Abstract
Amino acids, leucine in particular, are known to inhibit autophagy, at least in part by their ability to stimulate MTOR-mediated signaling. Evidence is presented showing that glutamate dehydrogenase, the central enzyme in amino acid catabolism, contributes to leucine sensing in the regulation of autophagy. The data suggest a dual mechanism by which glutamate dehydrogenase activity modulates autophagy, i.e., by activating MTORC1 and by limiting the formation of reactive oxygen species.
Introduction
(Macro)autophagy is responsible for the degradation of long-lived proteins and for the elimination of either damaged or functionally redundant intracellular structures under stress conditions.Citation1 In starvation, amino acids produced by autophagic proteolysis are recycled and used, at least in part, for the production of ATP to maintain cell survival. Amino acids are feedback inhibitors of autophagy so that autophagic flux decreases with increasing amino acid concentration.
Ever since the discovery that amino acids inhibit autophagy by means of their ability, at least in part, to stimulate MTOR-mediated signaling in the absence of insulin,Citation2 the nature of the amino acid sensor has remained enigmatic.Citation1,Citation3-Citation7 In most cell types, among the various amino acids, leucine, but not the other branched-chain amino acids valine and isoleucine, is most potent in inhibiting autophagy and in stimulating MTOR signaling.Citation2,Citation8-Citation11 Metabolism of leucine is not requiredCitation12 and nonmetabolizable analogs of leucine can mimic its effect.Citation8,Citation11 Therefore, any proposed mechanism of amino acid sensing must account for the high leucine specificity.
Very recently, it has been demonstrated that the v-ATPase in the lysosomal membrane, in addition to its role in proton pumping, functions as an intralysosomal amino acid sensor in MTOR signaling.Citation13 A rise in the intralysosomal amino acid concentration causes a conformational change of the ATPase which results in increased binding of Ragulator, a scaffolding protein complex that anchors RRAG to the lysosomal surface. This promotes the translocation of the MTORC1 protein complex (formed by MTOR and binding partners involved in the regulation of autophagy) to the extralysosomal surface, upon which MTOR becomes activated. In this mechanism, the rate of transport of amino acids out of the lysosomes controls the concentration of amino acids within the lysosomal lumen, and thus the extent of MTORC1 activation.Citation13
In other recent studies, it has been shown that leucyl-tRNA synthetase (LARS) can also act as an amino acid sensor.Citation14,Citation15 The enzyme directly binds to RRAG GTPase in a leucine-dependent manner and functions as a GTPase-activating protein for RRAG GTPase to activate MTORC1. Leucylation of the tRNA is not required: it is sufficient that leucine binds to the leucine-binding domain of LARS and activates the enzyme.Citation15
We have previously hypothesized that glutamate dehydrogenase (GLUD/GDH), in addition to its role in amino acid catabolism, may also be involved in amino acid sensing.Citation1,Citation16,Citation17 This mitochondrial enzyme is known to be specifically activated by leucine.Citation18 In pancreatic β-cells the ability of leucine (but not of valine or isoleucine) to stimulate production of insulin has been ascribed to stimulation of GLUD.Citation19,Citation20 Moreover, a combination of glutamine (a glutamate donor) and leucine, which maximizes the flux through GLUD, is most effective in stimulating phosphorylation of RPS6KB/S6K in β-cellsCitation19 and hepatocytes,Citation21 and in inhibiting autophagic proteolysis in hepatocytes,Citation22,Citation23 Jurkat cellsCitation24 and HeLa cells.Citation25
In the present study, we describe experiments with HeLa cells in which GLUD1, the major isoenzyme, was knocked down. The data strongly suggest that GLUD1 does, indeed, contribute to the control of autophagy by amino acids, especially leucine. In addition, we showed that GLUD1, at least in part, controls autophagy by virtue of its ability to modulate MTOR signaling and to protect against the production of reactive oxygen species (ROS), known to be involved in the initiation of autophagy.Citation26-Citation31
Results
Knockdown of GLUD1 promotes autophagy and inhibits MTORC1 activity
To evaluate a potential role for GLUD1 in the regulation of autophagy, we employed an RNA interference approach in HeLa cells, resulting in 80% to 90% reduction of GLUD1 protein (). We then assessed the effect of GLUD1 knockdown on the conversion of cytosolic LC3-I to the lipid-bound LC3-II upon autophagosome formation. The amount of LC3-II reflects the number of autophagosomes and is widely accepted as a reliable measurement of autophagic flux.Citation32 Because the steady-state level of autophagosomes is dependent on both “de novo” synthesis and consumption in the lysosomal compartment, the effects on autophagic flux are best determined in the presence of bafilomycin A1, which blocks the fusion and subsequent content degradation of autophagosomes in the lysosome. Silencing GLUD1 led to a significant increase of LC3-II, both in the presence and absence of bafilomycin A1 (). In agreement, we observed diminished levels of the autophagy cargo protein SQSTM1/p62 in GLUD1-depleted cells, indicative of increased autophagic degradation (). We next evaluated the effect of GLUD1 depletion on cellular LC3 distribution in HeLa cells stably transfected with GFP-LC3. In line with the results obtained with western blot analysis, GLUD1 knockdown led to a marked increase in GFP-positive punctate structures compared with control cells (), indicating that GLUD1 is involved in the regulation of autophagy.
Figure 1. Knockdown of GLUD1 stimulates autophagy and inhibits MTORC1 activity. (A) Immunoblot analysis of GLUD1, LC3-I and LC3-II levels in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA. The cells were cultured in complete medium. Where indicated, 200 nM of bafilomycin A1 (BAF) was present for 2 h to block the lysosomal degradation of LC3-II. Immunoblotting of ACTB was used as a loading control. The LC3-II/ACTB ratio was determined using Bio-1D quantification software. Columns: mean; bars: SEM (n = 3); *p < 0.05. (B) Immunoblot analysis of SQSTM1 levels in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA. Cells were cultured in complete medium. Immunoblotting of ACTB was used as a loading control. The SQSTM1/ACTB ratio was determined using Bio-1D quantification software. Columns: mean; bars: SEM (n = 3); *p < 0.05. (C) Representative images of GFP-LC3 staining in HeLa GFP-LC3 cells following a 72 h transfection with control (ct) siRNA or GLUD1 siRNA. Where indicated, 200 nM of bafilomycin A1 (BAF) was present for 2 h. The number of GFP-LC3 dots per cell was scored on 50 to 100 cells. Columns: mean; bars: SEM (n = 3); *p < 0.05. (D) Immunoblot analysis of phospho-RPS6 (P-RPS6), RPS6, phospho-EIF4EBP1 (P-EIF4EBP1) and EIF4EBP in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA, cultured in complete medium. Columns: mean; bars: SEM (n = 3); *p < 0.05.
