Abstract
Programmed nuclear death (PND) in the ciliate protozoan Tetrahymena thermophila is a novel type of autophagy that occurs during conjugation, in which only the parental somatic macronucleus is destined to die and is then eliminated from the progeny cytoplasm. Other coexisting nuclei, however, such as new micro- and macronuclei are unaffected. PND starts with condensation in the nucleus followed by apoptotic DNA fragmentation, lysosomal acidification, and final resorption. Because of the peculiarity in the process and the absence of some ATG genes in this organism, the mechanism of PND has remained unclear. In this study, we focus on the role of class III phosphatidylinositol 3-kinase (PtdIns3K, corresponding to yeast Vps34) in order to identify central regulators of PND. We identified the sole Tetrahymena thermophila ortholog (TtVPS34) to yeast Vps34 and human PIK3C3 (the catalytic subunit of PtdIns3K), through phylogenetic analysis, and generated the gene knockdown mutant for functional analysis. Loss of TtVPS34 activity prevents autophagosome formation on the parental macronucleus, and this nucleus escapes from the lysosomal pathway. In turn, DNA fragmentation and final resorption of the nucleus are drastically impaired. These phenotypes are similar to the situation in the ATG8Δ mutants of Tetrahymena, implying an inextricable link between TtVPS34 and TtATG8s in controlling PND as well as general macroautophagy. On the other hand, TtVPS34 does not appear responsible for the nuclear condensation and does not affect the progeny nuclear development. These results demonstrate that TtVPS34 is critically involved in the nuclear degradation events of PND in autophagosome formation rather than with an involvement in commitment to the death program.
Introduction
Tetrahymena thermophila (hereafter referred to as Tetrahymena) is a ciliate protozoan, which belongs to an independent eukaryotic kingdom, alveolates, which consists of 3 different phyla; dinoflagellates, apicomplexans, and ciliates. Ease in gene manipulation including gene knockout,Citation1 availability of the completed sequenced genome and microarray expression analysisCitation2 have enhanced its utility for important molecular and cellular model studies not only in closely related organisms but also in other protist groups. The most remarkable feature of ciliates including Tetrahymena is that they maintain specially differentiated germline and somatic nuclear genomes in the same cytoplasm.Citation3 The canonical germline genome is housed in the diploid micronucleus, while the polyploid (~50 copies in Tetrahymena) somatic genome is housed in the macronucleus. The micronuclear genome, which is transcriptionally silent in the asexual stage, is the repository of genetic information for sexual progeny. The macronuclear genome, on the other hand, is the primary source of gene transcripts in the cell, and its activity maintains the life of the cell and is responsible for the overall phenotype. The micro- and macronuclei have distinct nucleoporins on their surface, which characterize their functional and morphological features.Citation4
Both micro- and macronuclei are derived from a zygotic micronucleus during sexual reproduction called “conjugation.” In Tetrahymena, the progression of conjugation has been clearly illustrated by Cole and Sugai.Citation5 Briefly, conjugation is initiated by cell-to-cell interaction between matured different mating-types. Once cells begin mating, the micronucleus undergoes meiosis followed by reciprocal pronuclear exchange between the partners and formation of a zygotic micronucleus, corresponding to fertilization in metazoans. The zygotic micronucleus mitotically divides twice, resulting in 4 micronuclei. Two of these at the anterior of the cell differentiate into new macronuclei while the other 2 at the posterior of the cell remain as micronuclei. The new macronuclear development involves large-scale genome rearrangement and amplification that is accomplished by a mechanism involving RNAi-mediated, heterochromatin formation.Citation6-Citation11 During this process, at least 6,000 distinct sequences, likely remnants of transposable elements, called internal eliminated sequences, are deleted from the micronuclear genome in a site-specific manner.Citation12,Citation13
Concomitant with new macronuclear differentiation, the parental macronucleus starts to degrade and eventually disappears from the cytoplasm. This process is called programmed nuclear death (PND), in which only the parental macronucleus is highly condensed as a nucleus, being apoptotic, and then eliminated from the cytoplasm while other coexisting nuclei, such as new micro- and macronuclei are unaffected.Citation14-Citation18 In a previous report,Citation19 we demonstrated that PND was controlled by autophagy that differed from mammalian and yeast macroautophagy. When PND begins, the periphery of the parental macronuclear membrane changes its properties as if it is an autophagosomal membrane without fusion or accumulation of other membranous structures from the cytoplasm. In this process, the envelope uniformly becomes stainable with an autophagosome detector, monodansylcadaverine (MDC). The alteration of the membrane involves exposure of sugars and lipids to the outer surface, which are likely to attract small digestive vesicles including lysosomes toward the nucleus.Citation19 These vesicles fuse with the envelope and release their contents into the inside of the nucleus in a stepwise fashion. In the first step, a collaboration of apoptosis-inducing factor (TtAIF) and endonuclease G (EndoG)-like DNase at neutral pH degrade the genomic DNA into high molecular weight (1 to ~10 kb) fragments.Citation17,Citation18 Meanwhile, lysosomal enzymes including a nuclease similar to DNase II penetrate into the nucleus, which causes acidification of the nucleoplasm prior to the final resorption.Citation17,Citation19-Citation21 Because of these peculiarities and the absence of some autophagy-related (ATG) genes in Tetrahymena,Citation22,Citation23 much remains unknown about the molecular mechanism of PND.
Recently, 2 orthologs of the most well-characterized autophagosome markers, Atg8 whose mammalian orthologs include the MAP1LC3 (LC3) family, were found in Tetrahymena (TtATG8-2 and TtATG8-65) and the subcellular localizations and functions of their gene products were analyzed.Citation24 These exert their functions in the degradation of meiotic products (pronuclei) and in PND during conjugation. The targeted nuclei were surrounded by the EGFP- or mCherry-TtATG8s prior to degeneration as would occur during general macroautophagy. These phenomena were unlikely to be generated by random aggregations or other causes and happened concomitant with nuclear condensation prior to lysosome fusion with the nucleus.Citation24 This timing is consistent with the appearance of the MDC stainability on the parental macronuclear envelope,Citation19 suggesting critical roles of the TtATG8s in the biogenesis of the autophagosomal compartment at the early stage of PND. Knockout of the genes, TtATG8-2 in particular, caused delay in PND without DNA fragmentation and lysosomal acidification of the macronucleus, resulting in failure in the final resorption.Citation24 In addition, TtATG8-65 also played a role in survival under starvation conditions.Citation24 This report strongly suggests that PND is a new type of autophagy diverged from an evolutionarily conserved system and shares some molecular mechanisms with general macroautophagy.
In this study, we focus on the role of class III phosphatidylinositol 3-kinase in Tetrahymena in order to identify important components of PND. Class III PtdIns3K, containing the catalytic subunit Vps34 in yeast and PIK3C3 in humans, is a subgroup of the PtdIns3K family which activates a variety of cell signaling pathways such as for cell cycle and survival, protein synthesis, glucose uptake, and vesicle trafficking, as the precursor of second messengers.Citation25,Citation26 Vps34 was first described as a component of the vacuolar protein sorting system in the yeast Saccharomyces cerevisiaeCitation27 and is the sole PtdIns3K in that organism. The ortholog in human also plays a role in endocytic sorting.Citation28,Citation29 Vps34 is required in autophagosome nucleation together with the kinase complex composed of Vps15 (human PIK3R4)-VPS30/Atg6 (human BECN1)-Atg14 (human ATG14) when autophagy is induced.Citation29 This complex produces PtdIns3P at the site of autophagosome nucleation called the phagophore assembly site (PAS) in yeast, and then recruits Atg8 and other Atg proteins to start vesicle expansion.Citation30
Potential PtdIns3K activity has been detected in some Tetrahymena species through the use of pharmacological inhibitors such as wortmannin, 3-methyladenine, and LY294002.Citation31-Citation33 These reports suggest that PtdIns3K activity is important for regulation of proper phagocytotic activity and vesicular trafficking in the asexual stage. Yakisich and KaplerCitation31 suggest that during conjugation, wortmannin treatment blocks the final resorption of several types of nuclei such as pronuclei and the parental macronucleus, resulting in the accumulation of extra nuclei in the progeny cell. The remaining parental macronucleus shows no acidification of the nucleoplasm and actively incorporates bromodeoxyuridine, implying critical roles of PtdIns3K in the autophagic/lysosomal machinery followed by proper alteration of the generations in a single cytoplasm. However, because of the use of high concentration of these inhibitors, higher than the half maximal inhibitory concentration (IC50), the precise role of PtdIns3K upon PND remains unclear.
We have found a Tetrahymena ortholog to yeast VPS34 and its human ortholog PIK3C3 from the genome database and generated its somatic gene knockdown. Comparison between the knockdown mutant and wortmannin-treated wild-type cells at lower than IC50, we demonstrate that Tetrahymena utilizes class III PtdIns3K in PND as an essential factor controlling autophagic/lysosomal processes. We also discuss its relation to the TtATG8∆ mutants.
Results
Class III PtdIns3K in Tetrahymena
PtdIns3K is divided into 3 different classes: class I, class II, and class III, based on primary structure, regulation, and in vitro lipid substrate specificity.Citation25 Among the 3 classes, class I and III play critical roles in regulation and execution of macroautophagy.Citation29,Citation34 Class I PtdIns3K is responsible for producing PtdIns3P, PtdIns(3,4)P2 and PtdIns(3,4,5)P3, whereas class III can produce only PtdIns3P from PtdIns.Citation35 Four genes (TTHERM_0032302, TTHERM_00649380, TTHERM_00655270 and TTHERM_00951960) have been described as PtdIns3K homologs in the Tetrahymena genome database (http://ciliate.org) based on protein sequence similarities. However, it remained unclear as to which gene belongs to class III PtdIns3K. To identify the ortholog of yeast VPS34 in Tetrahymena, we used a phylogenetic analysis of PtdIns3Ks from major taxonomic groups including Tetrahymena (; Fig. S1).
