Abstract
Peroxisomes are highly dynamic organelles that have multiple functions in cellular metabolism. To adapt the intracellular conditions to the changing extracellular environment, peroxisomes undergo constitutive segregation and degradation. The segregation of peroxisomes is mediated by 2 dynamin-related GTPases, Dnm1 and Vps1, whereas, the degradation of peroxisomes is accomplished through pexophagy, a selective type of autophagy. During pexophagy, the size of the organelle is always a challenging factor for the efficiency of engulfment by the sequestering compartment, the phagophore, which implies a potential role for peroxisomal fission in the degradation process, similar to the situation with selective mitochondria degradation. In this study, we report that peroxisomal fission is indeed critical for the efficient elimination of the organelle. When pexophagy is induced, both Dnm1 and Vps1 are recruited to the degrading peroxisomes through interactions with Atg11 and Atg36. In addition, we found that specific peroxisomal fission, which is only needed for pexophagy, occurs at mitochondria-peroxisome contact sites.
Keywords: :
Introduction
In most eukaryotic cells, mitochondria and peroxisomes drive the catabolism of long-chain fatty acids through β-oxidation, and convert the hydrogen peroxide into water and oxygen; in yeasts, this process is restricted to peroxisomes. One byproduct of β-oxidation is the production of reactive oxygen species that have the potential to damage cellular components. Therefore, it is essential that proper peroxisomal function is maintained, and that these organelles are removed if damaged or no longer needed. To adapt to the changing extracellular environment, peroxisomes undergo a remarkable remodeling of their cellular pattern of expression, morphology, and abundance. Peroxisomes achieve this dynamic property through biogenesis, division, and degradation. Budding from the endoplasmic reticulum (ER) appears to be one source of the peroxisome membrane, which is generated in a Pex3- and Pex19-dependent manner; however, the organelle may be primarily generated through fission of a preexisting peroxisome followed by the acquisition of additional membrane and proteins.Citation1 The proliferation and replication of peroxisomes that is achieved by division is under the control of 2 dynamin-related GTPases, Dnm1 and Vps1.Citation1,Citation2 Superfluous, or extensively damaged peroxisomes are targeted for vacuole-dependent degradation through selective autophagy, which is termed pexophagy.Citation3,Citation4
Autophagy is a conserved lysosome/vacuole-dependent catabolic pathway degrading cytosol, protein aggregates, and organelles. Depending on the extracellular stress and the degrading targets, autophagy can occur in either nonselective or selective modes. Nonselective autophagy mediates bulk degradation and recycling of cytoplasm to support cell survival during nutrient deprivation.Citation5,Citation6 Selective autophagy recognizes and targets specific cargos or organelles, such as peroxisomes and mitochondria (mitophagy).Citation7
After a decade of study, a general model has been established for selective autophagy. A ligand on the degrading target binds to a specific receptor; the receptor in turn recruits a scaffold protein, which links the cargo-receptor complex with the autophagy machinery.Citation8 In yeast, Atg11 is the scaffold and its binding receptor varies depending on the degrading cargos. In the case of pexophagy, Atg36 serves as the receptor in Saccharomyces cerevisiae, and PpAtg30 in Pichia pastoris; Pex3/PpPex3 functions as the ligand, binding to either Atg36 or PpAtg30.Citation9,Citation10 The receptor Atg36 binds to Atg11 (as does PpAtg30 bind to PpAtg11), a scaffold, which will promote the attachment of the degrading peroxisomes with Atg8–PE, and subsequent engulfment of the organelles by the phagophore, the initial sequestering compartment. We recently showed that when mitophagy is induced, the Atg11 scaffold is recruited by the mitophagy receptor Atg32, and in turn links the fission complex containing Dnm1 to drive the division of those mitochondria that are destined for degradation.Citation11 Similar to mitochondria, the size of peroxisomes also makes it problematic to sequester this organelle by the phagophore, leading us to hypothesize that peroxisomal fission is necessary for pexophagy.
Here, we show that the 2 dynamin-related GTPases Dnm1 and Vps1 are important for pexophagy. The scaffold protein Atg11 interacts with both Dnm1 and Vps1 with the Atg11-Dnm1 interaction occurring on both mitochondria and peroxisomes, whereas the Atg11-Vps1 interaction takes place exclusively on peroxisomes. Unlike the mitophagy receptor Atg32, the pexophagy receptor Atg36 is able to interact directly with Dnm1 and Vps1, and these interactions occur on the peroxisomes. These interactions representing the process of pexophagy-specific fission always occur at mitochondria-peroxisome contact sites.