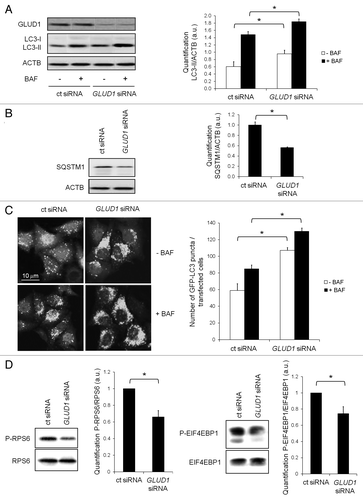
Because MTORC1 is the main repressor of autophagy under nutrient-rich conditions, we examined whether the increase in autophagy observed upon GLUD1 depletion was due to compromised MTORC1 activity. Indeed, silencing of GLUD1 reduced the phosphorylation of RPS6/S6 and EIF4EBP1/4E-BP1 (), two direct downstream substrates of MTORC1, indicating that GLUD1 negatively regulates autophagy, at least in part, through modulation of MTORC1 activity.
Leucine inhibits autophagy in a GLUD1-dependent fashion
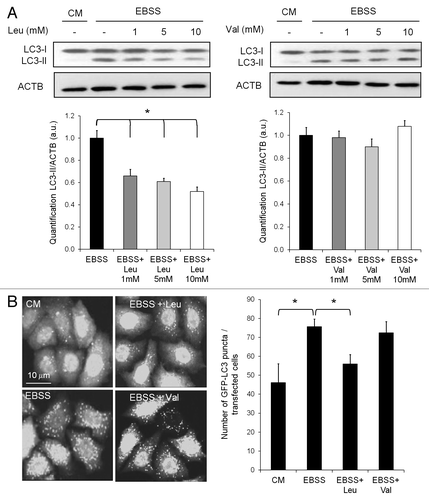
The activity of GLUD1 is specifically regulated by the amino acid leucine, which has been extensively studied as a potent suppressor of autophagy in many different cell types. In the first series of experiments, we confirmed the inhibitory effect of leucine on starvation-induced autophagy (). Leucine inhibited accumulation of LC3-II in a concentration-dependent manner upon nutrient starvation (). Similarly, leucine prevented the accumulation of GFP-LC3 punctate structures (). To verify that the inhibitory effect of leucine on autophagy was specific, we repeated the starvation experiments in the presence of the amino acid valine. The inhibitory effect of leucine on autophagy upon nutrient starvation could not be recapitulated by valine, as judged by the degree of LC3-II accumulation () and of GFP-LC3 puncta (). From these results, we concluded that leucine indeed strongly inhibits starvation-induced formation of autophagosomes.
Figure 2. Leucine, not valine, inhibits autophagy. (A) Immunoblot analysis of LC3-I and LC3-II levels in HeLa cells cultured in complete medium or EBSS supplemented with 1, 5 or 10 mM of leucine or valine for 4 h. Immunoblotting of ACTB was used as a loading control. The LC3-II/ACTB ratio was determined using Bio-1D quantification software. Columns: mean; bars: SEM (n = 3); *p < 0.05. (B) Representative images of GFP-LC3 staining in HeLa GFP-LC3 cells cultured in complete medium or EBSS supplemented with 10 mM of leucine or valine for 4 h. The number of GFP-LC3 dots per cell was scored on 100 cells. Columns: mean; bars: SEM (n = 3); *p < 0.05.
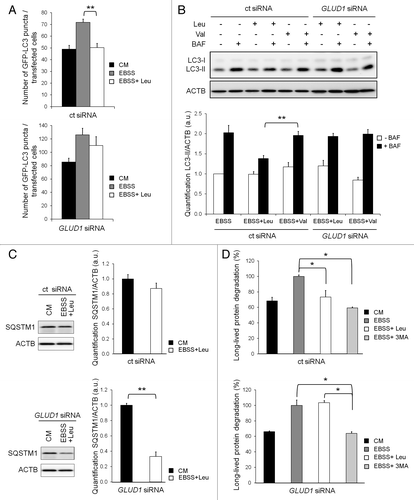
We next investigated whether the regulation of autophagy by leucine is dependent on the expression of GLUD1. For this purpose, we assessed the effect of leucine on autophagy in both control cells and in GLUD1-depleted cells (). Again, in nutrient-starved control cells, the presence of leucine was sufficient to inhibit the biosynthesis of autophagosomes as judged by the number of GFP-LC3 puncta (). In agreement, leucine normalized the autophagic flux to the level observed in cells cultured in complete medium, as shown by the absence of LC3-II accumulation () and impaired degradation of SQSTM1 (). Finally, leucine also prevented starvation-induced long-lived protein degradation (). Importantly, we found that knockdown of GLUD1 abolished the inhibitory effect of leucine on the accumulation of GFP-LC3 puncta (). The increase in autophagosomes correlated with a sustained autophagic flux and lysosomal activity as determined by the accumulation of LC3-II in bafilomycin A1-treated cells () and by the degradation of SQSTM1 (), which could no longer be suppressed by leucine in GLUD1-depleted cells. Similarly, the rate of starvation-induced long-lived protein degradation in GLUD1-depleted cells was shown to be insensitive to leucine addition, but remained sensitive to 3-MA (), an inhibitor of autophagosome formation by interfering with the activity of PIK3C3 (the class III PtdIns3K).Citation33 Taken together, these results strongly suggest that GLUD1 is required for the inhibitory effect of leucine on autophagy.