Figure 1. Characterization of phosphatidylinositol 3-kinases in Tetrahymena. (A) Phylogenetic tree of PtdIns3Ks. The tree was reconstructed with a maximum likelihood method (see Materials and Methods). Superscripts on the accession numbers stand for databases, which the protein sequences were taken from;†: dictybase, **: Tair, $: Saccharomyces Genome Database, #: WormBase, *:FlyBase, §: GeneDB, ##: OxyDB, ††: ParameciumDB, §§: IchDB, and $$:TGB. The accession numbers without superscript indicate the protein sequences that were taken from GenBank. Branches with different widths represent bootstrap values. Scale bar: 0.5, expected amino acid residue substitutions per site. (B) Multiple-sequence alignment of the whole amino acid sequence of class III PtdIns3Ks (human PIK3C3 or yeast Vps34) including Homo sapiens, Saccharomyces cerevisiae, Dictyostelium discoideum, Arabidopsis thaliana, and Tetrahymena thermophila. Each color box represents the conserved domain, which corresponds to the schematic representation of the primary structure of orthologs of yeast Vps34. Asterisks indicate identical amino acids. Colons and semicolons indicate amino acid similarity.
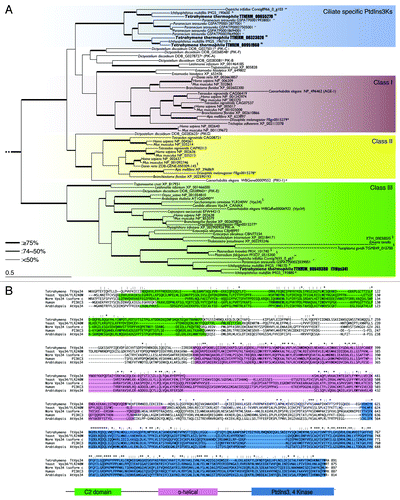
TTHERM_00649380 was the sole PtdIns3K in Tetrahymena which belonged to the class III clade (). The other 3 genes appeared between class I and II branches, which formed a ciliate-specific cluster as nonclass III PtdIns3Ks in Amoebozoa and Kinetoplastids.Citation35 Unlike this, class III PtdIns3K was widely conserved from protists to mammals and its phylogeny reflected actual evolutionary lineage (). Multiple sequence alignment of TTHERM_00649380 together with human PIK3C3 and its yeast ortholog Vps34 (), indicates the amino acid sequence of the gene shares 3 typical domains of class III PtdIns3K with its orthologs in other species (). The C2 domain in the N-terminal region is the putative membrane-bound domain, and the α-helical structure in the middle is common in all 3 classes of PtdIns3Ks as well as PtdIns4Ks.Citation25,Citation29 The domain in the C-terminal region plays an important role in catalysis.Citation29 We refer to this gene as TtVPS34. The expression profile of the gene (http://tfgd.ihb.ac.cn/search/detail/gene/TTHERM_00649380)Citation36 is presented in Figure S2. TtVPS34 is stably expressed but at a low level during growth. Expression increases up to triple the quantity under starvation condition, and decreases once conjugation starts. Expression increases again to the same level as under starvation in the later stages of conjugation.
To address questions of function of PND, we used particle bombardment to generate somatic knockout mutants of the TtVPS34 gene for mating-types II and VII (). The mutants obtained underwent phenotypic assortment through cultivation in the presence of paromomycin. Exogenous genes such as the Neo4 cassette can replace all 50 target gene copies if the target genes are not essential for vegetative growth, whereas incomplete replacement occurs if they are essential.Citation1 This assortment is attributed to the random distribution of allelic copies in a compound macronucleus by an amitotic nuclear division.Citation1 Southern blot analysis following phenotypic assortment showed incomplete replacement of the endogenous TtVPS34 by the Neo4 cassette (), implying its essential role in the vegetative stage for both the mating types. The Neo4 cassette replaced approximately 70% of the TtVPS34 copies ().
Figure 2. Generation of somatic TtVPS34 gene-knockdown mutants. (A) A schematic showing the TtVPS34 genomic locus (upper), the plasmid vector carrying Neo4 cassette (middle) and after homologous recombination (lower). (B) Southern blot analysis of AccI and SphI digested genomic DNA from wild-type cells and TtVPS34∆ mutants. Molecular weight of the signals against the probe corresponds to the prediction in (A). (C) Cell-growth curves in nutrient-rich conditions. The cells (0.5 ml) maintained in the medium for 1, 2, and 3 d were fixed with paraformaldehyde, diluted 100× and counted under a microscope. (D) Remaining cells in nutrient-deprivation conditions. The cells inoculated in 10 mM TRIS-HCl pH 7.2 were sampled (5 μl) every 24 h until day 10 and swimming (living) cells were counted under a microscope. Points and attached bars correspond to the mean of the 3 identical measurements and standard deviations.
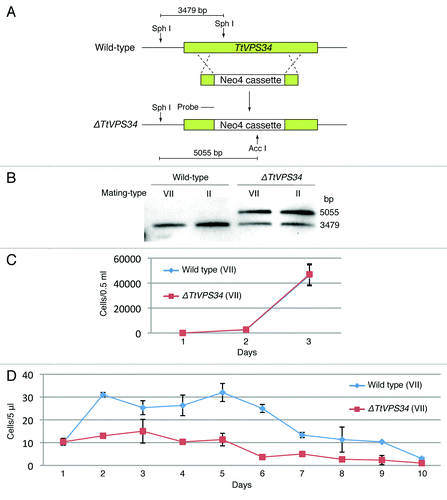
The reduced copy number of TtVPS34 (knockdown) did not affect growth rate in vegetatively growing cells (, P > 0.05, Student t test), whereas growth and viability under nutrient deprivation conditions were impaired; the wild-type cells were able to undergo fission at least one time until day 2 while the knockdown cells hardly grew (, P < 0.05, Student t test). Almost all the knockdown cells were gone at day 6 after induction of starvation, which was 4 d before the wild-type cells (). These results suggest that TtVPS34 plays roles in cell survival under starvation, similar to TtATG8-65.Citation24
Loss of PtdIns3K activity induces a phenotype similar to the TtATG8∆ mutants during PND.
Wortmannin has been used as an irreversible and membrane-permeable inhibitor of PtdIns3K activity. Human PIK3C3 is extremely sensitive to wortmannin (IC50 = 1 to 15 nM), whereas yeast Vps34 is relatively resistant (IC50 = 3 μM).Citation37 Yakisich and KaplerCitation31 treated conjugating Tetrahymena at 5 h post mixing with 25 to 250 nM wortmannin. This prevented loss of both the pronuclei and parental macronucleus from the cytoplasm. We have used 1 to 10 nM wortmannin in order to minimize nonspecific actions of the inhibitor on other kinases. In order to focus on PND, we added the inhibitor at 6 h after induction of conjugation, at which time most cells were undergoing post-zygotic nuclear division (). The cells were fixed for microscopy at 2 different time points (8 h and 14 h). The TtVPS34∆ crosses were also fixed at the same time points to compare the gene knockdowns to the wortmannin treatment.
Figure 3. Involvement of abnormal localization and lack of digestion of the parental macronucleus with addition of wortmannin or knockdown of TtVPS34. (A) Nuclear events during conjugation of wild-type Tetrahymena. MA, macronucleus; mi, micronucleus; A, progeny macronuclear anlagen; pMA, parental macronucleus; m, progeny micronucleus. (B) Conjugating cells at 8 h (upper) and 14 h (lower) stained with DAPI. White arrowheads, parental macronucleus; asterisks, progeny macronuclear anlagen; dollar signs, progeny micronuclei; yellow arrows, undigested pronuclei. Scale bars: 10 μm. (C) Effects of the treatments on appearance of the abnormalities. Wortmannin was used at a concentration range from 0 to 10 nM. Red and blue columns represent percentage of parental macronucleus (pMA) abnormally localized at 8 h and undigested at 14 h, respectively. The columns and attached bars correspond to the means of 4 identical measurements and standard deviations.
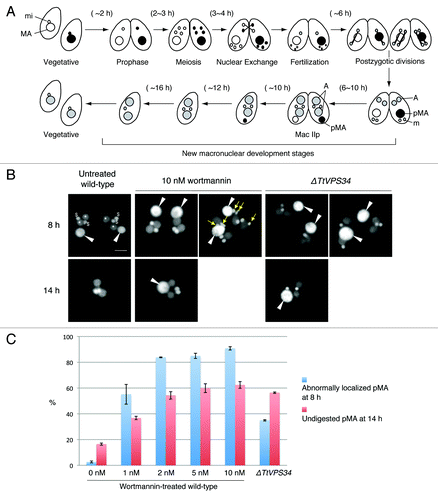
Almost all the conjugating cells reached the progeny macronuclear developmental stage called Mac IIp at 8 h irrespective of the presence of the inhibitor or knocking down of TtVPS34. In the untreated wild-type crosses, the parental macronucleus was condensed and localized at the posterior of the cytoplasm (). In the presence of wortmannin, however, the condensed parental macronucleus often abnormally localized at the anterior or middle region of the cytoplasm (, left panel of 10 nM wortmannin at 8 h). The percentage of the abnormal localization was dose dependent (), and 1 nM was enough to induce a significant difference from the wild-type crosses (P < 0.05, Wilcoxon U test). Undigested pronuclei due to the treatment occasionally remained in the cytoplasm as initially observed by Yakisich and KaplerCitation31 (, right panel of 10 nM wortmannin at 8 h) because the progression of conjugation is not perfectly synchronized in the population. The TtATG8∆ mutants behave similarly,Citation24 implying a relation between PtdIns3K activity and ATG8 function in pronuclear elimination and macronuclear localization. The TtVPS34∆ crosses also show abnormal localization of the macronucleus in 37% of cells (), significantly higher than the untreated wild-type crosses (P < 0.05, Wilcoxon U test). This suggests that TtVPS34 as well as TtATG8 is required at this stage. However, none of the meiotic pronuclei remained in the cytoplasm unlike with wortmannin treatment, implying involvement of other PtdIns3Ks in pronuclear elimination.