Results
Peroxisomal fission is a significant step during pexophagy
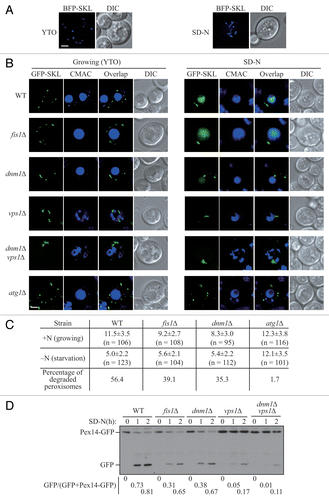
In Saccharomyces cerevisiae, peroxisomes proliferate with increasing number and size when cells are cultured in growth medium containing oleic acid as the sole carbon source. When these cells are subjected to conditions of nitrogen starvation in the presence of glucose, this elevated population of peroxisomes is no longer necessary, and pexophagy is induced to degrade the excess organelles.Citation3 To track the state and morphology of peroxisomes during pexophagy, we transformed a plasmid expressing the blue fluorescent protein (BFP) fused with a C-terminal type I peroxisomal targeting signal, serine-lysine-leucine (BFP-SKL).Citation12 When yeast cells were cultured in nutrient-rich medium with oleic acid (YTO), punctate peroxisomes appeared in the cytosol (, left). After we shifted the cells to nitrogen starvation medium with glucose (SD-N) for 2 h, which we have previously shown induces pexophagy,Citation3 the morphology of some peroxisomes changed and these organelles displayed an elongated pattern (, right). The elongation of peroxisomal membrane precedes the division of these organelles, which is significant for their replication and proliferation.Citation2 The occurrence of peroxisomal elongation under pexophagy-inducing conditions implied that the fission event also happened during the degradation of peroxisomes. Accordingly, we asked whether fission plays an important role for the progression of pexophagy.
Figure 1. Peroxisomal fission is required for pexophagy. (A) Wild-type yeast cells, transformed with pBFP-SKL (XLY049), were cultured in YTO as described in Materials and Methods to induce peroxisome proliferation, and shifted to SD-N for 2 h. (B) Wild-type (SEY6210), atg1∆ (WHY1), dnm1∆ (KDM1252), fis1∆ (XLY060), vps1∆ (XLY061), and dnm1∆ vps1∆ (XLY062) cells, transformed with pGFP-SKL, were cultured in YTO to induce peroxisome proliferation and shifted to SD-N for 7 h. CellTracker Blue CMAC was used to stain the vacuolar lumen. (C) Quantification of the numbers of peroxisomes in (B). Twelve Z-section images were projected and the number of peroxisomes per cell was determined. Standard deviation was calculated from 3 independent experiments. In (A and C), all of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar: 2 μm. (D) GFP was tagged at the C terminus of the PEX14 gene on the genome of wild-type (TKYM67), dnm1∆ (KDM1103), fis1∆ (KDM1104), vps1∆ (KDM1105), and dnm1∆ vps1∆ (XLY059) cells. These cells were cultured in YTO to induce peroxisome proliferation and shifted to SD-N for 1 and 2 h. Immunoblotting was done with anti-YFP antibody. The extent of pexophagy was estimated by calculating the amount of free GFP compared with the total amount of intact Pex14-GFP and free GFP.
The division of peroxisomes in budding yeast is mediated by 2 dynamin-related GTPases, Dnm1 and Vps1. Dnm1 is able to constrict membrane, and its proper function and localization on the peroxisomes requires a fission complex, which also includes Fis1, Mdv1, and Caf4.Citation13 Fis1 is a tail-anchored membrane receptor that interacts with the partially redundant proteins Mdv1 and Caf4, which in turn bind to Dnm1, to recruit the latter to the peroxisome. This fission complex is shared with mitochondria, and promotes mitochondrial fission through the same mechanism of Dnm1 recruitment.Citation2,Citation14 Vps1 also controls the segregation of peroxisomes, and is recruited to the organelle through interaction with Pex19.Citation15
In order to detect and quantify the number of peroxisomes during pexophagy, we used a plasmid containing a green fluorescent protein with the C-terminal type I peroxisomal targeting signal (GFP-SKL) and CellTracker Blue CMAC dye to mark the peroxisomes and vacuole lumen, respectively. The atg1∆ mutant served as a control, since autophagy and peroxisomal degradation are completely absent in the atg1∆ mutant. When the yeast cells were grown in YTO medium, wild-type and atg1∆ mutant cells had approximately 11.5 and 12.3 peroxisomes on average per cell, respectively; however, there were fewer peroxisomes in fis1∆ (9.2/cell) and dnm1∆ (8.3/cell) mutants (), which is consistent with the previous report that the replication of peroxisomes is compromised in these mutants.Citation16 In vps1∆ and dnm1∆ vps1∆ mutants, the peroxisomes were highly clustered making it difficult to differentiate and quantify individual peroxisomes (). After the wild-type cells were starved in SD-N medium for 7 h, peroxisomes were degraded in the vacuole; however, the fluorescent signal from GFP-SKL was relatively stable and was diffuse in the vacuolar lumen. At the same time, the number of peroxisomes in the cytosol was largely decreased (corresponding to 56.4% turnover). In fis1∆ and dnm1∆ mutants, a vacuolar GFP signal was also detected, but a reduced number of peroxisomes (39.1% and 35.3%, respectively) were degraded (), which indicated that pexophagy was partially blocked in these 2 mutants. In atg1∆, vps1∆ and dnm1∆ vps1∆ mutants, no vacuolar GFP was detected, which implied that pexophagy was largely defective in these mutants (). However, as noted above, peroxisomes were largely clustered in the vps1∆ and dnm1∆ vps1∆ mutants (), making the quantification of peroxisomal degradation difficult.