Figure 3. Knockdown of GLUD1 prevents the inhibition of autophagy by leucine. (A) The number of GFP-LC3 dots per cell was scored on 50 to 100 cells following a 72 h transfection with control (ct) siRNA (upper histogram) or GLUD1 siRNA (lower histogram) and a 4 h treatment with complete medium (CM), starved medium (EBSS) and starved medium supplemented with 10 mM Leucine (EBSS+Leu). Columns: mean; bars: SEM (n = 3); **p < 0.01. (B) Immunoblot analysis of LC3-I and LC3-II levels in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA for 72 h and cultured in EBSS supplemented with 10 mM leucine or valine for 4 h. Where indicated, 200 nM of bafilomycin A1 (BAF) was present for 2 h to block the lysosomal degradation of LC3-II. Immunoblotting of ACTB was used as a loading control. The LC3-II/ACTB ratio was determined using Bio-1D quantification software. Columns: mean; bars: SEM (n = 3); **p < 0.01. (C) Immunoblot analysis of SQSTM1 levels in HeLa cells transfected with control (ct) siRNA (upper histogram) or GLUD1 siRNA (lower histogram) and cultured for 4 h in complete medium or EBSS supplemented with 10 mM leucine. Immunoblotting of ACTB was used as a loading control. The SQSTM1/ACTB ratio was determined using Bio-1D quantification software. Columns: mean; bars: SEM (n = 3); **p < 0.01. (D) Measurement of the degradation of long-lived proteins degradation in HeLa cells transfected with control (ct) siRNA (upper western blot and histogram) or GLUD1 siRNA (lower western blot and histogram) for 72 h and cultured in complete medium or EBSS supplemented with 10 mM leucine or 10 mM 3-methyladenine (3-MA). Columns: mean; bars: SEM (n = 3); *p < 0.05.
Leucine requires GLUD1 for promoting MTORC1 activity
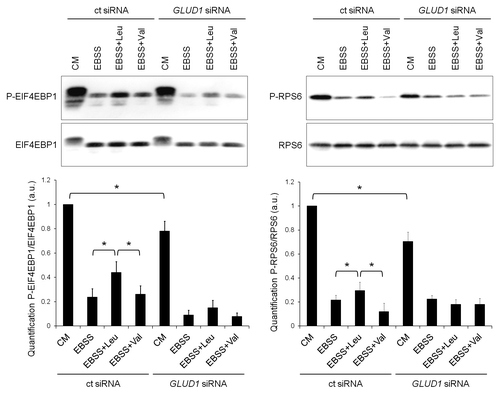
Of all amino acids, leucine is not only the most effective in inhibiting autophagy but it can also stimulate MTORC1. Because leucine apparently requires GLUD1 in order to repress autophagy, we addressed whether this was due to its ability to reactivate the MTORC1 pathway. Here we showed that the presence of leucine, at least in part, preserved MTORC1 activation in starved control cells (). However, this stimulatory effect was abolished upon GLUD1 knockdown. Valine, which does not control autophagy, was not able to stimulate the MTORC1 pathway in either condition. These results suggest that leucine inhibits autophagy, at least in part, by activating MTORC1 in a GLUD1-dependent fashion.
Figure 4. Knockdown of GLUD1 prevents the stimulation of MTORC1 signaling by leucine. Immunoblot analysis of phospho-EIF4EBP1 (P-EIF4EBP1), EIF4EBP1, phospho-RPS6 (P-RPS6) and RPS6 in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA and cultured in complete medium or EBSS supplemented with 10 mM leucine or valine for 4 h. Columns: mean; bars: SEM (n = 3); *p < 0.05.
Enhanced ROS production upon GLUD1-depletion contributes to autophagy induction
In addition to the gatekeeper function of MTORC1 in the regulation of autophagy, there is ample evidence suggesting that the intracellular levels of ROS may play a key role in the initiation of autophagy.Citation26-Citation31 Because GLUD1 activity results in the production of reducing equivalents, we evaluated whether GLUD1, in addition to its ability to promote MTORC1 activity, could also influence the formation of ROS. Initial experiments with the ROS probe DHE indicated that ROS production was higher in GLUD1-depleted cells (, left). Since DHE mainly detects cytosolic superoxide production,Citation34,Citation35 further experiments were performed with MitoSOX, the mitochondrial targeted form of DHE. The results presented in (right panel) show that mitochondrial ROS (mROS) production was also higher in GLUD1-depleted cells. These data indicate that flux through GLUD1 indeed contributes to the lowering of mROS production.
Figure 5. Flux through GLUD1 inhibits induction of autophagy by promoting the elimination of ROS. (A) Cellular ROS production in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA was measured with DHE as probe as described in Materials and Methods (left histogram). Mitochondrial ROS production in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA was measured with MitoSOX as probe as described in Materials and Methods (right histogram). Columns: mean; bars: SEM (n = 3); *p < 0.05. (B) Cellular ROS production in HeLa cells transfected with control (ct) siRNA (left histogram) or GLUD1 siRNA (right histogram) was measured with DHE as probe as described in Materials and Methods. Cells were incubated in the absence (dotted lines) or presence (continuous lines) of amino acids for 45 min after which DHE was added for another 15 min. (C) Mitochondrial ROS production in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA was measured with MitoSOX as probe as described in Materials and Methods. The extent of MitoSOX oxidation was determined after 60 min of incubation with the dye. Columns: mean; bars: SEM (n = 8); **p < 0.01. Background fluorescence at t = 0 amounted to MFI = 13.3 ± 0.5. (D) Immunoblot analysis of LC3-I and LC3-II levels in HeLa cells transfected with control (ct) siRNA or GLUD1 siRNA for 72 h and cultured in EBSS supplemented with 10 mM N-acetylcysteine (NAC) or 0.5 µM MitoQ for 24 h. Where indicated, 200 nM of bafilomycin A1 (BAF) was present for 2 h to block the lysosomal degradation of LC3-II. Immunoblotting of ACTB was used as a loading control. The LC3-II/ACTB ratio was determined using Bio-1D quantification software. Columns: mean; bars: SEM (n = 3); **p < 0.01.