Most cells completed pairing and formed exconjugants by 14 h irrespective of the treatments (). The parental macronucleus usually disappeared from the wild-type exconjugants, in which 2 of each progeny macro- and micronuclei existed (). In the presence of wortmannin, however, the parental macronucleus remained significantly in the exconjugants as seen with the TtATG8∆ mutantsCitation24 depending upon the concentration of the inhibitor (, P < 0.05, Wilcoxon U test). The macronucleus in the TtVPS34∆ exconjugants more often than not remained in the cytoplasm (, P < 0.05, Wilcoxon U test). These results strongly suggest that as for TtATG8s, TtVPS34 plays an important role in the macronuclear elimination from the exconjugant cytoplasm as well as the macronuclear arrangement at the Mac IIp stage. Although the TtVPS34∆ cells fail in macronuclear elimination at a higher frequency than the appearance of the abnormal localization (), it remains unclear if the abnormality is necessary for the parental macronucleus to be eliminated.
PtdIns3K activity is critical for DNA degradation during PND
During PND, genomic DNA degrades into high molecular weight (1 to ~10 kb) fragments and subsequently oligonucleosome-sized fragments in a stepwise fashion.Citation15,Citation16 DNA degradation was drastically reduced in the ATG8-2∆ mutant, in which the parental macronucleus remained TUNEL-negative until the exconjugant stage.Citation24 In order to address the question of the effect on DNA degradation, by loss of PtdIns3K activity, we performed the following experiments with 10 nM wortmannin-treated cells and TtVPS34∆ knockdown cells.
A TUNEL-assay was performed with fixed cells at 8 h after induction of conjugation to observe DNA fragmentation (). shows the effects of the parental macronucleus to the TUNEL-assay and its localization. In the untreated wild-type crosses as positive control, almost all macronuclei were TUNEL-positive and localized at the posterior of the cytoplasm (). In the presence of 10 nM wortmannin, approximately 80% of the nuclei were TUNEL-negative and most of these localized at the anterior or middle of the cytoplasm (). In the TtVPS34∆ crosses, a large fraction of the nuclei were also TUNEL-negative (). However, approximately half of these localized properly at the posterior of the cytoplasm, unlike with wortmannin treatment (, P < 0.05, Wilcoxon U test). In , the left panel of the TtVPS34∆ is an example where parental macronuclei are TUNEL-negative and localize at the anterior of the cytoplasm, while the right panel is another example that shows the coincidence of TUNEL-positive and -negative macronuclei at the posterior of the cytoplasm in a single pair. These observations suggest that the abnormal nuclear arrangement is unlikely to be necessary to impair the DNA degradation.
Figure 4. A critical role of PtdIns3K activity in DNA degeneration. (A) TUNEL assay results of wortmannin (10 nM)-treated cells and TtVPS34∆ crossings. The cells were sampled at 8 h (left panel) and 14 h (right panel) for the assay. White arrowheads, parental macronucleus; asterisks, developing new macronuclear anlagen; yellow arrowheads, TUNEL-positive parental macronucleus. Scale bars: 10 μm. (B) Relation between reactions of parental macronucleus to TUNEL assay and its localization. Measurements were done on more than 100 cells. (C) Agarose gel electrophoresis using fragmented macronuclear genome extracted from conjugating cells at 8 h. Arrows in the picture correspond to a DNA ladder pattern at ~180 bp intervals. M denotes a DNA sample marker.
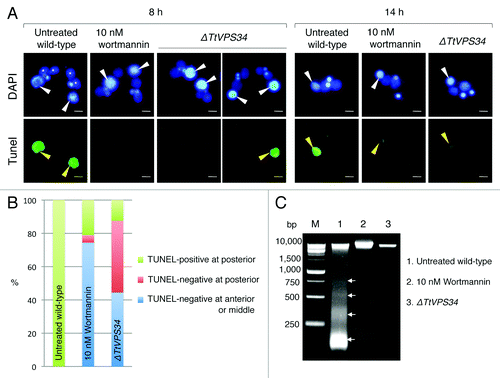
DNA fragments were extracted from the cells at the same time point and analyzed by agarose gel electrophoresis for further comparison of the DNA degradation. The macronuclear DNA in the untreated wild-type crosses was largely degraded into fragments ranging from larger than 10 kb to 180 bp (), indicating involvement of apoptosis-like DNA degradation. Addition of 10 nM wortmannin drastically impaired the degradation, and few nuclei were degraded into high molecular weight fragments and small pieces (). Knocking down of TtVPS34 further prevented the fragmentation, and small DNA pieces rarely appeared (). These results reveal that PtdIns3K activity is an essential factor for DNA degradation during PND.
TUNEL-positive parental macronuclei were detected in both the wortmannin-treated cells and the TtVPS34∆ mutant at the exconjugant stage (). This observation supports the idea proposed by Liu and YaoCitation24 that autophagy is not the only pathway responsible for PND. There may be other systems that contribute to DNA degradation in the later stage of PND.
Impact of wortmannin and knockdown of TtVPS34 on autophagic/lysosomal events during PND
Yakisich and KaplerCitation31 have shown that addition of the PtdIns3K inhibitors prevented acidification of the parental macronucleus, which caused failure in final resorption of the nucleus in the exconjugant stage. Similar to these treatments, the macronucleus in the TtATG8-2∆ crossings escaped from lysosomal acidification and then remained in the exconjugants.Citation24 Altogether, PND is most likely to require the TtATG and TtATG-related proteins in the autophagic/lysosomal events during PND.Citation19 In order to examine the impact of the treatment with 10 nM wotrmannin and knocking down of TtVPS34 on these events, the living conjugants at 8 h were stained with a combination of MDC and a lysosome indicator, LysoTracker Red (LTR), for fluorescence microscopy. Hoechest 33342 was also used to visualize the nuclei. In the untreated wild-type crosses, the parental macronucleus localized at the posterior of the cytoplasm and was uniformly covered by a greenish MDC signal (), corresponding to the generation of an autophagosomal characteristic on the envelope and thus representing autophagosome formation in PND.Citation19 Many small digestive vesicles attached on the nucleus and the nucleoplasm were strongly LTR-positive, indicating an acidic environment (, a). Addition of wortmannin at 2 nM or higher drastically prevented the autophagic/lysosomal events, in which the parental macronucleus showed neither MDC nor LTR signal at the anterior or middle of the cytoplasm (, b–E, e). Noteworthy, not only did the digestive vesicles not approach the nucleus but they also decreased in number in inverse proportion to the concentration of the inhibitor (, b–E, e), implying a critical role of PtdIns3K activity in autophagosome formation, as well as vesicular generation and migration.
Figure 5. Effects of wortmannin or knockdown of TtVPS34 on autophagic/lysosomal events during PND. Conjugating cells at 8 h were stained with a combination of MDC (upper) and LTR (lower). Hoechest was also used to visualize nuclei (upper panels). (A, a) Untreated wild-type cross. (B, b–E, e) Wild-type crosses treated with various concentrations of wortmannin ranging from 1 to 10 nM. (F, f) TtVPS34∆ crosses with both the parental macronuclei localized at the anterior region of the cells. (G, g) TtVPS34∆ cross with the parental macronuclei localized at middle (left) and posterior regions of the cytoplasm. White arrowheads, parental macronucleus; asterisks, developing new macronuclear anlagen. Scale bars: 10 μm.
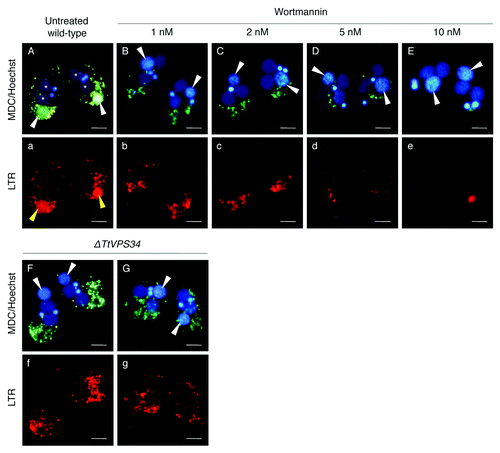
In the TtVPS34∆ crosses, on the other hand, the parental macronucleus escaped from the autophagic/lysosomal events wherever the nucleus localized (, f and G, g). Unlike the wortmannin treatment, the digestive vesicles were not reduced in number (, f and G, g). These observations suggest that TtVPS34 exerts its function in autophagosome formation at an early stage of PND, which is an essential event for progress of the subsequent digestive processes. The question still remained unclear, however, as to whether TtVPS34 controlled attraction of the vesicles to the nucleus.