To confirm the pexophagy defects in the fission mutants, and to more precisely evaluate the pexophagy activities in vps1∆ and dnm1∆ vps1∆ mutants, we took advantage of a second method for monitoring pexophagy, the Pex14-GFP processing assay. PEX14 encodes a peroxisomal integral membrane protein, and a chromosomally tagged version with GFP at the C terminus is correctly localized on this organelle. When pexophagy is induced, peroxisomes, along with Pex14-GFP, are delivered into the vacuole for degradation. Pex14 is proteolytically degraded, whereas the GFP moiety is relatively stable and accumulates in the vacuolar lumen. Thus, pexophagy can be monitored based on the amount of free GFP by immunoblot.Citation17 After 1 or 2 h of nitrogen starvation with glucose, a considerable amount of free GFP was detected in wild-type cells (); however, free GFP was hardly detectable in atg1∆ cells (Fig. S1). The amount of free GFP was reduced in fis1∆, and dnm1∆ mutants, and dramatically reduced in the vps1∆, and dnm1∆ vps1∆ mutants (). These results were consistent with a previous report and suggested that both Dnm1- and Vps1-dependent peroxisomal fission are important for pexophagy, with Vps1 playing a more significant role.Citation18
Atg11 interacts with Dnm1 and Fis1 on the degrading peroxisomes
In growing yeast cells, Atg11 is mostly diffuse in the cytosol; its translocation to various organelles depends on the presence of different receptor proteins. Our recent studies showed that upon translocation to mitochondria through binding to Atg32, Atg11 recruits the Dnm1-containing fission complex to these mitochondria that are being targeted for degradation by mitophagy.Citation11 The Atg11-bound population of Dnm1, which is specific for fission associated with mitophagy, is different from the previously characterized Atg11-free Dnm1, which controls homeostatic mitochondrial division. Pexophagy and mitophagy share the use of the Atg11 scaffold and the Dnm1-containing fission complex. Accordingly, we speculated that Atg11 is also able to recruit the Dnm1-containing fission complex to peroxisomes to facilitate their division prior to pexophagy.
To test this hypothesis, we used the bimolecular fluorescence complementation (BiFC) assay to determine where the Atg11-Dnm1 interaction occurs. Briefly, in the BiFC assay, the Venus yellow fluorescent protein (vYFP) is split into 2 fragments, VN (N terminus of vYFP) and VC (C terminus of vYFP).Citation19 We fused VN to Atg11 on the genome and transformed cells with a plasmid containing Dnm1-VC. Fluorescence from these 2 chimeras can only be detected when the 2 proteins interact and bring the 2 fragments of vYFP proximal to each other. Thus, for example, when mitophagy is induced, VN-Atg11 and Dnm1-VC form vYFP puncta on the mitochondrial network.Citation11 We performed the same strategy and also introduced a C-terminal mCherry at the PEX14 locus on the chromosome to be able to monitor peroxisomes. When we starved the yeast cells to induce pexophagy, the Atg11-Dnm1 interacting puncta colocalized with Pex14-mCherry (, arrowheads), which indicated that Atg11 recruited Dnm1 to the degrading peroxisomes when pexophagy was induced. To exclude the possibility that the YFP signal detected was due to the overexpression of VN-Atg11 under the control of the ribosomal protein promoter (RPL7Bp), we generated an RPL7Bp-driven VN strain and transformed cells with a plasmid containing Dnm1-VC. No YFP fluorescence was detected in this control experiment, which suggested the YFP puncta represented Atg11-Dnm1 interaction (Fig. S2A).
Figure 2. Atg11 recruits the Dnm1 fission complex to peroxisomes. (A) VN-ATG11 PEX14-mCherry (Pex14-mChe; XLY070) cells, transformed with pDnm1-VC, were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 1 h. The arrowheads mark colocalizing BiFC and mCherry puncta. (B and D) The plasmids pBFP-SKL and pDnm1-VC were transformed into the VN-ATG11 (KDM1535) cells (B), and pBFP-SKL was transformed into VN-ATG11 VC-FIS1 (KDM1545) cells (D). The arrows mark BiFC puncta colocalizing with MitoTracker Red, arrowheads denote BiFC puncta colocalizing with peroxisomes that are proximal to mitochondria, and asterisks indicate BiFC puncta colocalizing with both peroxisomes and mitochondria. Cells were cultured as described in the Materials and Methods to induce peroxisome proliferation and shifted to SD-N for 1 h. MitoTracker Red was used to stain mitochondria. (C and E) Quantification of the localization of Atg11-Dnm1 interacting puncta (B) and Atg11-Fis1 interacting puncta (D); percentage was calculated based on the number of puncta at a specific location compared with the total number of puncta. In (A, B, and D), all of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar: 2 μm.