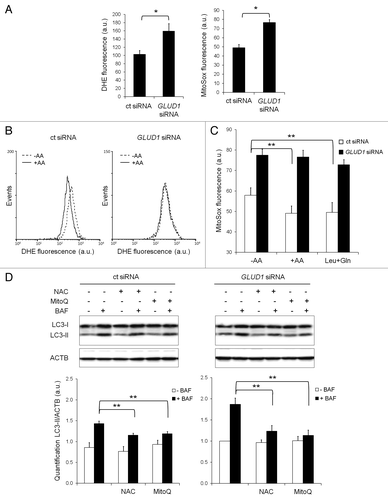
Furthermore, cytosolic and mitochondrial ROS were measured in the presence or absence of amino acids to determine whether the GLUD1-dependent inhibitory effect of amino acids on autophagy was mediated by ROS. As shown in , a complete mixture of amino acids significantly lowered cytosolic ROS and mROS production in control cells, but not in GLUD1-depleted cells. Also the relative effect of the combination of leucine and glutamine (to maximize flux through GLUD1) on ROS production was much lower in the GLUD1-depleted cells (). Taken together, these data indicate that amino acids, and especially leucine, lower mROS by enhancing the flux through GLUD1.
To determine whether the elevated level of mROS observed in GLUD1-depleted cells contributes to the increased autophagosome formation, we performed short-term starvation experiments in the presence or absence of the general ROS scavenger N-acetylcysteine (NAC) and the mitochondrial specific ROS scavenger MitoQ.Citation36 Addition of these scavengers to control cells slightly affected the degree of LC3-II accumulation in the presence of bafilomycin A1 (). In contrast, the observed increase in autophagosome formation in GLUD1 knockdown cells was highly sensitive to both scavengers (), indicating that the increased production of ROS upon GLUD1 depletion promotes the initiation of autophagy. These results are in line with a very recent report by Li et al.Citation37 in which it is shown that NAC stimulates MTOR activity.
Discussion
Collectively, our data suggest a dual mechanism by which GLUD1 flux modulates autophagy, both by relaying cellular amino acid availability to MTORC1 and by generating reducing equivalents that interfere with the accumulation of ROS. Involvement of GLUD in amino acid sensing provides an explanation for the leucine specificity in the control of both processes, because this amino acid acts as an allosteric activator of GLUD.Citation18 Although the enzyme catalyzes a reversible reaction, the occurrence of hyperammonemia in hyperinsulinism/hyperammonia syndrome, when GLUD is overactivated,Citation38 suggests that in vivo the enzyme operates in the direction of deamination of glutamate.
A possible connection between GLUD and the regulation of autophagy and MTOR may be related to NADPH, one of its reaction products.Citation1 The mitochondrial respiratory chain is an important producer of ROSCitation39 and can potentially harm the cell by oxidizing lipid, DNA and proteins. Autophagy can protect against these deleterious effects. There is ample evidence that autophagy is initiated by ROS in a manner that is sensitive to antioxidantsCitation26,Citation28-Citation31 and amino acids,Citation27,Citation31 perhaps because of the oxidation of a critical cysteine residue in ATG4.Citation31 NADPH, derived from the GLUD reaction, may be used to scavenge ROS through, e.g., the glutathione-glutathione reductase system, and/or to prevent ROS production at complex I of the mitochondrial respiratory chain.Citation40 It should be pointed out that this mechanism is MTORC1-independent and accounts for the disparity between the degree of MTORC1 activation and inhibition of autophagy ( and ). This is in agreement with the fact that in the presence of amino acids, complete inhibition of MTORC1 by rapamycin never fully restores autophagic activity (see ref. Citation1 for a review).
GLUD can react with both NADP+ and NAD+ 38 and the rate of their utilization is kinetically determined by their concentrations in the mitochondria.Citation41 In case the enzyme would react with NAD+, NADPH can still be formed by transfer of hydrogen from NADH to NADP+, catalyzed by the energy-linked nicotinamide nucleotide transhydrogenase, which then also results in suppression of mitochondrial ROS production.Citation40 The data of appear to support the view that GLUD can protect against ROS and in this way can contribute to the control of autophagy.
Another mechanism by which GLUD may contribute to amino acid sensing is related to the production of 2-oxoglutarate which, upon further metabolism, produces GTP in the succinylCoA synthetase reaction.Citation1 This GTP may then be used to activate RHEB (Ras homolog enriched in brain) and RRAG, which would lead to autophagy inhibition, as hypothesized previously.Citation1,Citation16,Citation17
Involvement of GLUD in amino acid sensing may have its implications for the interpretation of some data on autophagy published in the past. For example, autophagy has been shown to be stimulated pharmacologically by the green tea component epigallocatechin gallate,Citation42 a compound which, in addition to having other effects,Citation43 is also a powerful inhibitor of GLUD.Citation44 Chloroquine, often used in autophagic flux measurements because of its ability to raise the intralysosomal pH,Citation32 is also known to inhibit GLUD.Citation45 This may lead to overestimation of the autophagic flux.
The interpretation of our data on GLUD is not compatible with previous studies on redox regulation of MTOR activity in which chemical compounds were used to manipulate the cellular redox state.Citation46 For example, British anti-Lewisite (BAL), a strong reducing agent, inhibits, rather than stimulates, MTOR.Citation46 However, it must be stressed that the effect of BAL on MTOR activity is probably unrelated to the physiological redox regulation of MTOR activity, because this compound inhibits mitochondrial electron transport between cytochrome b and c.Citation47 The possibility that AMPK (which stimulates autophagyCitation1) is activated under these conditions cannot be ruled out.
As mentioned in the Introduction, recent information indicates that the v-ATPase in the lysosomal membrane acts as an amino acid sensor and that it responds to the level of amino acids within the lysosomal lumen.Citation13 This level is determined by the cytosolic amino acid concentration, the rate of proteolysis within the lysosome (e.g., by autophagy, which was not considered in these studies) and the rate of amino acid transport out of the lysosomes. LARS has been proposed as another amino acid sensor,Citation15 in this case of the cytosolic amino acid pool. If, as our studies imply, GLUD also contributes to amino acid sensing in the regulation of autophagy and of MTOR signaling, the question arises as to how these various amino acid sensing mechanisms fit together. An interesting possibility would be that the efflux of amino acids from the lysosomes is redox controlled and becomes inhibited by increased reduction. Because GLUD generates NADPH in the mitochondria, the reducing equivalents then need to be transferred to the cytosol. The isocitrate-2-oxoglutarate shuttle mechanismCitation48 can take care of this. As far as we are aware, redox control of lysosomal amino acid transport has never been studied and remains to be explored. Likewise, the activity of LARS may be redox controlled. Finally, it is also possible that the three mechanisms of amino acid sensing, v-ATPase, LARS and GLUD, act in parallel, independent of each other.
