Abstract
Peroxisomes are critical organelles housing various, often oxidative, reactions. Pexophagy, the process by which peroxisomes are selectively targeted for destruction via autophagy, is characterized in yeast and mammals but had not been reported in plants. In this article, we describe how the peroxisome-related aberrations of a mutant defective in the LON2 peroxisomal protease are suppressed when autophagy is prevented by mutating any of several key autophagy-related (ATG) genes. Our results reveal that plant peroxisomes can be degraded by selective autophagy and suggest that pexophagy is accelerated when the LON2 protease is disabled.
LON proteins are ATP-dependent proteases that degrade damaged and short-lived proteins in bacteria. Eukaryotic LON proteases are targeted to organelles; plant LON isoforms reside in chloroplasts, mitochondria, and peroxisomes. LON2/At5g47040 is the sole peroxisomal LON isoform in the reference plant Arabidopsis thaliana. LON2 is positioned to degrade peroxisomal matrix proteins, but data supporting this role were elusive. In fact, glyoxylate cycle enzymes, peroxisomal proteins that are normally degraded a few days after germination, are degraded more quickly in lon2 mutants than in wild type, rather than more slowly as expected if LON2 facilitated matrix protein degradation. We found that peroxisomal matrix proteins localize normally in young lon2 seedlings but become increasingly cytosolic as seedlings age. In addition, peroxisomes become less abundant and appear as large orbs rather than the small puncta that characterize peroxisomes in wild type. Moreover, lon2 mutants seem to β-oxidize less efficiently as seedlings mature. The progressive decline in peroxisome function in lon2 contrasts with various other Arabidopsis peroxisome-defective mutants, which often appear to recover some function as plants mature.
To explore genetic interactions with LON2, we screened for suppressors of lon2 physiological defects and used whole-genome sequencing to identify the causal mutations. We were surprised to find multiple independent mutations in key autophagy genes (ATG2, ATG3, and ATG7) that each fully suppresses lon2 defects in peroxisome matrix protein import, physiology, morphology, and abundance. This discovery implies that lon2 physiological and molecular defects are not primarily caused by loss of the LON2 protease per se but instead that peroxisomes are targeted for autophagy when LON2 is dysfunctional. This increased pexophagy may eventually result in an insufficient number of peroxisomes to efficiently import matrix proteins or conduct peroxisomal metabolism (), explaining the aberrant phenotypes of lon2 mutants.
Figure 1. Pexophagy is enhanced when the peroxisomal protease LON2 is dysfunctional. In addition to core peroxisomal proteins, wild-type (LON2 ATG) peroxisomes house glyoxylate cycle enzymes in germinating seedlings and house several photorespiration enzymes in photosynthetic seedlings. When autophagy is prevented (LON2 atg), peroxisomes function normally, but certain glyoxylate cycle enzymes are slightly stabilized, suggesting that pexophagy plays a minor role in degrading seedling peroxisomes. In the lon2 mutant, peroxisomes are present and functional shortly after germination but are sparse and fail to efficiently import matrix proteins in older seedlings. These lon2 defects are fully suppressed by mutating any of several autophagy genes (ATG2, ATG3, or ATG7), suggesting that increased numbers of peroxisomes are targeted for pexophagy when LON2 is mutated. Although lon2 atg double mutant peroxisomes appear to import matrix proteins normally, glyoxylate cycle enzymes are inefficiently degraded, suggesting that LON2 normally promotes turnover of obsolete matrix proteins. The pexophagy trigger in lon2 mutants is not identified.
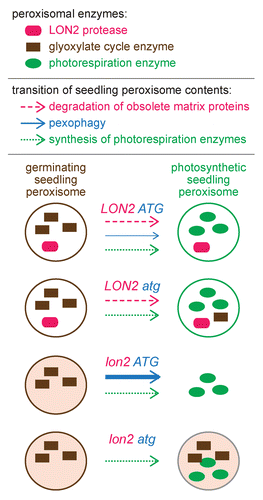
In addition to uncovering plant pexophagy, analysis of lon2 atg mutants revealed a role for LON2 in matrix protein turnover. In contrast to the accelerated matrix protein degradation observed in lon2 single mutants, lon2 atg double mutants display delayed degradation of obsolete glyoxylate cycle enzymes during seedling development. We propose that LON2 facilitates protein degradation during peroxisome content remodeling and that pexophagy is enhanced when this degradation is impaired. It remains to be determined whether LON2 directly degrades matrix proteins or functions indirectly, for example by facilitating disruption of protein complexes in the peroxisome matrix to allow protein retrotranslocation for cytosolic degradation.
Many of the >30 ATG genes first discovered in yeasts are conserved in plants and other eukaryotes, suggesting common core autophagy mechanisms. Reverse-genetic approaches targeting conserved Arabidopsis ATG genes have revealed similar mutant phenotypes; autophagy is required for plant survival of nitrogen and fixed-carbon starvation as well as drought, salinity, and oxidative stresses. Our screen for suppressors of a lon2 peroxisomal protease mutant, which resulted in isolation of atg2 (2 alleles), atg3 (1 allele), and atg7 (5 alleles) mutants, is the first facile forward-genetic screen for essential autophagy components in plants; very few plant atg mutants (and no atg3 alleles) have emerged from previous forward-genetic screens. This discovery provides an opportunity to subject plant autophagy (and pexophagy) to homology-independent genetic dissection, which has been spectacularly successful in yeast to uncover both conserved and novel autophagy components. Homology-independent approaches may be needed to discover pexophagy-specific components as plants lack homologs of yeast pexophagy receptors (e.g., Atg30 and Atg36).
The observation that peroxisomes are destroyed via autophagy when LON2 is dysfunctional demonstrates the existence of pexophagy in plants and prompts additional questions: What triggers heightened pexophagy in the lon2 mutant? Why do peroxisomes appear enlarged in the lon2 mutant? What is the pexophagy receptor in plants? What does this receptor target on the peroxisome surface? Is ubiquitination involved? How does pexophagy contribute to peroxisome remodeling during seedling development and during other developmental transitions? Can disabling pexophagy improve functioning in other peroxisome-defective mutants? These questions will be the subject of future studies, which will be facilitated by the developmentally modulated increased pexophagy in the Arabidopsis lon2 mutant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the NIH (R01GM079177), the National Science Foundation (MCB-0745122 and MCB-1244182), and the Robert A Welch Foundation (C-1309).