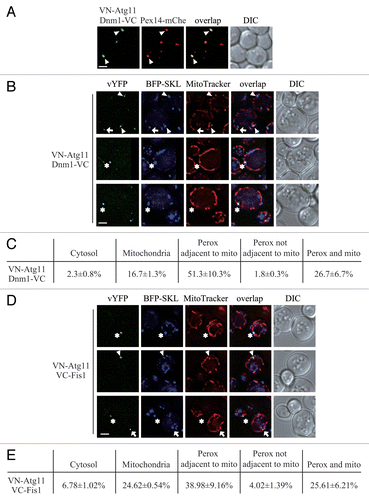
Atg11 and the Dnm1-containing fission complex are shared by peroxisomes and mitochondria. To carefully analyze the location of the Atg11-Dnm1 interaction, we transformed a plasmid harboring BFP-SKL to display peroxisomes and stained the mitochondria with MitoTracker Red dye. After pexophagy was induced by nitrogen starvation with glucose, the localization of Atg11-Dnm1 interacting puncta could be classified into 3 major different classes. The class I puncta appeared on the mitochondrial network (, arrow; ), and thus correspond to complexes that are involved in mitophagy-specific fission.Citation11 Mitophagy is highly induced when yeast are grown solely in the presence of a nonfermentable carbon source and then switched to nitrogen starvation medium containing a fermentable carbon source.Citation20 Because oleic acid is a nonfermentable carbon source, these culture conditions (i.e., switching from YTO to SD-N) likely induce mitophagy. Class II puncta were localized on peroxisomes (, arrowhead; ), and are presumed to represent the fission complexes that mediate pexophagy-specific fission. Unexpectedly, these puncta were found to be mostly proximal to mitochondria. The class III puncta colocalized with both peroxisomes and mitochondria (, asterisk; ). At the same time, we also found that a small number of puncta localized in the cytosol or on peroxisomes that were not adjacent to mitochondria (). The cellular pattern of both class II and class III puncta implied that the Dnm1-mediated peroxisomal fission might occur at the mitochondria-peroxisome contact sites.
To test this hypothesis, we also examined the sites of the Atg11-Fis1 interaction. Based on our previous work, Atg11 interacts with Fis1 in addition to Dnm1 on the targeted mitochondria.Citation11 We chromosomally tagged ATG11 and FIS1 with VN and VC, respectively, and used BFP-SKL and MitoTracker Red to track peroxisomes and mitochondria, respectively. The distribution of Atg11-Fis1 puncta was extremely similar to that of Atg11-Dnm1 puncta, with the existence of distinct populations localized to mitochondria (, arrow; ), peroxisomes that were proximal to mitochondria (, arrowhead; ), and puncta localized on both organelles (, asterisks; ). Of note, the peroxisomes marked by the Atg11-Fis1 interaction were primarily close to or associated with mitochondria. We also performed a control experiment to make sure that the YFP signal detected was due to Atg11-Fis1 interaction; indeed, we could only detect a BiFC signal when both halves of vYFP were fused to either Atg11 or Fis1 (Fig. S2B). After the analysis of both Atg11-Dnm1 and Atg11-Fis1 puncta, we concluded that Atg11 recruited Dnm1-containing fission complexes to peroxisomes being targeted for degradation, and this fission event happened at mitochondria-peroxisome contact sites. We propose that this distinct localization might be due to the fact that mitochondria and peroxisomes share the Dnm1-containing fission complex.
Atg11 interacts with Vps1 on peroxisomes
We noticed that compared with Dnm1, inhibition of the Vps1-mediated peroxisomal fission had a stronger effect on pexophagy (). Therefore, we wondered if there is also an Atg11-bound form of Vps1, which localizes on the peroxisomes that will become targeted for sequestration and regulates pexophagy-specific division. We chromosomally tagged ATG11 with VN and PEX14 with mCherry, and transformed the cells with a plasmid expressing Vps1-VC. VN-Atg11 and Vps1-VC formed puncta in pexophagy-inducing conditions, and these puncta localized on the peroxisomes marked by Pex14-mCherry (, arrowhead). At the same time these puncta were not due to the overexpression of Vps1-VC (Fig. S3A).
Figure 3. Atg11 recruits Vps1 to peroxisomes that are targeted for degradation. (A) VN-ATG11 PEX14-mCherry (Pex14-mChe; XLY070) cells, transformed with pVps1-VC, were cultured in YTO to induce peroxisome proliferation and subsequently shifted to SD-N for 1 h. (B) The plasmid pCuHA-Atg11 was transformed into atg11∆ (untagged, YTS147), atg11∆ VPS1-PA (KDM1269), atg11∆ DNM1(C30∆)-PA (KDM1249), and atg11∆ VPS1(C30∆)-PA (XLY073) cells. The cells were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 2 h. Cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation. The eluted proteins were separated by SDS-PAGE and detected with monoclonal anti-HA antibody and an antibody that binds to PA. (C) The plasmids pBFP-SKL and pVps1-VC were transformed into VN-ATG11 (KDM1535) cells. Cells were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 1 h. MitoTracker Red was used to stain mitochondria. (D) Quantification of the localization of Atg11-Vps1 interacting puncta in (C); the percentage was calculated based on the number of puncta at a specific location compared with the total number of puncta. In (A and C) all of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar: 2 μm.