In some of our experiments, we have used the v-ATPase inhibitor bafilomycin A1 to monitor the appearance of LC3-II as a measure of autophagic flux, as recommended in reference Citation32. Because inhibition of the v-ATPase may have interfered with the amino acid-sensing mechanism as proposed previously,Citation13 and thus with the autophagic process itself, we have checked its effect on MTOR activity in control experiments. However, in the presence of amino acids, we failed to observe an effect of bafilomycin A1 on MTOR activity as measured by the phosphorylation of RPS6KB and EIF4EBP1 (data not shown). These results differ from the results obtained with concanamycin A and salicylhalamide A, two other v-ATPase inhibitors,Citation13 but at least indicate that bafilomycin A1 in our study for some reason had not interfered with the v-ATPase amino acid-sensing mechanism proposed by Zoncu et al.Citation13
In our studies we have used starvation to induce autophagy. Of course it remains to be seen whether other stimuli of autophagy, also acting through MTOR, such as hypoxiaCitation49 and ER stress,Citation50 will be similarly affected by leucine addition and GLUD depletion. Future studies may answer this question.
Upon completion of the present study, an article by Duran et al.Citation51 was published, showing that metabolism of glutamine, through glutaminase and GLUD, stimulates MTOR signaling by enhancing GTP-loading of RRAG and reduces the steady-state level of autophagosomes within the cell (as predicted in refs. Citation1,Citation16,Citation17). The effect of leucine in this system could be ascribed to an activation of GLUD. Concomitant silencing of both glutaminase and GLUD blocked the ability of glutamine and of leucine to decrease the steady-state level of autophagosomes.Citation51 Our results on the role of GLUD in the regulation of autophagic flux and MTOR signaling are in line with the data of Duran et al.,Citation51 and further extend their observations by demonstrating that flux through GLUD1 exerts control over autophagy by limiting the accumulation of ROS.
Materials and Methods
Reagents and antibodies
For cell treatments, bafilomycin A1 (B1793), 3-methyladenine (M9281), N-acetylcysteine (A7250), leucine (L8912) and valine (V0500) were all purchased from Sigma. MitoQ ([10-(4,5-Dimethoxy-2-methyl-3,6-dioxo-cyclohexa-1,4-dienyl)-decyl]-triphenyl-phosphonium methanesulfonate) was a kind gift of Dr. M.P. Murphy (MRC, Cambridge, UK). Dyes used for ROS detection were dihydroethidium (DHE) and MitoSOX, both purchased from Invitrogen (D1168 and M36008, respectively). The following antibodies were used for immunoblotting: anti-LC3B (Sigma, L7543), anti-SQSTM1 lck ligand (BD Biosciences, 610833), anti-β-ACTB/β-actin (Millipore, MAB1501), anti-phospho-EIF4EBP1/4EBP (Cell Signaling, 9459), anti-EIF4EBP (Cell Signaling, 9452), anti-phospho-RPS6/S6 ribosomal protein (Cell Signaling, 2211), anti-RPS6 ribosomal protein (Cell Signaling, 2317), anti-mouse IgG-HRP (Bio-Rad, 170-6516), anti-rabbit IgG-HRP (Amersham, NA9340V).
Cell culture
The human cervical cancer cell line HeLa was grown in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C under 5% CO2. The HeLa-GFP-LC3 cell line was obtained from Dr. Aviva M. Tolkovsky (University of Cambridge, UK), and cultured in RPMI 1640 supplemented with 10% fetal bovine serum and 500 μg/ml geneticin at 37°C under 5% CO2. For serum and amino acid deprivation, cells were cultured in Earle’s Balanced Salt Solution, fortified with 25 mM HEPES, pH 7.4. To test the effect of leucine and valine on autophagy, cells were cultured in Earle’s Balanced Salt Solution with 25 mM HEPES plus 10 mM leucine or 10 mM valine, unless otherwise indicated.
GFP-LC3 assay
The assay was performed in HeLa cells stably transfected with rat GFP-LC3. Prior to analysis, cells were treated as described in the text. Autophagy was then measured using a confocal microscope (LSM 510/Zeiss) by counting the number of GFP-LC3 puncta per cell with Imaris 6.3 software. At least 50 to 100 cells were counted per experiment.
Western blot analysis
Cellular extracts were prepared in 10 mM Tris pH 7.4, 1% SDS, 1 mM sodium vanadate, treated with benzon nuclease (Sigma, E1014) for 5 min at room temperature, and boiled for 3 min. Fractions (20 μg) of the cellular extracts were subjected to SDS-PAGE. After electrophoresis, proteins were transferred to a protean nitrocellulose transfer membrane (Amersham Biosciences, RPN303E). Before blocking membranes (with 5% nonfat milk protein in PBS/0.1% Tween-20), protein loading was assessed by Ponceau staining. Blots were then incubated with primary antibodies using the manufacturer’s protocol followed by the appropriate horseradish peroxidase-conjugated secondary antibody (anti-mouse IgG-HRP, Bio-Rad, 170-6516; anti-rabbit IgG-HRP, Amersham, NA9340V). Immunostained proteins were revealed using a chemoluminescent substrate (Millipore). Densitometric analysis on the immunoblots was quantified by Bio-1D software. The experiments were repeated at least three times, and representative autoradiograms are shown.