A possible “attack-me” signal is exposed on the parental macronuclear surface in the absence of PtdIns3K activity
In a previous report we demonstrated that the initial stage of PND involved exposure of sugars and lipids, which were normally restricted in the inner leaflet of the membrane, on the parental maconuclear envelope.Citation19 As “eat-me” signals on apoptotic cells,Citation38,Citation39 these molecules were not present on the normal nuclei such as progeny micro- and macronuclei. Therefore we postulated that these molecules might act as an “attack-me” signal, which is responsible for the attraction of the digestive vesicle complexes to the parental macronucleus.Citation19,Citation40 Since the vesicles did not migrate to the parental macronucleus without PtdIns3K activity (), we suggest that the nucleus was unable to expose such molecules on the envelope under these conditions.
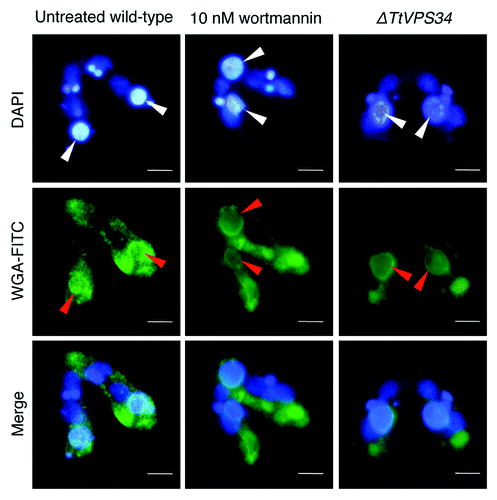
To examine if loss of PtdIns3K activity affects exposing molecules on the surface, the conjugating cells at 8 h were fixed and then used for cytological analysis with FITC-labeled wheat germ agglutinin (WGA) that binds to N-acetyl-D-glucosamine or sialic acid on the PND nuclei.Citation19 Noticeable differences between the parental macronucleus and other nuclei were found with WGA-FITC in the untreated wild-type crosses, in which the lectin bound to the parental macronuclear envelope but not to the other nuclear envelopes (). Contrary to our expectation, the parental macronucleus possessed the ability to bind to the lectin in spite of the addition of 10 nM wortmannin or knocking down of TtVPS34 (). This observation strongly suggests exposing of the “attack-me” signal does not coincide with autophagosome formation.
Figure 6. Binding of lectin to the parental macronucleus. Conjugating cells at 8 h were fixed and stained with DAPI (upper) and FITC-labeled WGA (middle). The lower parts show a merged image. White arrowheads, parental macronucleus; red arrowheads, concentrated FITC-signal on the nuclear surface. Scale bars: 10 μm.
Deficiency in PND and its relation to progeny macronuclear differentiation
Tetrahymena uses a variety of histone posttranslational modifications in both micro- and macronuclei throughout the life cycle.Citation41-Citation43 Akematsu et al.Citation19 have shown an example of the modifications during PND with an antibody against dimethylation in histone H3 at lysine 4 (dimeH3K4), a hallmark for active transcription. Once PND starts, the signal of dimeH3K4 disappears from the parental macronucleus while it coincidentally appears in the progeny macronuclear anlagen. Similarly, RNA polymerase II, the central component for basal transcription, also disappears from the parental macronucleus while it rapidly appears in the anlagen.Citation44 Even more important, Twi1, which is a Piwi family member of argonaute proteins in Tetrahymena and localized to the parental macronucleus until 7 h (before the Mac IIp stage), translocated to the anlagen at 8 h (Mac IIp; see example in ) accompanied with siRNAs called scnRNA for removal of internal eliminated sequences from the progeny macronuclear genome in the later stage of conjugation.Citation12
Figure 7. Effects of wortmannin or knockout of TtVPS34 on behavior of Twi1 and hallmarks for active transcription. Conjugating cells were fixed and used for indirect immunofluorescence observations using FITC-labeled antibodies (middle). DAPI was also used to stain nuclei (upper). The lower parts show a merged image. (A) Twi1. (B) Phosphorylated RNA polymerase II at Ser 2 of the C-terminal domain repeat (RNApol-II S2ph). (C) Dimethylation of histone H3 at Lys4 (H3K4 dime). White arrowheads, parental macronucleus; asterisks, developing new macronuclear anlagen. Scale bars: 10 μm.
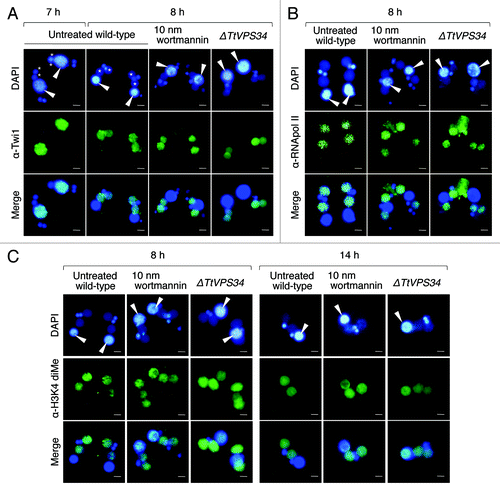
TtATG8 likely plays a role in progeny macronuclear development.Citation24 However, we suppose that failure in PND may involve some harmful influences on development since PND is not homeostatic autophagy such as starvation-induced autophagy and plays an important role to provide alternate genotypes from parents to progeny in a single cytoplasm. In order to address this question, the conjugating cells at 8 h were fixed and then analyzed by immunofluorescence with antibodies against each marker.
The Twi1 signal disappeared entirely from the parental macronucleus and exclusively appeared in the developing anlagen in the untreated wild-type crosses (). It was not affected with 10 nM wortmannin and knocking down of TtVPS34 (), implying independence of the event from autophagosome formation. Similarly, the signal of phosphorylated RNA polymerase II at the C-terminal domain repeats, which represents the activated form of this enzyme, also entirely disappeared from the parental macronucleus and appeared in the developing anlagen in every cross (). This strongly suggests that the progeny genotype replaces the transcriptional activity of the parental stage by the early stage of PND irrespective of autophagosome formation.
In contrast, the signal of dimeH3K4 in the parental macronucleus was not entirely replaced in the anlagen at 8 h in the presence of wortmannin and in the TtVPS34∆ mutant, resulting in overlapping of the signal in both generations of the macronuclei (). These signals still overlapped with almost equal intensities even in the exconjugant stage at 14 h (). Since phosphorylated RNA polymerase II was already moved to the anlagen at 8 h (), the parental macronucleus might no longer transcribe mRNAs after this stage. This observation provided important data that the parental macronucleus retained the epigenetic status for active transcription unless PND was successful, which might affect some pathways that contribute proper alteration of the generations in a single cytoplasm. If so, autophagy is the only way in which the parental stage completely collapses. In other words, loss of the epigenetic mark might depend on enzymes coming from outside of the nucleus similar to the situation seen in diacetylation of parental macronuclear histone H4 during PND.Citation45
Discussion
PtdIns3Ks in Tetrahymena
Phylogenetic analysis revealed that class I PtdIns3Ks were mainly found in metazoans with a single isoform in invertebrates but not in protist groups including ciliates (). Instead, some protist groups possessed specific PtdIns3Ks that belonged to neither metazoan class I or II. Class I PtdIns3K are usually activated by G-protein-coupled receptors and tyrosine kinase receptors, both of which relay their signal to protein kinase B (AKT/PKB), resulting in negatively regulating autophagy.Citation34 Tyrosine kinases have been found in some nonmetazoan species such as Chlamydomonas reinhardtii, Monosiga brevicolis, and Entamoeba histolyticaCitation46-Citation48 and there is some evidence for their presence in Tetrahymena (Miao, personal communication; Pearlman, manuscript in preparation). In Tetrahymena, only PtdIns3P and PtdIns(3,5)P2 have been detected as products of PtdIns3K activities, although PtdIns4P and PtdIns(4,5)P2 also exist in the cytoplasm.Citation33,Citation49 These data suggest that ciliates as well as Amoebozoa and Kinetoplastids have independently evolved novel PtdIns3Ks that may control distinct signaling pathways from metazoan class I- and II-mediated pathways.
On the other hand, class III PtdIns3K or orthologs of yeast Vps34 occur in all major taxonomic groups and are the sole PtdIns3K encoded in the genomes of plants, yeasts and Tetrahymena (). Vps34 participates in not only autophagy but also endocytosis, both of which are remarkable cell functions and conserved in all eukaryotes.Citation50,Citation51 In the endocytotic pathway, PIK3C3/VPS34 forms a complex with UVRAG/VPS38 (UV-irradiation-resistance-associated gene) instead of ATG14.Citation52 Other than survival for starvation, PIK3C3 is indispensable for maintaining cell homeostasis. In native T cells, for instance, PIK3C3-dependent canonical autophagy promotes the cell’s survival through quality control of damaged mitochondria that produce reactive oxygen species.Citation53 Although there may be differences in its interacting proteins among organisms, TtVPS34 exhibits considerable sequence similarity in the 3 conserved domains to yeast Vps34 and human PIK3C3 (), implying essential functions of the protein in Tetrahymena as well. In fact, the Neo4 cassette did not replace all the TtVPS34 copies in the macronuclear genome (). The reduced copy number of the gene caused a delay in growth/survival of the cells under nutrient-deprivation conditions (). Consistent with Engelman et al.,Citation54 these results suggest that class III is the origin of PtdIns3Ks that is acquired in the early history of eukaryotic evolution, and that this enzyme subsequently differentiated into a family of enzymes to regulate a variety of aspects of cellular metabolism.
TtVPS34 in PND
Liu and YaoCitation24 provided the first evidence that the Tetrahymena orthologs to the diverse mammalian orthologs of yeast ATG8 (such as the MAP1LC3 isoforms) played a role in degradation of the parental macronucleus. Tetrahymena has at least 2 functional ATG8 genes in the macronuclear genome. One of these (TtATG8-2) has an exclusive function in nuclear degradation while another (TtATG8-65) has a significant role in resistance under starvation condition as well as in nuclear degradation.Citation24 This information provides new insights into the question of whether PND is a specialized type of macroautophagy diverted from the conserved autophagy system such as starvation-induced macroautophagy due to development of the nuclear dimorphism course of ciliate evolution.