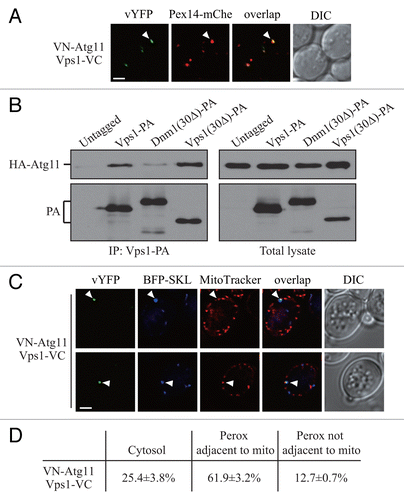
To verify the interaction between Atg11 and Vps1, we performed protein A affinity isolation with IgG-Sepharose. In our previous work, we showed that the Dnm1 mutant lacking the last 30 amino acids [Dnm1(30Δ)] is unable to interact with Atg11. The C terminus of Vps1 has a high similarity at the amino acid sequence level compared with Dnm1 (Fig. S3B). We suspected that the Vps1 mutant lacking the last 30 amino acids [Vps1(30Δ)] would also lose interaction with Atg11. Protein A (PA)-tagged wild-type Vps1 and Dnm1 co-precipitated a substantial amount of the available HA-Atg11, whereas Dnm1(30Δ) precipitated a very small amount of this protein (; Fig. S3C). In contrast to our expectation, Vps1(30Δ) precipitated a similar level of HA-Atg11 relative to that affinity isolated by wild-type (i.e., full-length) Vps1 (). Therefore, Vps1 appears to interact with Atg11 through a different mechanism/binding motif from the one used by Dnm1.
We showed that the Dnm1-regulated peroxisomal division occurred extremely close to mitochondria (), which we propose may be due to the fact that the Dnm1-containing fission complex is shared by mitochondria and peroxisomes. Up until now, there have not been any published data indicating a role for Vps1 in mitochondria segregation. Accordingly, we decided to determine whether Vps1-mediated peroxisomal fission, in contrast to that driven by Dnm1, occurred at a site that was distal from mitochondria. We transformed cells with the BFP-SKL plasmid and used MitoTracker Red to allow us to monitor both peroxisomes and mitochondria, respectively. Puncta corresponding to the sites of Atg11-Vps1 interaction did not appear to localize directly on the mitochondrial network; however, the majority of peroxisomes marked by these puncta were still closely associated with mitochondria (, arrowhead; ). This result suggested that pexophagy-specific fission, whether mediated by Dnm1 or Vps1, might require the participation of mitochondria.
Atg36 interacts with Dnm1 and Vps1
Atg32 and Atg36 are receptors that bind to Atg11 during mitophagy and pexophagy, respectively; however, their properties vary in several respects. For example, overexpression of Atg36 results in a high level of basal pexophagy independent of nitrogen starvation, whereas overexpression of Atg32 does not induce mitophagy in nutrient-rich conditions.Citation10,Citation21 Atg32 is localized on the mitochondrial outer membrane and interacts with the Dnm1-containing fission complex through Atg11. In contrast, Atg36 is a cytosolic protein and is targeted to peroxisomes through binding to Pex3,Citation10,Citation21 which raises the possibility that Atg36 is also able to interact with Dnm1 or Vps1, which are also cytosolic proteins, to recruit the fission complex to peroxisomes; this would be in contrast to Atg32, which binds Dnm1 indirectly through Atg11.
To test this hypothesis, and to gain further insight into the mechanism of Vps1-mediated pexophagy, we generated a series of strains expressing VN-Atg36 combined with Dnm1-VC, VC-Fis1, Mdv1-VC, Caf4-VC, or Vps1-VC, and performed the BiFC assay to search for the possible interactions of these chimeric pairs. VN-Atg36 formed vYFP puncta with Dnm1-VC, or Vps1-VC, but not with the other fusion proteins (). Therefore, unlike Atg32, Atg36 interacts with the 2 dynamin-related GTPases Dnm1 and Vps1.
Figure 4. Atg36 interacts with both Dnm1 and Vps1 on the degrading peroxisomes. (A) The plasmid pDnm1-VC or pVps1-VC was transformed into VN-ATG36 (XLY063) cells. These cells together with cells from strains VN-ATG36 VC-FIS1 (XLY064), VN-ATG36 CAF4-VC (XLY065), and VN-ATG36 MDV1-VC (XLY066) were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 1 h. (B and D) The plasmid pDnm1-VC was transformed into ATG36-VN pRS405-BFP-SKL (XLY072) cells (B), and pVps1-VC was transformed into ATG36-VN pRS405-BFP-SKL (XLY072) cells (D). Cells were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 1 h. MitoTracker Red was used to stain mitochondria. (C and E) Quantification of the localization of Atg36-Dnm1 interacting puncta in (B) and Atg36-Vps1 interacting puncta in (D); the percentage was calculated based on the number of puncta at a specific location compared with the total number of puncta. In (A, B, and D), all of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar: 2 μm.
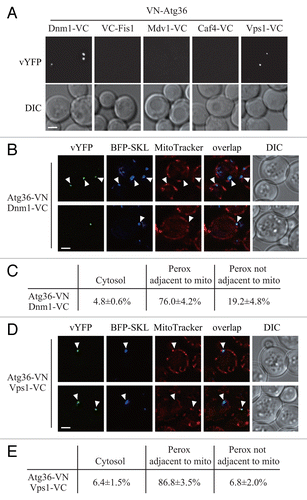
To determine where Atg36 interacts with Dnm1 and Vps1, we used BFP-SKL and MitoTracker Red to track peroxisomes and mitochondria, respectively. Both Atg36-Dnm1 and Atg36-Vps1 interacting puncta were largely localized on peroxisomes extremely close to mitochondria (, arrowheads; ). This location was similar to that seen for Atg11-Dnm1, Atg11-Fis1, and Atg11-Vps1, which reinforced our finding that the pexophagy-specific fission happened at the mitochondrial periphery.