Analysis of degradation of long-lived proteins
This assay was adapted from Bauvy et al.Citation52 72 h after siRNA transfection, HeLa cells were incubated for 24 h at 37°C with 0.2 μCi/ml l-[14C] valine in complete medium (RPMI1640 supplemented with 10% fetal bovine serum and 25 mM HEPES). At the end of the radiolabeling period, the cells were washed three times with phosphate-buffered saline, pH 7.4. Cells were then incubated in complete medium supplemented with 10 mM unlabeled valine for 1 h (pre-chase period by which short-lived proteins are degraded). The medium was then replaced by either complete medium or Earle’s Balanced Salt Solution with 25 mM HEPES (starved medium) supplemented or not with 10 mM leucine. The incubation was continued for an additional 4 h (chase period). Cells and radiolabeled proteins from the 4 h chase medium were precipitated in trichloroacetic acid at a final concentration of 10% (vol/vol) at 4°C. The precipitated proteins were separated from the soluble radioactivity by centrifugation at 600 × g for 10 min and then dissolved in 0.5 ml of 0.2 N NaOH. Radioactivity was determined by liquid scintillation counting. Protein degradation was calculated by dividing the acid-soluble radioactivity recovered from both cells and medium by the radioactivity contained in precipitated proteins from both cells and medium.
Transfections of cells with small interfering RNAs
In humans, there are two genes encoding GLUD: GLUD1 and GLUD2. GLUD1 occurs in all mammals but GLUD2 is only present in humans, where it is expressed in brain (astrocytes) and testis (Sertoli cells) only.Citation53 For this reason, we decided to perform experiments in which small interfering RNAs against GLUD1 were used (Invitrogen). Control siRNA was synthesized by Eurogentec (Seraing, Belgium). Cells cultured in 6-well plates were transfected with siRNA at 200nM final concentration by using oligofectamine reagent (Invitrogen, 12252011). Cells were then incubated for 8 h at 37°C prior to the addition of 5% fetal bovine serum and then left for another 72 h. Cells were then treated as described in the text and subjected to western blotting analysis, long-lived protein degradation or GFP-LC3 assays.
Measurement of ROS production
Cells treated with control siRNA or GLUD1 siRNA were trypsinized and washed twice with incubation medium, consisting of 132 mM NaCl, 20 mM HEPES, 5 mM KCl, 1.2 mM KPi, 1 mM MgCl2, 1.3 mM CaCl2 and 0.2% (w/v) dialyzed BSA (final pH 7.4). Cells were resuspended at a concentration of 0.5 × 106/ml and preincubated for 45 min at 37°C in the absence or presence of a complete amino acid (AA) mixture or with a combination of leucine (1 mM) and glutamine (2 mM). A 25-fold concentrated AA mixture was prepared from a 100× stock solution of non-essential AA (Gibco/Invitrogen, 11140) and a 50× stock solution of essential AA (Gibco/Invitrogen, 11130) with glutamine (20 mM) and HEPES (50 mM) added and sufficient NaOH to neutralize the pH of this acidic mixture. After the preincubation plus or minus AA, the cells (in 300 µl aliquots) were transferred to tubes containing 1 µl DHE (1.5 mM, dissolved in DMSO) or, in subsequent experiments, to tubes containing 1 µl MitoSOX (300 µM, dissolved in DMSO). Samples were analyzed without washing on a FACS Calibur (BD BioSciences) and results of 6000 events were expressed as mean fluorescence intensity (MFI) in the red (FL3) channel. The increase in red fluorescence with MitoSOX (a measure for mitochondrial superoxide productionCitation54) proved to be linear over a period of at least 90 min. Cell debris and cells showing a very high red fluorescence (< 5% of the total population) were excluded from the analysis. High MitoSox staining appeared to be due to binding to the DNA of dead cells as demonstrated in control experiments by double staining with Sytox Green (Invitrogen, S7020).
Statistical analysis
Data are expressed as means ± standard errors of the means (SEM) of at least three experiments. The statistical significance was assessed by a Student’s t-test. p < 0.05 was considered statistically significant.
| Abbreviations: | ||
| 3-MA | = | 3-methyladenine |
| ACTB | = | β-actin |
| AMPK | = | AMP-activated protein kinase |
| DHE | = | dihydroethidium |
| EBSS | = | Earle’s Balanced Salt Solution |
| EIF4EBP1/4E-BP1 | = | eukaryotic translation initiation factor 4E binding protein 1 |
| GFP-LC3 | = | green fluorescent protein-microtubule-associated protein 1 light chain 3 |
| GLUD/GDH | = | glutamate dehydrogenase |
| LARS | = | leucyl-tRNA synthetase |
| MitoQ | = | [10-(4,5-dimethoxy-2-methyl-3,6-dioxo-cyclohexa-1,4-dienyl)-decyl]-triphenyl-phosphonium methanesulfonate |
| MTOR | = | mechanistic target of rapamycin |
| MTORC1 | = | mechanistic target of rapamycin complex 1 |
| ROS | = | reactive oxygen species |
| RPS6/S6 | = | ribosomal protein S6 |
| RPS6KB/S6K | = | 70 kDa ribosomal protein S6 kinase |
| RRAG/RAG | = | RAS-related GTPase |
| SQSTM1/p62 | = | sequestosome 1 |
| v-ATPase | = | vacuolar H+-adenosine triphosphatase |
Acknowledgments
This paper is dedicated to the memory of Prof. Joseph M. Tager, the teacher of two of us (A.J.M., A.J.V.), who died in February 2011. His studies on glutamate dehydrogenase in the 1960s have been fundamental to our idea that this enzyme might influence signaling in a manner that differs from its traditional function in ammonia metabolism.
The authors are grateful to Dr. S.P.J. Albracht for sharing his ideas concerning the role of the mitochondrial respiratory chain, and that of Complex I in particular, in the production of ROS. The authors also thank Dr. M.P. Murphy (MRC, Cambridge, UK) for his kind gift of, and expert advice on, MitoQ, and V. Keizer for her contribution to the ROS measurements.
Work in P. Codogno's laboratory is supported by institutional funding from the Institut National de la Santé et de la Recherche Médicale (INSERM), grants from the Agence Nationale de la Recherche (ANR) and the Institut National du Cancer (INCa).
Disclosure of Potential Conflicts of Interest
There are no conflicts of interest.