Loss of TtVPS34 activity caused failure in PND as was the case with the TtATG8∆ mutants, in which the DNA fragmentation and subsequent nuclear elimination were drastically prevented ( and ). The live-cell imaging using a combination of MDC and LTR revealed that loss of TtVPS34 activity prevented autophagosome formation on the parental macronucleuar envelope and consequent nuclear acidification with the digestive vesicles (), which is reminiscent of typical macroautophagy that starts with engulfment of target components followed by a lysosomal pathway. Although the nature of the MDC stainability on the envelope still remains unclear, it is generally thought that MDC is incorporated into multilamellar bodies such as autophagosomes by both an ion-trapping mechanism and the interaction with membrane lipids.Citation55 Together with localizing TtATG8 during PND,Citation24 the envelope probably directly changes its nature into an autophagosomal structure in a PtdIns3P- and TtATG8-dependent manner as occurs during general macroautophagy. In starved HEK-293 cells, as an analogous example, PtdIns3P is enriched in specific endoplasmic reticulum subdomains that plays an essential role in autophagosome formation.Citation56 During the process, the autophagosome is generated through invagination and direct maturation of endoplasmic reticulum enriched in PtdIns3P.Citation57
Even more important, failure in autophagosome formation resulted in preventing fusion of the digestive vesicles to the macronucleus despite the fact that a putative “attack-me” signal was exposed on the surface and the nucleoplasm did not become acidic ( and ). These results suggest that there may be other recognition systems rather than the “attack-me” signal to ensure selective nuclear degradation. If TtVPS34 substantially played a role in the discrimination of the parental macronucleus from the healthy nuclei rather than the “attack-me” signal, a high affinity between PtdIns3P and FYVE zinc finger domain-containing proteins would be a reasonable candidate contributing to membrane trafficking. In mammalian cells, this type of affinity controls specific recruitment of proteins to membranous structures such as endosomes, multivesicular bodies, and autophagosomes.Citation58-Citation60 Another hypothesis could be that the digestive vesicles might possess some FYVE domain-containing proteins, through which PtdIns3P embedded in the nuclear envelope was targeted. Alternatively, the “attack-me” signal might be essential but require PtdIns3P as an essential collaborator to exert its function in a discrimination step.
Addition of wortmannin decreased the number of digestive vesicles including lysosomes in a concentration-dependent manner, which was unlikely affected by inhibition of TtVPS34 activity (). Some evidence for participation of PtdIns3K activity in maturation of lysosomes has been reported through the use of PtdIns3K inhibitors. For instance, addition of wortmannin or LY294002 to rat hepatocytes causes replacement of dense lysosomes with swollen vesicles.Citation61 In other instances, wortmannin impairs transport of CTSD (cathepsin D) and PDGFRA (platelet-derived growth factor receptor-α) to lysosomes in mammalian cells.Citation62-Citation64 This evidence implies that other PtdIns3Ks of Tetrahymena play critical roles in maturation and stability of the digestive vesicles.
Role of TtVPS34 in positioning of the parental macronucleus during PND
Similar to the situations in the atg8-deficient mutants,Citation24 loss of TtVPS34 activity occasionally caused abnormal positioning of the parental macronucleus during PND, in which the nucleus localized at either the anterior or the middle region of the cytoplasm (). Based on our observations, the abnormal positioning of the nucleus was not responsible for failure of PND ( and ). It remains unclear how the parental macronucleus migrates to the posterior region of the cytoplasm at the Mac IIp stage. However, there seemed to be a novel function of the autophagy-mediated proteins in the positioning.
There is a possibility that a membrane-trafficking system or equivalent controls the positioning. In fact, it is accepted that autophagy shares some proteins with endocytosis, which is a membrane-trafficking system.Citation65 Tetrahymena has a cytoproct (cell anus) at the posterior region of the cytoplasm where lysosomes are abundantly localized.Citation66 In the vegetatively growing cell, the food vacuole, called the phagosome, is formed from the oral apparatus in the cell anterior, and the nascent phagosome is finally delivered to the cytoproct with an actin-based myosin motor.Citation67 Around the cytoproct, small GTPases called RABs are abundant and localized. These have been considered as key determinants of compartmental specificity by recruiting endocytic structures to the cytoproct.Citation68 In mammals, RAB5 isoforms (RABA, B and C) are downstream of PtdIns3K signaling, which play roles as endosome organizers that transport a cargo from the plasma membrane to lysosomes.Citation69 Moreover, RAB5 plays a role for not only membrane trafficking but also for autophagy as a member of a complex that also contains the proteins BECN1 and PIK3C3, and that associates with autophagosomal precursors.Citation70,Citation71
It would be reasonable to assume that some autophagy-mediated molecules in Tetrahymena including TtVPS34 and TtATG8s are utilized in the positioning besides executing the autophagic/lysosomal pathway. Loss of the proteins might disturb the proper positioning. These findings could bring new insights to the question about nuclear alignment during Tetrahymena conjugation.
Relation between PND and new macronuclear development
Genetic analysis of PNDCitation15 using a Tetrahymena mutant pair nullisomic for chromosome 3 of the micronucleus (NULLI 3) showed that the exconjugants failed to resorb the degenerated parental macronucleus, because of a lack of zygotic gene expression from the nullisomic progeny macronucleus. This suggests that the early stage of autophagic events and the following final resorptive stage are genetically distinct. The behavior of Twi1 and markers for active transcription indicated that the progeny genotype had already started its transcription at the Mac IIp stage irrespective of TtVPS34 activity (). However, the progeny genotype did not rescue the autophagic/lysosomal processes () despite the fact that the progeny genotype derived from the zygotic micronucleus was intact and distinct from the parental genome.Citation1,Citation72 These results suggest that autophagosome formation through the parental TtVPS34 activity plays the principal role in executing downstream of the autophagic/lysosomal processes, in which progeny genotype does not play any role except the final resorption. To insure the proper regulation of PND, the amount of TtVPS34 expression from the parental macronucleus and its timing might be strictly controlled by upstream signaling of PND.
The behavior of the markers also implies that the commitments to both the developmental and PND programs, are made earlier than the Mac IIp stage. Observations suggesting that time point have been made in Tetrahymena through the use of some inhibitors and spontaneous mutants. Neither blocking of the pronuclear exchange with nocodazoleCitation73 nor of fertilization with vinblastineCitation74 affects the programs, in which development starts with haploid micronuclei. On the other hand, mixing with an amicronucleate mutantCitation75 or certain lines called “star” strainsCitation76,Citation77 abolishes conjugation in response to imperfections in meiosis events in the mutants. In another instance, our laboratory and collaborators have revealed that an essential histone chaperone, ASF1, in Tetrahymena, which forms a complex with KPNB1 for histone transport to nuclei, no longer localizes in the parental macronucleus once meiosis is complete.Citation78 We postulate that Tetrahymena triggers both development and PND between meiosis and pronuclear exchange. Once the commitments are made to the programs, the downstream events may automatically progress without any checkpoint arrests.
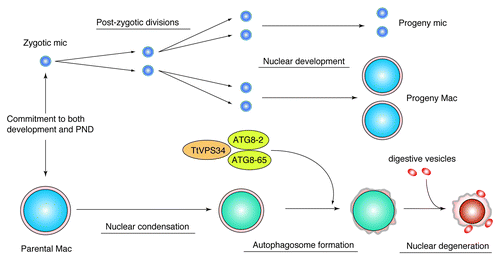
In summary, TtVPS34 is the sole ortholog to human PIK3C3 and yeast VPS34 in Tetrahymena. TtVPS34 plays important roles in growing/survival under starvation conditions and autophagosome formation on the parental macronucleus at the early stage of PND. Autophagosome formation involves subsequent events of PND such as DNA fragmentation, lysosomal acidification, and final resorption of the nucleus. On the one hand, the TtVPS34∆ mutant shows similar phenotypes to the TtATG8∆ mutants, implying collaboration or sequential roles of the proteins in autophagy events (). On the other hand, TtVPS34 as well as TtATG8 do not play a role in induction of PND, which is not responsible for triggering progeny macronuclear differentiation/development. This study offers new insights into addressing questions on not only “the specific autophagy system in ciliate PND” but also a “common mechanism for autophagosome formation in any types of autophagy across eukaryotes.”
Figure 8. Diagram illustrating a possible role of TtVPS34 in Tetrahymena PND. Once a commitment is made to both development and PND, the zygotic micronucleus and parental macronucleus start to divide and condense, respectively. TtVPS34 plays a critical role in autophagosome formation on the parental macronucleus together with two TtATG8s, which allows digestive vesicles to incorporate with the macronuclear envelope. Both TtVPS34 and TtATG8s do not appear responsible for nuclear condensation and progeny nuclear differentiation.
Materials and Methods
Culture methods and the induction of conjugation
The wild-type Tetrahymena strains, CU428 (mating type VII) and B2086 (mating type II), were distributed from the National Tetrahymena Stock Center (Cornell University; https://tetrahymena.vet.cornell.edu/). Cells were cultured at 30 °C in SPP medium containing 2% proteose peptone (Difco, 211684), 0.1% yeast extract (Difco, 212750), 0.2% glucose and 0.003% EDTA (Bioshop, 2868A84) with gentle shaking. To induce mating activity, the cells at mid-log phase (approximately 106 cells/ml) were washed with 10 mM TRIS-HCl (pH 7.2) and incubated overnight. To induce conjugation, equal numbers of both strains were mixed and kept at 30 °C without shaking.