The phosphorylation of Atg36 is important for its interaction with Atg11 and Atg8; specifically, phosphorylation of serine 31 is required for interaction with Atg8, and phosphorylation of serine 97 is required for interaction with Atg11.Citation22 Therefore, we tested whether phosphorylation of Atg36 is required for interaction with Dnm1 and Vps1 by using the yeast 2-hybrid assay. BD-Atg36 was able to interact with both AD-Dnm1 and AD-Vps1 (). BD-Atg36 (S31A), which was reported to lose interaction with Atg8, still interacted with both AD-Dnm1 and AD-Vps1. Similarly, BD-Atg36 (S97A), which was reported to lose interaction with Atg11, interacted with both AD-Dnm1 and AD-Vps1. These results suggested that the Atg36-Atg11 interaction is not required for Atg36-Dnm1 and Atg36-Vps1 interactions. Therefore, Atg36 might be able to recruit Dnm1 and Vps1 to the peroxisomes independent of Atg11.
Figure 5. Overexpression of Vps1 induces pexophagy. (A) Yeast 2-hybrid analysis of Atg36 and the indicated mutants interacting with Dnm1 and Vps1. (B and C) GFP was tagged at the C terminus of the PEX14 gene in the genome of wild-type (TKYM67), GAL1p-3HA-ATG36 (XLY088), and GAL1p-3HA-VPS1 (XLY089) cells. These cells were cultured in YTO to induce peroxisome proliferation and shifted to YPG for 3, 6, and 9 h. Immunoblotting was done with anti-HA antibody in (B) to monitor protein expression following galactose induction, and with anti-HA or anti-YFP antibody in (C).
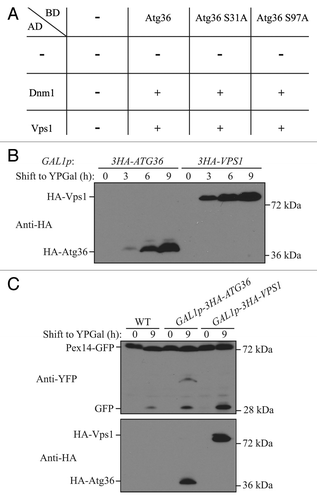
Overexpression of Atg36 enhances pexophagy without nitrogen starvation.Citation10 We showed that Atg36 interacted with Vps1, which raised a possibility that overexpression of Atg36 increased the level of Vps1 recruitment to the peroxisomes. If this is the case, we would expect that overexpression of Vps1 might enhance pexophagy independent of nitrogen starvation. We constructed 2 yeast strains containing either HA-Atg36 or HA-Vps1 under the control of the GAL1 promoter, and used Pex14-GFP processing to detect pexophagy. Yeast cells were cultured in YTO, and shifted to nutrient-rich medium with galactose (YPG). We observed a robust increase of the protein level of HA-Atg36 and HA-Vps1 by 6 h of galactose induction (). When we monitored pexophagy, we detected a clear increase of free GFP after culturing the cells for 9 h in YPG when Atg36 was overexpressed, which agreed with the previous report;Citation10 however, the pexophagy level was even higher when Vps1 was overexpressed (). These results suggested that overexpression of Atg36 might accelerate peroxisomal fission due to its direct interaction with Vps1.
Discussion
The degradation of organelles is an energy- and time-consuming process in eukaryotic cells. In addition, the size of the organelle can present steric challenges, and smaller fragments of organelles are presumably easier for sequestering. Autophagy is the primary mechanism responsible for the bulk degradation of cytoplasmic components and the selective removal of organelles. In nonselective autophagy, the protein level of Atg8 controls the size of the autophagosomes.Citation23 In contrast, during selective types of autophagy the phagophore membrane is closely apposed to the cargo, excluding bulk cytoplasm. Therefore, the factors that determine the curvature of the phagophore and the ultimate size of the autophagosomes might be somewhat distinct between these 2 modes of sequestration. The cytoplasm-to-vacuole targeting (Cvt) pathway is a biosynthetic route that delivers resident hydrolases to the vacuole. Overexpression of the primary cargo of the Cvt pathway, precursor aminopeptidase I, results in the formation of larger complexes, which are sequestered less efficiently.Citation24 Mitochondria exist as a highly extended, reticular structure, which is even more difficult for sequestering within phagophores. Our recent work suggested that during mitophagy a selective and specific fission event occurs on the mitochondria that will become substrates for degradation.Citation11 Although the peroxisomes may be dispersed in the cytosol as individual compartments, the size of this organelle appears to be close to the limit for sequestration by smaller phagophores, such as those generated in the absence of Atg17.Citation25 This may be a particular problem after these organelles proliferate following growth in an oleic acid-containing medium. Therefore, we hypothesized that peroxisomes have to divide prior to degradation, and that only the small fragments of the peroxisomes would be targeted by autophagy.