References
- Meijer AJ, Codogno P. Autophagy: regulation and role in disease. Crit Rev Clin Lab Sci 2009; 46:210 - 40; http://dx.doi.org/10.1080/10408360903044068; PMID: 19552522
- Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 1995; 270:2320 - 6; http://dx.doi.org/10.1074/jbc.270.5.2320; PMID: 7836465
- Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab 2009; 296:E592 - 602; http://dx.doi.org/10.1152/ajpendo.90645.2008; PMID: 18765678
- Dann SG, Thomas G. The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett 2006; 580:2821 - 9; http://dx.doi.org/10.1016/j.febslet.2006.04.068; PMID: 16684541
- Kim J, Guan K-L. Amino acid signaling in TOR activation. Annu Rev Biochem 2011; 80:1001 - 32; http://dx.doi.org/10.1146/annurev-biochem-062209-094414; PMID: 21548787
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274 - 93; http://dx.doi.org/10.1016/j.cell.2012.03.017; PMID: 22500797
- van Sluijters DA, Dubbelhuis PF, Blommaart EF, Meijer AJ. Amino-acid-dependent signal transduction. Biochem J 2000; 351:545 - 50; http://dx.doi.org/10.1042/0264-6021:3510545; PMID: 11042107
- Lynch CJ, Fox HL, Vary TC, Jefferson LS, Kimball SR. Regulation of amino acid-sensitive TOR signaling by leucine analogues in adipocytes. J Cell Biochem 2000; 77:234 - 51; http://dx.doi.org/10.1002/(SICI)1097-4644(20000501)77:2<234::AID-JCB7>3.0.CO;2-I; PMID: 10723090
- Patti ME, Brambilla E, Luzi L, Landaker EJ, Kahn CR. Bidirectional modulation of insulin action by amino acids. J Clin Invest 1998; 101:1519 - 29; http://dx.doi.org/10.1172/JCI1326; PMID: 9525995
- Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell 2011; 19:613 - 28; http://dx.doi.org/10.1016/j.ccr.2011.03.012; PMID: 21575862
- Shigemitsu K, Tsujishita Y, Miyake H, Hidayat S, Tanaka N, Hara K, et al. Structural requirement of leucine for activation of p70 S6 kinase. FEBS Lett 1999; 447:303 - 6; http://dx.doi.org/10.1016/S0014-5793(99)00304-X; PMID: 10214966
- Lynch CJ, Halle B, Fujii H, Vary TC, Wallin R, Damuni Z, et al. Potential role of leucine metabolism in the leucine-signaling pathway involving mTOR. Am J Physiol Endocrinol Metab 2003; 285:E854 - 63; PMID: 12812918
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334:678 - 83; http://dx.doi.org/10.1126/science.1207056; PMID: 22053050
- Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell 2012; 46:105 - 10; http://dx.doi.org/10.1016/j.molcel.2012.02.009; PMID: 22424774
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012; 149:410 - 24; http://dx.doi.org/10.1016/j.cell.2012.02.044; PMID: 22424946
- Meijer AJ. Amino acid regulation of autophagosome formation. Methods Mol Biol 2008; 445:89 - 109; http://dx.doi.org/10.1007/978-1-59745-157-4_5; PMID: 18425444
- Meijer AJ, Codogno P. Nutrient sensing: TOR’s Ragtime. Nat Cell Biol 2008; 10:881 - 3; http://dx.doi.org/10.1038/ncb0808-881; PMID: 18670446
- Fahien LA, Teller JK, Macdonald MJ, Fahien CM. Regulation of glutamate dehydrogenase by Mg2+ and magnification of leucine activation by Mg2+. Mol Pharmacol 1990; 37:943 - 9; PMID: 2359406
- Xu G, Kwon G, Cruz WS, Marshall CA, McDaniel ML. Metabolic regulation by leucine of translation initiation through the mTOR-signaling pathway by pancreatic beta-cells. Diabetes 2001; 50:353 - 60; http://dx.doi.org/10.2337/diabetes.50.2.353; PMID: 11272147
- Carobbio S, Frigerio F, Rubi B, Vetterli L, Bloksgaard M, Gjinovci A, et al. Deletion of glutamate dehydrogenase in beta-cells abolishes part of the insulin secretory response not required for glucose homeostasis. J Biol Chem 2009; 284:921 - 9; http://dx.doi.org/10.1074/jbc.M806295200; PMID: 19015267
- Krause U, Bertrand L, Maisin L, Rosa M, Hue L. Signalling pathways and combinatory effects of insulin and amino acids in isolated rat hepatocytes. Eur J Biochem 2002; 269:3742 - 50; http://dx.doi.org/10.1046/j.1432-1033.2002.03069.x; PMID: 12153571
- Leverve XM, Caro LH, Plomp PJ, Meijer AJ. Control of proteolysis in perifused rat hepatocytes. FEBS Lett 1987; 219:455 - 8; http://dx.doi.org/10.1016/0014-5793(87)80271-5; PMID: 3301406
- Caro LH, Plomp PJ, Leverve XM, Meijer AJ. A combination of intracellular leucine with either glutamate or aspartate inhibits autophagic proteolysis in isolated rat hepatocytes. Eur J Biochem 1989; 181:717 - 20; http://dx.doi.org/10.1111/j.1432-1033.1989.tb14782.x; PMID: 2567237
- Fumarola C, La Monica S, Guidotti GG. Amino acid signaling through the mammalian target of rapamycin (mTOR) pathway: Role of glutamine and of cell shrinkage. J Cell Physiol 2005; 204:155 - 65; http://dx.doi.org/10.1002/jcp.20272; PMID: 15605414
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009; 136:521 - 34; http://dx.doi.org/10.1016/j.cell.2008.11.044; PMID: 19203585
- Azad MB, Chen Y, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid Redox Signal 2009; 11:777 - 90; http://dx.doi.org/10.1089/ars.2008.2270; PMID: 18828708
- Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ 2009; 16:1040 - 52; http://dx.doi.org/10.1038/cdd.2009.49; PMID: 19407826
- Djavaheri-Mergny M, Amelotti M, Mathieu J, Besançon F, Bauvy C, Souquère S, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem 2006; 281:30373 - 82; http://dx.doi.org/10.1074/jbc.M602097200; PMID: 16857678