Phylogenetic analysis
Tetrahymena orthologs to human PtdIns3Ks and PtdIns4Ks were identified with gene description in the Tetrahymena genome database (http://www.ciliate.org). The protein sequences of other organisms were obtained from GenBank or databases for each species through the use of BLASTp search against human PtdIns3Ks and PtdIns4Ks. Identical numbers of the proteins are shown in as well as in Figure S1. We employed the amino acid sequences of the catalytic domain at the C-terminal region for the multiple alignments with Clustal Omega (Ver. 1.1.0),Citation79 which were predicted by using PROSITE in ExPASy (http://prosite.expasy.org/). All gap regions appearing after the alignments were eliminated from the sequences, and the resulting 216 amino acid residues were used for reconstructing the phylogenetic tree. TreefinderCitation80 equipped in the software AminosanCitation81 provided LG+I+G+F as the best evolutionary model for this data set. The phylogenetic tree was finally constructed using the maximum likelihood method in RAxML (Ver. 7.3.0).Citation82 Confidence in the phylogeny was estimated by using the bootstrap method in 100 replications.
Construction of TtVPS34 disruption vector
Approximately 1-kb sequences upstream (5′) and downstream (3′) of TtVPS34 genomic locus were amplified from CU428 genomic DNA with High-Fidelity DNA Polymerases (BioLabs, M0530) using the following primer sets: TtVPS34-5F (GAGCTCacct gcgcaactga gcat)—TtVPS34-5R (GCGGCCGCtg acatttacat cttacccaa) for the 5′ sequence and TtVPS34-3F (CTCGAGactg ctaccatctt cgtc)—TtVPS34-3R (GGTACCcagc aataggcata agcca) for the 3′ sequence. The amplified PCR products were cloned into pT7 blue T-vector (Invitrogen, 45-0071) and then retrieved from the plasmids by using SacI-NotI for the 5′ sequence and XhoI-KpnI for the 3′ sequence. These fragments were sequentially integrated into the backbone vector pNeo4 (GenBank: EU606202.1). The resulting vector named pKOTtVps34 was linearized with KpnI before biolistic bombardment.
Tetrahymena transformation
For Tetrahymena transformation, mid-log phase cells were harvested by centrifugation and incubated overnight in 10 mM TRIS-HCl (pH 7.2). The cells were centrifuged and packed in 1 ml of 10 mM TRIS-HCl (pH 7.2) at a final concentration of 108 cells/ml. The cell solution was spread on a sterile filter paper (Fisher Scientific, 09-795C). Transformation was achieved using a Biolistic Particle Delivery System (Bio-Rad, PDS-1000). Gold particles 0.6 μm in size (Bio-Rad, 165-2262: 10 mg/ml in sterile glycerol) were coated with 5 μg linearized DNA/50 μl particles. Cells were bombarded with the DNA-coated gold particles at 900 psi. Following bombardment, the cells were resuspended in SPP and incubated for 3 h. Ten milligrams/milliliter of CdCl2 was added to allow expression of the Neo4 gene under MTT1 promoter control. After 1 h incubation, the transformants were screened with 100 μg/ml paromomycin (Calbiochem, 512731). The paromomycin-resistant cells appeared in 3 d and were grown in SPP containing increasing concentrations of paromomycin (from 100 to 5000 μg/ml) to allow the allelic assortment.
Southern blotting
Genomic DNA extracted from both the wild-type and TtVPS34∆ mutants were digested with a combination of AccI and SphI. The same amount of the digested DNA samples (5 μg) were electrophoresized through an 0.8% agarose gel, capillary transferred onto a Hybond-nylon membrane (GE Healthcare, RPN203B) and baked 2 h at 80 °C under vacuum. The membrane was hybridized with probes overnight at 42 °C in the hybridization buffer as described in the Gene Images AlkPhos Direct Labeling and Detection System (GE Healthcare, RPN3680). Alkaline phosphatase-labeled probes were made by using the system with PCR products from genomic DNA. Hybridized DNA probe was detected by using CDP-Star detection reagent (GE Healthcare, NIF1229) as substrate. The membrane was exposed to X-ray film for 1 h to 6 h.
Wortmannin treatment
Wortmannin (Sigma, W3144) was dissolved in dimethyl sulfoxide (DMSO; Sigma, D2650) for the stock solution (1 μM). The stock solution was diluted with DMSO to prepare the desired concentration before use, and then added to the media containing conjugating Tetrahymena at 5 h after induction of conjugation. The final concentration of DMSO was 1%. Untreated wild-type cells were exposed to 1% DMSO.
DAPI staining
Cells were fixed with 4% paraformaldehyde, and kept on ice for 30 min. Paraformaldehyde was removed by centrifugation, and the samples were resuspended in 10 mM TRIS-HCl (pH 7.2) containing 1% Nonidet P-40 (Santa Cruz Biotech, sc-29102) and kept at room temperature for 30 min followed by the addition of 1 μg/μl of 4-6-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen, D1306) in distilled water.
TUNEL assay
Cells were fixed with 4% paraformaldehyde for 10 min and then membrane permeabilized with 0.1% sodium citrate and 0.1% Triton X-100 for 2 min on ice. Terminal transferase reaction to free 3′-OH DNA ends was made by using an In Situ Cell Death Detection Kit, Fluorescein (Roche, 1-684-795) for 1 h at 37 °C. After washing with PBS (0.2 g KCl, 0.24 g KH2PO4, 8 g NaCl, 1.44 g Na2HPO4, adjusted to pH 7.4 and brought to 1 L with distilled water) twice, the samples were resuspended into PBS containing 1 μg/μl of DAPI to stain nuclei and then analyzed using fluorescence microscopy.
Fragmental DNA extraction
Fragmental DNA during PND, such as kb-sized and oligonucleosome-sized DNA, was extracted from conjugating cells at 8 h. In the following procedure, intact genomic DNA is not generally recovered.Citation18 Fifty microliters of cell lysis buffer consisting of 100 mM EDTA, 5% Triton-X 100 and 100 mM TRIS-HCl buffer (pH 7.2) was added to 450 μl of cell solution. After 10 min incubation on ice, lysates were centrifuged at 13500 g for 10 min and the collected supernatants were incubated with 200 μg/ml proteinase K (Sigma, P6556) for 30 min at 50 °C. 100 μl of 5 M NaCl and 600 μl of 2-propanol were added, and the samples were incubated overnight at −20 °C. Fragmented DNA was recovered by centrifugation at 13500 g for 10 min, and the precipitate was dissolved in 15 μl of TE buffer (10 mM TRIS-HCL, pH 7.5, 1 mM EDTA) containing 10 μg/ml ribonuclease A (Sigma, R4642). After incubation for 30 min at 37 °C, the DNA samples were electrophoresed through a 2% agarose gel.
Labeling of autophagosomal structures and lysosomes
Autophagic vacuoles and acidic compartments were labeled with 0.1 mM MDC (BioChemika, 30432) and 1 mM LTR (Molecular Probes Inc., L-7528), respectively. Hoechst 33342 (Dojindo, H342; 5 mg/ml) was also used to stain nuclei. Living cells in 10 mM TRIS-HCl, pH 7.2 were incubated with these fluorescent compounds at 30 °C for 30 min in the dark. After incubation, the cells were washed with 10 mM TRIS-HCl (pH 7.2) and immediately analyzed by fluorescence microscopy. For photography, cells were anesthetized with NiCl2 (Sigma, N6136; 15 mM).
Lectin binding
Cells were fixed and membrane-permeabilized by successive treatment with 4% paraformaldehyde and 0.1% Tween 20 on ice for 10 min. The samples were washed with PBS and incubated with 10 μg/ml of FITC-labeled WGA (Sigma, L4895) on ice for 1 h. One micrograms/microliter of DAPI was also used to stain nuclei. After incubation, the samples were washed with PBS and analyzed by fluorescence microcopy.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde and membrane-permeabilized with cold methanol on ice for 10 min. After washing with PBS, the cells were blocked with 0.1% BSA (Sigma, A9647) and incubated with the primary antibodies (1:500) overnight at 4 °C. Twi1 antibody derived from mouse was kindly supplied by Dr. Kazafumi Mochizuki (Institute of Molecular Biotechnology, Vienna, Austria). Both anti-phosphorylated RNApol-II at Ser2 of the C-terminal domain (Abcam, ab5095) and anti-dimethylated histone H3 at Lys4 (Millipore, 07-030) were derived from rabbit. After washing twice with PBS, the cells were incubated with FITC-labeled secondary antibodies (1:500 dilution), goat anti-mouse (Millipore, AP181F) or goat anti-rabbit (Millipore, AP132F) for 1 h at room temperature in the dark. After washing with PBS, the cells were stained with 1 μg/μl of DAPI and observed with fluorescence microcopy.