Here, we showed that deletion of the DNM1 and VPS1 genes, which encode 2 dynamin-related GTPases, resulted in substantially lower efficiency of peroxisomal degradation. Previous work indicated that even though the amino acid sequences of Dnm1 and Vps1 are quite similar, deletion of either individual gene has different effects on peroxisomal division. Deletion of VPS1 dramatically abolishes peroxisomal fission when yeast cells are cultured in both glucose and oleic acid media. However, deletion of DNM1 affects the division of peroxisomes only in oleic acid, and the defect is not as strong as that of the vps1∆ mutant.Citation16 Consistent with these results, pexophagy was significantly blocked in a vps1∆ strain, whereas there was only an intermediate effect in the dnm1∆ mutant (). Overall, it is clear that peroxisomal fission, whether mediated by Dnm1 or Vps1, is important for pexophagy, which is in agreement with our hypothesis. When we were preparing our manuscript, Manivannan et al. published a paper showing that Dnm1 and Vps1 are important for the removal of intra-peroxisomal protein aggregates and pexophagy in Hansenula polymorpha and Saccharomyces cerevisiae.Citation18 Here, we extend that analysis by providing evidence indicating how the fission machinery is recruited to the peroxisomes to facilitate peoxphagy.
As a scaffold protein, Atg11 binds to a variety of cargo receptors to mediate different types of selective autophagy. For example, Atg11 binds to Atg19, to Atg32, and to Atg36 for cargo selection during the Cvt pathway, mitophagy, and pexophagy, respectively.Citation10,Citation12,Citation21,Citation26,Citation27 Atg11 also interacts with Atg1 and Atg17, which connects the step of cargo selection to the initiation of autophagosome formation.Citation27 Here, and with our previous work, we add a new piece to the puzzle: during pexophagy and mitophagy, Atg11 recruits the fission machineries to the organelles and promotes their specific segregation, resulting in smaller fragments of these compartments that can be easily degraded.
It is surprising that pexophagy-specific fission, mediated either by Dnm1 or Vps1, always occurred in proximity to mitochondria. The division of mitochondria requires the participation of the ER,Citation28 and the fission of peroxisomes apparently involves mitochondria. The crosstalk that occurs between organelles is attracting increasing attention. The peroxisome represents an interesting organelle to study in this regard, having connections with the ER, the mitochondria, and the vacuole.
Materials and Methods
Strains, media, and growth conditions
Yeast strains are listed in . Yeast cells were grown in rich (YPD; 1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal (SMD; 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as needed) media. For peroxisome proliferation, cells were grown in YPD to approximately OD600 = 0.5 and shifted to glycerol medium (SGd; 0.67% yeast nitrogen base, 0.1% glucose, and 3% glycerol) for 16 h. The cells were then incubated for 4 h with the addition of yeast extract and peptone into the SGd medium. The cells were ultimately shifted to oleic acid medium (YTO; 0.67% yeast nitrogen base, 0.1% oleic acid, 0.1% Tween 40 and auxotrophic amino acids as needed) for 20 h. Pexophagy was induced by shifting the cells to nitrogen starvation medium containing glucose (SD-N; 0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% glucose). For GAL1 promoter-driven overexpression, the cells were cultured in galactose medium (YPG; 1% yeast extract, 2% peptone, and 2% galactose).
Table 1. List of strains used in this study
Plasmids
pBFP-SKL(405), pCuGFP-SKL(416), pCuHA-Atg11(416), and pDnm1-VC(416) have been reported previously.Citation11,Citation12,Citation21 For constructing pVps1-VC(416), the DNM1 promoter and ORF were removed by XbaI and XmaI from pDnm1-VC(416), and the resulting linearized vector was ligated in the presence of the DNA fragment containing the VPS1 promoter and ORF, which was amplified by PCR from the genome of yeast strain SEY6210 and digested with XbaI and XmaI. For constructing pAD-Dnm1, pAD-Vps1, and pBDU-Atg36, the DNM1, VPS1 and ATG36 ORFs were amplified by PCR from the genome of strain SEY6210. XmaI and BamHI were used to digest DNM1 and ATG36, and EcoRI and PstI to digest VPS1. The digested DNM1 and VPS1 fragments were inserted into pAD-C1, and ATG36 into pBDU-C1.Citation31 pBDU-Atg36 (S31A) and pBDU-Atg36 (S97A) were generated by site-directed mutagenesis from pBDU-Atg36
Fluorescence microscopy
For fluorescence microscopy, yeast cells were grown as described above to induce peroxisome proliferation and shifted to SD-N for nitrogen starvation. Samples were then examined by microscopy (Delta Vision, Applied Precision) using a 100× objective and pictures were captured with a CCD camera (CoolSnap HQ; Photometrics). For each microscopy picture, 12 Z-section images were captured with a 0.3-μm distance between 2 neighboring sections. MitoTracker Red CMXRos (Molecular Probes/Invitrogen, M7512) was used to stain the mitochondria.
