes were isolated from 10 to 12-week-old mouse brains or from ~1.4 × 108 HeLa cells by density gradient centrifugation. Isolated lysosomes (25 to 50 μg of protein) were then incubated with 5 or 10 μg of purified total RNA at 37°C in 30 μl of 0.3 M sucrose containing 10 mM MOPS buffer (pH 8.0) with or without energy regeneration system (10 mM ATP, 10 mM MgCl2, 2 mM phosphocreatine, and 50 μg/ml creatine phosphokinase). After the incubation, lysosomes were precipitated by centrifugation and the levels of RNA remaining in the supernatant were analyzed. To assess the presence of uptaken RNA inside lysosomes, RNA outside of lysosomes was degraded by 200 μg of RNase A after the incubation, and the levels of RNase A-resistant RNA were analyzed. Degradation of RNA by isolated lysosomes was investigated by analyzing the total levels of RNA in the samples.
Immunogold electron microscopy
Isolated lysosomes were fixed in 0.1% glutaraldehyde and 4% paraformaldehyde in PBS for 16 h at 4°C. Samples were then dehydrated in a series of water/ethanol mixtures to 100% ethanol, and embedded in LR White (Nisshin EM Co., Ltd., 3962). The embedded samples were sectioned at 100 nm, and collected on 400-mesh nickel grids. Immunogold labeling was performed using an anti-rRNA antibody followed by anti-mouse IgG coupled with 10 nm of gold particles, and samples were viewed using a Tecnai Spirit transmission electron microscope (FEI) at 80 kV.
Statistical analyses
For comparisons between two groups, the statistical significance of differences was determined by Student’s t test. For comparisons of more than two groups, Dunnett’s multiple comparison test was used. * and ** indicate p < 0.05 and p < 0.01, respectively.
Further details on materials and methods are provided in Supplementary Materials and Methods.
| Abbreviations: | ||
| CMA | = | chaperone-mediated autophagy |
| RBPs | = | RNA-binding proteins LAMP, lysosome-associated membrane protein |
| LMP | = | LAMP/CD68-like membrane protein |
| HSPA8 | = | heat shock 70 kD protein 8 |
| Hsc70 | = | 70 kD heat shock cognate protein |
| RP | = | ribosomal protein |
| HNRNP | = | heterogenous nuclear ribonucleoprotein |
| GAPDH | = | glyceraldehyde-3-phosphatedehydrogenase |
| RNase | = | ribonuclease |
| ATPase | = | adenosine triphosphatase |
| WT | = | wild type |
| KO | = | knockout |
| Hs | = | Homo sapiens |
| Ce | = | Caenorhabditis elegans |
| Dm | = | Drosophila melanogaster |
Additional material
Download Zip (740.8 KB)Acknowledgments
We thank Yohei Fujimoto, Hiromi Fujita, Yoshiko Hara and Tomoko Okada for technical assistance, Drs. Yoshitaka Nagai, Hirohiko Hohjoh and Toshihide Takeuchi for helpful discussions, and Drs. Judith Blanz and Paul Saftig for the LAMP2-deficient mice. This work was supported by a Grant-in-Aid for Young Scientists (A) (24680038 to T.K.) and Grants-in-Aid for Scientific Research (to K.W.) from the Japan Society for the Promotion of Science; by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO), Japan (to K.W.); and partly by a Grant-in-Aid for Young Scientists (B) (22790838 to T.K.) and Grants-in-Aid for Scientific Research on Nervous and Mental Disorders from the Ministry of Health, Labour, and Welfare of Japan (21-9, 23-1 and 24-11 to T.K.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/23002
References
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147:728 - 41; http://dx.doi.org/10.1016/j.cell.2011.10.026; PMID: 22078875
- Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 2011; 20:131 - 9; http://dx.doi.org/10.1016/j.devcel.2010.12.003; PMID: 21238931
- Eskelinen E-L, Cuervo AM, Taylor MR, Nishino I, Blum JS, Dice JF, et al. Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic 2005; 6:1058 - 61; http://dx.doi.org/10.1111/j.1600-0854.2005.00337.x; PMID: 16190986
- Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 2000; 406:906 - 10; http://dx.doi.org/10.1038/35022604; PMID: 10972294
- Tanaka Y, Guhde G, Suter A, Eskelinen E-L, Hartmann D, Lüllmann-Rauch R, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 2000; 406:902 - 6; http://dx.doi.org/10.1038/35022595; PMID: 10972293
- Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 1996; 273:501 - 3; http://dx.doi.org/10.1126/science.273.5274.501; PMID: 8662539
- Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci 2000; 113:4441 - 50; PMID: 11082038
- Dice JF. Chaperone-mediated autophagy. Autophagy 2007; 3:295 - 9; PMID: 17404494
- Kabuta T, Furuta A, Aoki S, Furuta K, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem 2008; 283:23731 - 8; http://dx.doi.org/10.1074/jbc.M801918200; PMID: 18550537
- Merrill BM, Stone KL, Cobianchi F, Wilson SH, Williams KR. Phenylalanines that are conserved among several RNA-binding proteins form part of a nucleic acid-binding pocket in the A1 heterogeneous nuclear ribonucleoprotein. J Biol Chem 1988; 263:3307 - 13; PMID: 2830282
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 2005; 272:2118 - 31; http://dx.doi.org/10.1111/j.1742-4658.2005.04653.x; PMID: 15853797
- Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 1989; 246:382 - 5; http://dx.doi.org/10.1126/science.2799391; PMID: 2799391
- Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A 2006; 103:5805 - 10; http://dx.doi.org/10.1073/pnas.0507436103; PMID: 16585521
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy 2007; 3:181 - 206; PMID: 17224625
- Aniento F, Roche E, Cuervo AM, Knecht E. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J Biol Chem 1993; 268:10463 - 70; PMID: 8486700
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 2004; 305:1292 - 5; http://dx.doi.org/10.1126/science.1101738; PMID: 15333840
- Dice JF, Chiang HL, Spencer EP, Backer JM. Regulation of catabolism of microinjected ribonuclease A. Identification of residues 7-11 as the essential pentapeptide. J Biol Chem 1986; 261:6853 - 9; PMID: 3700419
- Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci 1990; 15:305 - 9; http://dx.doi.org/10.1016/0968-0004(90)90019-8; PMID: 2204156
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell 2009; 136:763 - 76; http://dx.doi.org/10.1016/j.cell.2009.01.019; PMID: 19239894
- Futai M, Miyata S, Mizuno D. Acid ribonucleases of lysosomal and soluble fractions from rat liver. J Biol Chem 1969; 244:4951 - 60; PMID: 4980956
- Saha BK, Graham MY, Schlessinger D. Acid ribonuclease from HeLa cell lysosomes. J Biol Chem 1979; 254:5951 - 7; PMID: 36388
- Pisoni RL. Lysosomal nucleic acid and phosphate metabolism and related metabolic reactions. Subcell Biochem 1996; 27:295 - 330; http://dx.doi.org/10.1007/978-1-4615-5833-0_9; PMID: 8993164
- Stoykova AS, Dabeva MD, Dimova RN, Hadjiolov AA. Ribosome biogenesis and nucleolar ultrastructure in neuronal and oligodendroglial rat brain cells. J Neurochem 1985; 45:1667 - 76; http://dx.doi.org/10.1111/j.1471-4159.1985.tb10521.x; PMID: 3850925
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell 2009; 136:777 - 93; http://dx.doi.org/10.1016/j.cell.2009.02.011; PMID: 19239895
- Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem Pharmacol 2008; 75:589 - 602; http://dx.doi.org/10.1016/j.bcp.2007.07.043; PMID: 17868652
- Ho MK, Springer TA. Tissue distribution, structural characterization, and biosynthesis of Mac-3, a macrophage surface glycoprotein exhibiting molecular weight heterogeneity. J Biol Chem 1983; 258:636 - 42; PMID: 6336756
- Garner RE, Malick AP, Yurochko AD, Elgert KD. Shifts in macrophage (M phi) surface phenotypes during tumor growth: association of Mac-2+ and Mac-3+ M phi with immunosuppressive activity. Cell Immunol 1987; 108:255 - 68; http://dx.doi.org/10.1016/0008-8749(87)90211-5; PMID: 2957065