Abstract
Autophagy is an evolutionarily conserved process in eukaryotes by which cytoplasmic components including macromolecules and organelles are degraded by the lysosome/vacuole. Autophagy is implicated in a number of physiological processes important for human health and disease. Although primarily cytoprotective, autophagy can also contribute to cell death; it is thus important to understand what distinguishes the life or death decision in autophagic cells. Despite the fact that the execution of autophagy includes a unique set of cytoplasmic events, nuclear events, in particular transcriptional programs, have emerged as an important regulator of this process. In addition, a critical linkage was recently unveiled between specific histone posttranslational modifications and the transcriptional regulation of autophagy-related genes, which initiates a regulatory feedback loop, and serves as a key determinant of survival versus death responses upon autophagy induction.
Macroautophagy, hereafter referred to as autophagy, is a catabolic process that results in the autophagosome-dependent lysosomal degradation of bulk cytoplasmic contents, abnormal protein aggregates, and excess or damaged organelles. Paradoxically, although autophagy is primarily a protective process for the cell, it can also play a role in cell death; however, it is not clear what distinguishes the autophagy-mediated survival or death decision. Autophagy is associated with various physiological as well as pathological processes. Therefore, understanding the pathways regulating the autophagic life and death decision and its cellular long-term effects might help to improve autophagy-based clinical treatments.
Until recently, nuclear events were not considered of importance for autophagy, as reports indicated that enucleated cells are still able to accumulate LC3 puncta in response to autophagic stimuli. Probably as a consequence, autophagy is commonly seen as a set of primarily cytoplasmic events. Nevertheless, compelling evidence indicates that a transcriptional program controls major steps of the autophagic pathway. Gene expression patterns not only heavily rely on the transcriptional factor networks, but also on the properties of the chromatin within the cells. It is becoming clear that covalent post-translational modifications of the histones by histone modifying enzymatic complexes are involved in the orchestration of the transcriptional program in cells. However, hitherto, no changes in the chromatin per se have been recognized upon the induction of autophagy.
We recently discovered that induction of autophagy using various stimuli (amino acid starvation, Torin 1, or rapamycin treatment) is coupled in various mammalian cell types (MEF, HEK 293, U1810, HeLa, and U2OS) to reduction of histone H4 lysine 16 acetylation (H4K16ac) through the downregulation of the histone acetyltransferase KAT8/hMOF/MYST1 (). Furthermore, the expression of Sas2, the yeast homolog of KAT8, together with H4K16 acetylation levels, were also found to be severely repressed upon autophagy induction in yeast, revealing the conservation of this process through evolution. Our studies indicate that the KAT8 downregulation is part of the autophagy program, and it is rescued when autophagy is inhibited by 3MA or chloroquine. Worth noting, whereas many histone acetyltransferases show either little substrate specificity or preference for other residues, KAT8 is necessary and sufficient for the bulk of H4K16 acetylation and thereby antagonizes the enzymatic activity of SIRT1 (sirtuin 1), a NAD+-dependent deacetylase, which has a wide range of nonhistone targets, with H4K16 being its primary histone target. This single histone modification is of particular interest, as H4K16 acetylation influences higher order chromatin structure and plays an important role in transcription.
Figure 1. Scheme illustrating the potential epigenetic regulatory feedback loop regulating the autophagic flux via the KAT8-SIRT1 control of H4K16 acetylation/deacetylation.
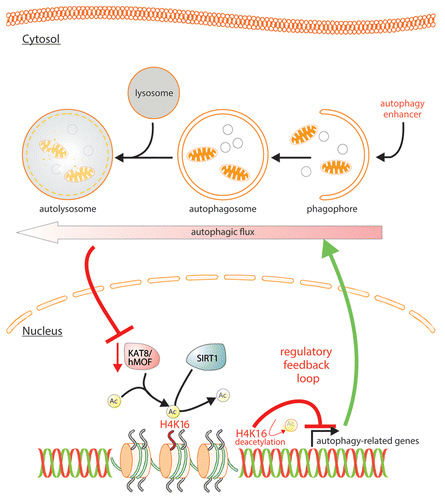
At a genome-wide level we found that H4K16 deacetylation is associated predominantly with the downregulation of autophagy-related genes (). Indeed, chromatin immunoprecipitation targeting H4K16ac, followed by high-throughput sequencing (ChIP-Seq) performed to elucidate the genome-wide occurrence of this histone mark in U1810 cells undergoing rapamycin-induced autophagy, revealed reduced H4K16ac occupancy. A global run-on-sequencing (GRO-Seq) assay used to generate a genome-wide view of the location, orientation, and density of nascent RNA transcripts unveiled a significant alteration of the cell transcriptome with the identification of an unexpectedly large fraction of genes found to be related to the autophagic process. Remarkably, there is an overall coincidence across the autophagy-related genes between the alteration of the GRO-Seq signal and the absence of H4K16 acetylation. Indeed, 55 autophagy-related genes (including genes belonging to the autophagic core machinery, such as ATG9A, GABARAPL2, MAP1LC3B, ULK1, ULK3, VMP1) identified by GRO-Seq analysis, were found to exhibit reduced H4K16ac tag counts upon rapamycin treatment. These data are consistent with the reported elevated H4K16 acetylation in the promoter and transcribed regions of active genes. Collectively, these genome-wide deep-sequencing analyses indicate that the observed deacetylation of H4K16 during rapamycin-induced autophagy results in transcriptional regulation of autophagy-related genes.
The observed dramatic changes in levels of H4K16 acetylation and associated transcriptional gene regulation suggested that there may be a functional role for this epigenetic change during autophagy. The autophagic flux was thus investigated in detail (by LC3 immunoblotting and making use of the tandem reporter mRFP-GFP-LC3). This investigation revealed that inhibition of H4K16 deacetylation does not inhibit autophagy, but on the contrary increases autophagic flux. The observed increase in the autophagic flux is in accord with the genome-wide investigations, which provide compelling evidence that H4K16 deacetylation is associated with the downregulation of autophagy-related genes. We extended our investigation to the analysis of cell death. Antagonizing H4K16ac downregulation through overexpression of KAT8 or antagonizing the activity of the H4K16ac deacetylase SIRT1, upon autophagy induction results in the promotion of cell death.
In summary, we uncovered a feedback regulatory loop occurring during autophagy, associated with the downregulation of the H4K16 acetyltranferase KAT8, and the H4K16-deacetylation dependent regulation of autophagy-related genes, which we propose acts to prevent an overstimulation of autophagic flux (). If this feedback loop is inhibited using SIRT1 knockdown with siRNA, VPA, Ex527 or KAT8 overexpression (all resulting in an increase of H4K16ac) the autophagic flux is increased and the cells undergo cell death. In conclusion, KAT8 is a substrate for autophagy, and its degradation and the consequent reduction of H4K16 acetylation, in turn, regulates the autophagic flux. This provides a built-in mechanism to prevent excessive autophagy, which would otherwise lead to cell death.
Collectively, these data indicate that alteration in a specific histone post-translational modification during autophagy affects the transcriptional regulation of autophagy-related genes and initiates a regulatory feedback loop, which serves as a key determinant of survival versus death responses upon autophagy induction. The identification of tightly regulated histone modifications associated with the autophagic process offers an attractive conceptual framework both to understand the short-term transcriptional response to stimuli eliciting autophagy, as well as constituting a potential aspect of long-term responses to autophagy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.