Micrograph
A digital camera (Olympus, DP70) equipped with a fluorescent microscope (Reichert-Jung, POLYVER) was used for all photography. Photographs were processed with ImageJ64 to change brightness/contrast and to merge 2 different images.
| Abbreviations: | ||
| AIF | = | apoptosis-inducing factor |
| AKT/PKB | = | protein kinase B |
| Atg | = | autophagy-related |
| BECN1 | = | Beclin 1 |
| CTSD | = | cathepsin D |
| DAPI | = | 4-6-diamidino-2-phenylindole dihydrochloride |
| dimeH3K4 | = | dimethylation in histone H3 at lysine 4 |
| DMSO | = | dimethyl sulfoxide |
| EGFP | = | enhanced green fluorescent protein |
| EndoG | = | endonuclease G |
| ER | = | endoplasmic reticulum |
| PIK3C3/VPS34 | = | phosphatidylinositol 3-kinase, catalytic subunit type 3 (human ortholog of yeast Vps34) |
| IC50 | = | half maximal inhibitory concentration |
| KPNB1 | = | karyopherin (importin) beta 1 |
| LTR | = | LysoTracker Red |
| MAP1LC3 | = | microtubule-associated protein 1 light chain 3 |
| MDC | = | monodansylcadaverine |
| PAS | = | phagophore assembly site |
| PDGF-R | = | platelet-derived growth factor receptors |
| Piwi | = | P-element induced wimpy testis |
| PND | = | programmed nuclear death |
| PtdIns | = | phosphatidylinositol |
| PtdIns3K | = | PtdIns 3-kinase |
| PtdIns4K | = | PtdIns 4-kinase |
| RAB | = | member RAS oncogene family |
| Twi1 | = | Tetrahymena Piwi |
| TtVPS34 | = | Tetrahymena thermophila VPS34 |
| UVRAG | = | UV radiation-resistance associated |
| Vps | = | vacuolar protein sorting |
| WGA | = | wheat germ agglutinin |
Additional material
Download Zip (1.7 MB)Acknowledgments
We thank Dr Kazufumi Mochizuki for providing Twi1 antibody and Anita Samardzic for media and buffer preparation. We thank Jyoti Garg, Masaaki Iwamoto, Albana Kume, Susanna Marquez, Ajay Matta, and Yoshiomi Takagi for their technical suggestions and invaluable discussions. This study was supported by a Banting Postdoctoral Fellowship through the Natural Sciences and Engineering Research Council of Canada to TA, Institutional Program for Young Researcher Overseas Visits through Japan Society for the Promotion of Science to YF and grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes for Health Research to REP.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Chalker DL. Transformation and strain engineering of Tetrahymena.. Methods Cell Biol 2012; 109:327 - 45; http://dx.doi.org/10.1016/B978-0-12-385967-9.00011-6; PMID: 22444150
- Coyne RS, Stover NA, Miao W. Whole genome studies of Tetrahymena.. Methods Cell Biol 2012; 109:53 - 81; http://dx.doi.org/10.1016/B978-0-12-385967-9.00004-9; PMID: 22444143
- Orias E, Cervantes MD, Hamilton EP. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res Microbiol 2011; 162:578 - 86; http://dx.doi.org/10.1016/j.resmic.2011.05.001; PMID: 21624459
- Iwamoto M, Mori C, Kojidani T, Bunai F, Hori T, Fukagawa T, Hiraoka Y, Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena.. Curr Biol 2009; 19:843 - 7; http://dx.doi.org/10.1016/j.cub.2009.03.055; PMID: 19375312
- Cole E, Sugai T. Developmental progression of Tetrahymena through the cell cycle and conjugation. Methods Cell Biol 2012; 109:177 - 236; http://dx.doi.org/10.1016/B978-0-12-385967-9.00007-4; PMID: 22444146
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena.. Cell 2002; 110:689 - 99; http://dx.doi.org/10.1016/S0092-8674(02)00909-1; PMID: 12297043
- Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena.. Cell 2002; 110:701 - 11; http://dx.doi.org/10.1016/S0092-8674(02)00941-8; PMID: 12297044
- Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena.. Curr Opin Genet Dev 2004; 14:181 - 7; http://dx.doi.org/10.1016/j.gde.2004.01.004; PMID: 15196465
- Yao MC, Yao CH, Halasz LM, Fuller P, Rexer CH, Wang SH, Jain R, Coyne RS, Chalker DL. Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena.. J Cell Sci 2007; 120:1978 - 89; http://dx.doi.org/10.1242/jcs.006502; PMID: 17519286
- Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K. The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell 2010; 140:692 - 703; http://dx.doi.org/10.1016/j.cell.2010.02.010; PMID: 20211138
- Chalker DL, Yao MC. DNA elimination in ciliates: transposon domestication and genome surveillance. Annu Rev Genet 2011; 45:227 - 46; http://dx.doi.org/10.1146/annurev-genet-110410-132432; PMID: 21910632
- Mochizuki K. DNA rearrangements directed by non-coding RNAs in ciliates. Wiley Interdiscip Rev RNA 2010; 1:376 - 87; http://dx.doi.org/10.1002/wrna.34; PMID: 21956937
- Bracht JR, Fang W, Goldman AD, Dolzhenko E, Stein EM, Landweber LF. Genomes on the edge: programmed genome instability in ciliates. Cell 2013; 152:406 - 16; http://dx.doi.org/10.1016/j.cell.2013.01.005; PMID: 23374338
- Weiske-Benner A, Eckert WA. Differentiation of nuclear structure during the sexual cycle in Tetrahymena thermphila; II. Degeneration and autolysis of macro-and micronuclei. Differentiation 1987; 34:1 - 12; http://dx.doi.org/10.1111/j.1432-0436.1987.tb00044.x
- Davis MC, Ward JG, Herrick G, Allis CD. Programmed nuclear death: apoptotic-like degradation of specific nuclei in conjugating Tetrahymena.. Dev Biol 1992; 154:419 - 32; http://dx.doi.org/10.1016/0012-1606(92)90080-Z; PMID: 1426647
- Kobayashi T, Endoh H. Caspase-like activity in programmed nuclear death during conjugation of Tetrahymena thermophila.. Cell Death Differ 2003; 10:634 - 40; http://dx.doi.org/10.1038/sj.cdd.4401216; PMID: 12761572
- Kobayashi T, Endoh H. A possible role of mitochondria in the apoptotic-like programmed nuclear death of Tetrahymena thermophila.. FEBS J 2005; 272:5378 - 87; http://dx.doi.org/10.1111/j.1742-4658.2005.04936.x; PMID: 16218967
- Akematsu T, Endoh H. Role of apoptosis-inducing factor (AIF) in programmed nuclear death during conjugation in Tetrahymena thermophila.. BMC Cell Biol 2010; 11:13; http://dx.doi.org/10.1186/1471-2121-11-13; PMID: 20146827
- Akematsu T, Pearlman RE, Endoh H. Gigantic macroautophagy in programmed nuclear death of Tetrahymena thermophila.. Autophagy 2010; 6:901 - 11; http://dx.doi.org/10.4161/auto.6.7.13287; PMID: 20798592
- Mpoke S, Wolfe J. DNA digestion and chromatin condensation during nuclear death in Tetrahymena.. Exp Cell Res 1996; 225:357 - 65; http://dx.doi.org/10.1006/excr.1996.0186; PMID: 8660924
- Lu E, Wolfe J. Lysosomal enzymes in the macronucleus of Tetrahymena during its apoptosis-like degradation. Cell Death Differ 2001; 8:289 - 97; http://dx.doi.org/10.1038/sj.cdd.4400807; PMID: 11319612
- Duszenko M, Ginger ML, Brennand A, Gualdrón-López M, Colombo MI, Coombs GH, Coppens I, Jayabalasingham B, Langsley G, de Castro SL, et al. Autophagy in protists. Autophagy 2011; 7:127 - 58; http://dx.doi.org/10.4161/auto.7.2.13310; PMID: 20962583
- King JS. Autophagy across the eukaryotes: is S. cerevisiae the odd one out?. Autophagy 2012; 8:1159 - 62; http://dx.doi.org/10.4161/auto.20527; PMID: 22722653
- Liu ML, Yao MC. Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila.. Eukaryot Cell 2012; 11:494 - 506; http://dx.doi.org/10.1128/EC.05296-11; PMID: 22366125
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem 1998; 67:481 - 507; http://dx.doi.org/10.1146/annurev.biochem.67.1.481; PMID: 9759495
- Foster FM, Traer CJ, Abraham SM, Fry MJ. The phosphoinositide (PI) 3-kinase family. J Cell Sci 2003; 116:3037 - 40; http://dx.doi.org/10.1242/jcs.00609; PMID: 12829733
- Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae.. Mol Cell Biol 1990; 10:6742 - 54; PMID: 2247081
- Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, Zvelebil MJ, Domin J, Panaretou C, Waterfield MD. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J 1995; 14:3339 - 48; PMID: 7628435
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J 2008; 410:1 - 17; http://dx.doi.org/10.1042/BJ20071427; PMID: 18215151
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107 - 32; http://dx.doi.org/10.1146/annurev-cellbio-092910-154005; PMID: 21801009
- Yakisich JS, Kapler GM. The effect of phosphoinositide 3-kinase inhibitors on programmed nuclear degradation in Tetrahymena and fate of surviving nuclei. Cell Death Differ 2004; 11:1146 - 9; http://dx.doi.org/10.1038/sj.cdd.4401473; PMID: 15257301
- Kovács P, Pállinger É. Phosphatidylinositol 3-kinase-like activity in Tetrahymena. Effects of wortmannin and LY 294002. Acta Protozool 2003; 42:277 - 85
- Leondaritis G, Tiedtke A, Galanopoulou D. D-3 phosphoinositides of the ciliate Tetrahymena: characterization and study of their regulatory role in lysosomal enzyme secretion. Biochim Biophys Acta 2005; 1745:330 - 41; http://dx.doi.org/10.1016/j.bbamcr.2005.06.011; PMID: 16081170
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6:463 - 77; http://dx.doi.org/10.1016/S1534-5807(04)00099-1; PMID: 15068787