- Huang J, Lam GY, Brumell JH. Autophagy signaling through reactive oxygen species. Antioxid Redox Signal 2011; 14:2215 - 31; http://dx.doi.org/10.1089/ars.2010.3554; PMID: 20874258
- Kissová I, Deffieu M, Samokhvalov V, Velours G, Bessoule JJ, Manon S, et al. Lipid oxidation and autophagy in yeast. Free Radic Biol Med 2006; 41:1655 - 61; http://dx.doi.org/10.1016/j.freeradbiomed.2006.08.012; PMID: 17145553
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007; 26:1749 - 60; http://dx.doi.org/10.1038/sj.emboj.7601623; PMID: 17347651
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4:151 - 75; PMID: 18188003
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 1997; 243:240 - 6; http://dx.doi.org/10.1111/j.1432-1033.1997.0240a.x; PMID: 9030745
- Rothe G, Valet G. Flow cytometric analysis of respiratory burst activity in phagocytes with hydroethidine and 2′,7′-dichlorofluorescin. J Leukoc Biol 1990; 47:440 - 8; PMID: 2159514
- Dickinson BC, Srikun D, Chang CJ. Mitochondrial-targeted fluorescent probes for reactive oxygen species. Curr Opin Chem Biol 2010; 14:50 - 6; http://dx.doi.org/10.1016/j.cbpa.2009.10.014; PMID: 19910238
- Kelso GF, Porteous CM, Coulter CV, Hughes G, Porteous WK, Ledgerwood EC, et al. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J Biol Chem 2001; 276:4588 - 96; http://dx.doi.org/10.1074/jbc.M009093200; PMID: 11092892
- Li L, Chen Y, Gibson SB. Starvation-induced autophagy is regulated by mitochondrial reactive oxygen species leading to AMPK activation. Cell Signal 2013; 25:50 - 65; http://dx.doi.org/10.1016/j.cellsig.2012.09.020; PMID: 23000343
- Li M, Li C, Allen A, Stanley CA, Smith TJ. The structure and allosteric regulation of mammalian glutamate dehydrogenase. Arch Biochem Biophys 2012; 519:69 - 80; http://dx.doi.org/10.1016/j.abb.2011.10.015; PMID: 22079166
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 2009; 417:1 - 13; http://dx.doi.org/10.1042/BJ20081386; PMID: 19061483
- Albracht SP, Meijer AJ, Rydström J. Mammalian NADH:ubiquinone oxidoreductase (Complex I) and nicotinamide nucleotide transhydrogenase (Nnt) together regulate the mitochondrial production of H₂O₂--implications for their role in disease, especially cancer. J Bioenerg Biomembr 2011; 43:541 - 64; http://dx.doi.org/10.1007/s10863-011-9381-4; PMID: 21882037
- Papa S, Tager JM, Francavilla A, de Haan EJ, Quagliariello E. Control of glutamate dehydrogenase activity during glutamate oxidation in isolated rat-liver mitochondria. Biochim Biophys Acta 1967; 131:14 - 28; http://dx.doi.org/10.1016/0005-2728(67)90027-8
- Li W, Zhu S, Li J, Assa A, Jundoria A, Xu J, et al. EGCG stimulates autophagy and reduces cytoplasmic HMGB1 levels in endotoxin-stimulated macrophages. Biochem Pharmacol 2011; 81:1152 - 63; http://dx.doi.org/10.1016/j.bcp.2011.02.015; PMID: 21371444
- Van Aller GS, Carson JD, Tang W, Peng H, Zhao L, Copeland RA, et al. Epigallocatechin gallate (EGCG), a major component of green tea, is a dual phosphoinositide-3-kinase/mTOR inhibitor. Biochem Biophys Res Commun 2011; 406:194 - 9; http://dx.doi.org/10.1016/j.bbrc.2011.02.010; PMID: 21300025
- Li C, Allen A, Kwagh J, Doliba NM, Qin W, Najafi H, et al. Green tea polyphenols modulate insulin secretion by inhibiting glutamate dehydrogenase. J Biol Chem 2006; 281:10214 - 21; http://dx.doi.org/10.1074/jbc.M512792200; PMID: 16476731
- Jarzyna R, Lenarcik E, Bryła J. Chloroquine is a potent inhibitor of glutamate dehydrogenase in liver and kidney-cortex of rabbit. Pharmacol Res 1997; 35:79 - 84; http://dx.doi.org/10.1006/phrs.1996.0108; PMID: 9149320
- Sarbassov DD, Sabatini DM. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J Biol Chem 2005; 280:39505 - 9; http://dx.doi.org/10.1074/jbc.M506096200; PMID: 16183647
- Slater EC, de Vries S. Identification of the BAL-labile factor. Nature 1980; 288:717 - 8; http://dx.doi.org/10.1038/288717a0; PMID: 6256640
- Papa S. Control of the utilization of mitochondrial reducing equivalents. In: Papa S, Tager JM, Quagliariello E, Slater EC, editors. The Energy Level and Metabolic Control in Mitochondria. Bari, Italy: Adriatica Editrice, 1969: 401-409.
- Blagosklonny MV. Hypoxia, MTOR and autophagy: Converging on senescence or quiescence. Autophagy 2013; 9:260 - 2; http://dx.doi.org/10.4161/auto.22783; PMID: 23192222
- Appenzeller-Herzog C, Hall MN. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol 2012; 22:274 - 82; http://dx.doi.org/10.1016/j.tcb.2012.02.006; PMID: 22444729
- Durán RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, et al. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell 2012; 47:349 - 58; http://dx.doi.org/10.1016/j.molcel.2012.05.043; PMID: 22749528
- Bauvy C, Meijer AJ, Codogno P. Assaying of autophagic protein degradation. Methods Enzymol 2009; 452:47 - 61; http://dx.doi.org/10.1016/S0076-6879(08)03604-5; PMID: 19200875
- Spanaki C, Zaganas I, Kleopa KA, Plaitakis A. Human GLUD2 glutamate dehydrogenase is expressed in neural and testicular supporting cells. J Biol Chem 2010; 285:16748 - 56; http://dx.doi.org/10.1074/jbc.M109.092999; PMID: 20194501
- Mukhopadhyay P, Rajesh M, Yoshihiro K, Haskó G, Pacher P. Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem Biophys Res Commun 2007; 358:203 - 8; http://dx.doi.org/10.1016/j.bbrc.2007.04.106; PMID: 17475217