Immunoprecipitation
Immunoprecipitation was performed as described previously.Citation21 Immunoprecipitation was performed with monoclonal anti-YFP antibody clone JL-8 (Clontech, 632381), monoclonal anti-HA antibody clone-HA7 (Sigma-Aldrich, H3663), and an antibody that binds to protein A with high affinity (no longer commercially available).
| Abbreviations: | ||
| BFP | = | blue fluorescent protein |
| GFP | = | green fluorescent protein |
| SKL | = | serine-lysine-leucine |
| YTO | = | synthetic yeast medium containing oleic acid |
Additional material
Download Zip (807.4 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NIH grant GM053396 to DJK, and by a University of Michigan Rackham Predoctoral Fellowship to KM.
References
- Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol 2007; 178:399 - 410; http://dx.doi.org/10.1083/jcb.200702167; PMID: 17646399
- Schrader M, Bonekamp NA, Islinger M. Fission and proliferation of peroxisomes. Biochim Biophys Acta 2012; 1822:1343 - 57; http://dx.doi.org/10.1016/j.bbadis.2011.12.014; PMID: 22240198
- Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci 1999; 112:4079 - 87; PMID: 10547367
- Sakai Y, Oku M, van der Klei IJ, Kiel JAKW. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta 2006; 1763:1767 - 75; http://dx.doi.org/10.1016/j.bbamcr.2006.08.023; PMID: 17005271
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102 - 9; http://dx.doi.org/10.1038/ncb1007-1102; PMID: 17909521
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069 - 75; http://dx.doi.org/10.1038/nature06639; PMID: 18305538
- Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 2013; 194:341 - 61; http://dx.doi.org/10.1534/genetics.112.149013; PMID: 23733851
- Mijaljica D, Nazarko TY, Brumell JH, Huang W-P, Komatsu M, Prescott M, Simonsen A, Yamamoto A, Zhang H, Klionsky DJ, et al. Receptor protein complexes are in control of autophagy. Autophagy 2012; 8:1701 - 5; http://dx.doi.org/10.4161/auto.21332; PMID: 22874568
- Farré JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell 2008; 14:365 - 76; http://dx.doi.org/10.1016/j.devcel.2007.12.011; PMID: 18331717
- Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae.. EMBO J 2012; 31:2852 - 68; http://dx.doi.org/10.1038/emboj.2012.151; PMID: 22643220
- Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell 2013; 26:9 - 18; http://dx.doi.org/10.1016/j.devcel.2013.05.024; PMID: 23810512
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA Jr., Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 2001; 153:381 - 96; http://dx.doi.org/10.1083/jcb.153.2.381; PMID: 11309418
- Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci 2008; 121:1633 - 40; http://dx.doi.org/10.1242/jcs.026344; PMID: 18445678
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet 2005; 39:503 - 36; http://dx.doi.org/10.1146/annurev.genet.38.072902.093019; PMID: 16285870
- Vizeacoumar FJ, Vreden WN, Fagarasanu M, Eitzen GA, Aitchison JD, Rachubinski RA. The dynamin-like protein Vps1p of the yeast Saccharomyces cerevisiae associates with peroxisomes in a Pex19p-dependent manner. J Biol Chem 2006; 281:12817 - 23; http://dx.doi.org/10.1074/jbc.M600365200; PMID: 16520372
- Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae.. J Cell Sci 2006; 119:3994 - 4001; http://dx.doi.org/10.1242/jcs.03166; PMID: 16968746
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae.. J Cell Biol 2011; 193:755 - 67; http://dx.doi.org/10.1083/jcb.201102092; PMID: 21576396
- Manivannan S, de Boer R, Veenhuis M, van der Klei IJ. Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events. Autophagy 2013; 9:1044 - 56; http://dx.doi.org/10.4161/auto.24543; PMID: 23614977
- Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae.. Yeast 2007; 24:767 - 75; http://dx.doi.org/10.1002/yea.1504; PMID: 17534848
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem 2008; 283:32386 - 93; http://dx.doi.org/10.1074/jbc.M802403200; PMID: 18818209
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 2009; 17:98 - 109; http://dx.doi.org/10.1016/j.devcel.2009.06.014; PMID: 19619495
- Farré JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep 2013; 14:441 - 9; http://dx.doi.org/10.1038/embor.2013.40; PMID: 23559066
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 2008; 19:3290 - 8; http://dx.doi.org/10.1091/mbc.E07-12-1292; PMID: 18508918
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol 1997; 139:1687 - 95; http://dx.doi.org/10.1083/jcb.139.7.1687; PMID: 9412464
- Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell 2005; 16:3438 - 53; http://dx.doi.org/10.1091/mbc.E04-10-0894; PMID: 15901835
- Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 2002; 3:825 - 37; http://dx.doi.org/10.1016/S1534-5807(02)00373-8; PMID: 12479808
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell 2005; 16:1593 - 605; http://dx.doi.org/10.1091/mbc.E04-11-1035; PMID: 15659643
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science 2011; 334:358 - 62; http://dx.doi.org/10.1126/science.1207385; PMID: 21885730
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol 1988; 8:4936 - 48; PMID: 3062374
- Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, Yip CK, Klionsky DJ. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A 2013; 110:E2875 - 84; http://dx.doi.org/10.1073/pnas.1300064110; PMID: 23858448
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1996; 144:1425 - 36; PMID: 8978031