- Brown JR, Auger KR. Phylogenomics of phosphoinositide lipid kinases: perspectives on the evolution of second messenger signaling and drug discovery. BMC Evol Biol 2011; 11:4; http://dx.doi.org/10.1186/1471-2148-11-4; PMID: 21208444
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One 2009; 4:e4429; http://dx.doi.org/10.1371/journal.pone.0004429; PMID: 19204800
- Stack JH, Emr SD. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem 1994; 269:31552 - 62; PMID: 7989323
- Depraetere V. “Eat me” signals of apoptotic bodies. Nat Cell Biol 2000; 2:E104; http://dx.doi.org/10.1038/35014098; PMID: 10854338
- Savill J, Gregory C. Apoptotic PS to phagocyte TIM-4: eat me. Immunity 2007; 27:830 - 2; http://dx.doi.org/10.1016/j.immuni.2007.12.002; PMID: 18093535
- Akematsu T, Kobayashi T, Osada E, Fukuda Y, Endoh H, Pearlman RE. Programmed nuclear death and its relation to apoptosis and autophagy during sexual reproduction in Tetrahymena thermophila.. Jpn J Protozool 2012; 45:1 - 15
- Vavra KJ, Allis CD, Gorovsky MA. Regulation of histone acetylation in Tetrahymena macro- and micronuclei. J Biol Chem 1982; 257:2591 - 8; PMID: 7061439
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena.. Proc Natl Acad Sci U S A 1998; 95:7480 - 4; http://dx.doi.org/10.1073/pnas.95.13.7480; PMID: 9636175
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena.. Proc Natl Acad Sci U S A 1999; 96:14967 - 72; http://dx.doi.org/10.1073/pnas.96.26.14967; PMID: 10611321
- Mochizuki K, Gorovsky MA. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot Cell 2004; 3:1233 - 40; http://dx.doi.org/10.1128/EC.3.5.1233-1240.2004; PMID: 15470252
- Slade KM, Freggiaro S, Cottrell KA, Smith JJ, Wiley EA. Sirtuin-mediated nuclear differentiation and programmed degradation in Tetrahymena.. BMC Cell Biol 2011; 12:40; http://dx.doi.org/10.1186/1471-2121-12-40; PMID: 21933443
- King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci U S A 2001; 98:15032 - 7; http://dx.doi.org/10.1073/pnas.261477698; PMID: 11752452
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science 2003; 301:361 - 3; http://dx.doi.org/10.1126/science.1083853; PMID: 12869759
- Shiu SH, Li WH. Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol Biol Evol 2004; 21:828 - 40; http://dx.doi.org/10.1093/molbev/msh077; PMID: 14963097
- Leondaritis G, Sarri T, Dafnis I, Efstathiou A, Galanopoulou D. Biochemical and genetic evidence for the presence of multiple phosphatidylinositol- and phosphatidylinositol 4,5-bisphosphate-specific phospholipases C in Tetrahymena.. Eukaryot Cell 2011; 10:412 - 22; http://dx.doi.org/10.1128/EC.00272-10; PMID: 21169416
- Samaj J, Baluska F, Voigt B, Schlicht M, Volkmann D, Menzel D. Endocytosis, actin cytoskeleton, and signaling. Plant Physiol 2004; 135:1150 - 61; http://dx.doi.org/10.1104/pp.104.040683; PMID: 15266049
- Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci 2011; 124:161 - 70; http://dx.doi.org/10.1242/jcs.064576; PMID: 21187343
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360 - 72; http://dx.doi.org/10.1091/mbc.E08-01-0080; PMID: 18843052
- Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A 2012; 109:8670 - 5; http://dx.doi.org/10.1073/pnas.1205305109; PMID: 22592798
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 2006; 7:606 - 19; http://dx.doi.org/10.1038/nrg1879; PMID: 16847462
- Niemann A, Takatsuki A, Elsässer HP. The lysosomotropic agent monodansylcadaverine also acts as a solvent polarity probe. J Histochem Cytochem 2000; 48:251 - 8; http://dx.doi.org/10.1177/002215540004800210; PMID: 10639491
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685 - 701; http://dx.doi.org/10.1083/jcb.200803137; PMID: 18725538
- Simonsen A, Stenmark H. Self-eating from an ER-associated cup. J Cell Biol 2008; 182:621 - 2; http://dx.doi.org/10.1083/jcb.200807061; PMID: 18725534
- Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci 2004; 117:4239 - 51; http://dx.doi.org/10.1242/jcs.01287; PMID: 15292400
- Lee SA, Eyeson R, Cheever ML, Geng J, Verkhusha VV, Burd C, Overduin M, Kutateladze TG. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc Natl Acad Sci U S A 2005; 102:13052 - 7; http://dx.doi.org/10.1073/pnas.0503900102; PMID: 16141328
- Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci 2006; 119:605 - 14; http://dx.doi.org/10.1242/jcs.02855; PMID: 16467569
- Mousavi SA, Brech A, Berg T, Kjeken R. Phosphoinositide 3-kinase regulates maturation of lysosomes in rat hepatocytes. Biochem J 2003; 372:861 - 9; http://dx.doi.org/10.1042/BJ20021136; PMID: 12646047
- Brown WJ, DeWald DB, Emr SD, Plutner H, Balch WE. Role for phosphatidylinositol 3-kinase in the sorting and transport of newly synthesized lysosomal enzymes in mammalian cells. J Cell Biol 1995; 130:781 - 96; http://dx.doi.org/10.1083/jcb.130.4.781; PMID: 7642697
- Davidson HW. Wortmannin causes mistargeting of procathepsin D. evidence for the involvement of a phosphatidylinositol 3-kinase in vesicular transport to lysosomes. J Cell Biol 1995; 130:797 - 805; http://dx.doi.org/10.1083/jcb.130.4.797; PMID: 7642698
- Shpetner H, Joly M, Hartley D, Corvera S. Potential sites of PI-3 kinase function in the endocytic pathway revealed by the PI-3 kinase inhibitor, wortmannin. J Cell Biol 1996; 132:595 - 605; http://dx.doi.org/10.1083/jcb.132.4.595; PMID: 8647891
- Lamb CA, Dooley HC, Tooze SA. Endocytosis and autophagy: Shared machinery for degradation. Bioessays 2013; 35:34 - 45; http://dx.doi.org/10.1002/bies.201200130; PMID: 23147242
- Sugita M, Nakano K, Sato M, Toyooka K, Numata O. The roles of actin cytoskeleton and microtubules for membrane recycling of a food vacuole in Tetrahymena thermophila.. Cell Motil Cytoskeleton 2009; 66:371 - 7; http://dx.doi.org/10.1002/cm.20374; PMID: 19418560
- Hosein RE, Williams SA, Gavin RH. Directed motility of phagosomes in Tetrahymena thermophila requires actin and Myo1p, a novel unconventional myosin. Cell Motil Cytoskeleton 2005; 61:49 - 60; http://dx.doi.org/10.1002/cm.20065; PMID: 15810016
- Nusblat AD, Bright LJ, Turkewitz AP. Conservation and innovation in Tetrahymena membrane traffic: proteins, lipids, and compartments. Methods Cell Biol 2012; 109:141 - 75; http://dx.doi.org/10.1016/B978-0-12-385967-9.00006-2; PMID: 22444145
- Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, Epstein-Barash H, Kuchimanchi S, Peng CG, Ruda VM, et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature 2012; 485:465 - 70; http://dx.doi.org/10.1038/nature11133; PMID: 22622570
- Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 2008; 121:1649 - 60; http://dx.doi.org/10.1242/jcs.025726; PMID: 18430781
- Bento CF, Puri C, Moreau K, Rubinsztein DC. The role of membrane-trafficking small GTPases in the regulation of autophagy. J Cell Sci 2013; 126:1059 - 69; http://dx.doi.org/10.1242/jcs.123075; PMID: 23620509
- Orias E. Tetrahymena thermophila genetics: concepts and applications. Methods Cell Biol 2012; 109:301 - 25; http://dx.doi.org/10.1016/B978-0-12-385967-9.00010-4; PMID: 22444149
- Kaczanowski A, Ramel M, Kaczanowska J, Wheatley D. Macronuclear differentiation in conjugating pairs of Tetrahymena treated with the antitubulin drug nocodazole. Exp Cell Res 1991; 195:330 - 7; http://dx.doi.org/10.1016/0014-4827(91)90381-4; PMID: 2070816
- Hamilton EP, Suhr-Jessen PB, Orias E. Pronuclear fusion failure: an alternate conjugational pathway in Tetrahymena thermophila, induced by vinblastine. Genetics 1988; 118:627 - 36; PMID: 3366365
- Kaney AR. A transmissible developmental block in Tetrahymena thermophila.. Exp Cell Res 1985; 157:315 - 21; http://dx.doi.org/10.1016/0014-4827(85)90116-8; PMID: 3979442
- Allen SL. Cytogenetics of genomic exclusion in Tetrahymena.. Genetics 1967; 55:797 - 822; PMID: 6036954
- Allen SL, File SK, Koch SL. Genomic exclusion in tetrahymena.. Genetics 1967; 55:823 - 37; PMID: 17248381
- Garg J, Lambert JP, Karsou A, Marquez S, Nabeel-Shah S, Bertucci V, Retnasothie DV, Radovani E, Pawson T, Gingras AC, et al. Conserved Asf1-importin β physical interaction in growth and sexual development in the ciliate Tetrahymena thermophila.. J Proteomics 2013; 94C:311 - 26; http://dx.doi.org/10.1016/j.jprot.2013.09.018; PMID: 24120531
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7:539; http://dx.doi.org/10.1038/msb.2011.75; PMID: 21988835
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 2004; 4:18; http://dx.doi.org/10.1186/1471-2148-4-18; PMID: 15222900
- Tanabe AS. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour 2011; 11:914 - 21; http://dx.doi.org/10.1111/j.1755-0998.2011.03021.x; PMID: 21592310
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006; 22:2688 - 90; http://dx.doi.org/10.1093/bioinformatics/btl446; PMID: 16928733
