Abstract
DIRAS3 is an imprinted tumor suppressor gene that is downregulated in 60% of human ovarian cancers. Re-expression of DIRAS3 at physiological levels inhibits proliferation, decreases motility, induces autophagy, and regulates tumor dormancy. Functional inhibition of autophagy with choroquine in dormant xenografts that express DIRAS3 significantly delays tumor regrowth after DIRAS3 levels are reduced, suggesting that autophagy sustains dormant ovarian cancer cells. This study documents a newly discovered role for DIRAS3 in forming the autophagosome initiation complex (AIC) that contains BECN1, PIK3C3, PIK3R4, ATG14, and DIRAS3. Participation of BECN1 in the AIC is inhibited by binding of BECN1 homodimers to BCL2. DIRAS3 binds BECN1, disrupting BECN1 homodimers and displacing BCL2. Binding of DIRAS3 to BECN1 increases the association of BECN1 with PIK3C3 and ATG14, facilitating AIC activation. Amino acid starvation of cells induces DIRAS3 expression, reduces BECN1-BCL2 interaction and promotes autophagy, whereas DIRAS3 depletion blocks amino acid starvation-induced autophagy. In primary ovarian cancers, punctate expression of DIRAS3, BECN1, and the autophagic biomarker MAP1LC3 are highly correlated (P < 0.0001), underlining the clinical relevance of these mechanistic studies. Punctate expression of DIRAS3 and MAP1LC3 was detected in only 21–23% of primary ovarian cancers but in 81–84% of tumor nodules found on the peritoneal surface at second-look operations following primary chemotherapy. This reflects a 4-fold increase (P < 0.0001) in autophagy between primary disease and post-treatment recurrence. We suggest that DIRAS3 not only regulates the AIC, but induces autophagy in dormant, nutrient-deprived ovarian cancer cells that remain after conventional chemotherapy, facilitating their survival.
Introduction
Autophagy is a highly conserved catabolic process in which organelles and long-lived intracellular proteins are engulfed within double-membrane vesicles, termed autophagosomes. Lysosomes then fuse with autophagosomes, forming autolysosomes.Citation1,Citation2 Depending upon the cellular context, induction of autophagy can either sustain or eliminate metabolically active cancer cells. In the short-term, autophagy can facilitate survival of cancer cells in nutrient-deprived microenvironments with inadequate blood supply. Consistent with a protective role for autophagy, inhibition of autophagic function with chloroquine can induce cancer cell death through necroptosis.Citation3 Persistent autophagy can, however, eliminate cancer cells. Enhanced mammary tumorigenesis has been observed in heterozygous Becn1+/− mice with reduced expression of an essential component required for autophagy.
Formation of autophagosomes is a highly regulated and complex process that includes phases of initiation, nucleation, membrane extension, maturation, and fusion.Citation2,Citation4 Factors regulating formation of autophagosomes in normal and malignant cells are not completely understood. Autophagy is initiated during starvation when MTOR activity is reduced, leading to the formation of a ULK1-ATG13-RB1CC1 complex. Further induction of autophagy depends upon formation of a second autophagosome initiation complex that contains BECN1.Citation5,Citation6 Following nutrient deprivation, BECN1 dimers dissociate from their negative regulator BCL2. BECN1 dimers are then disrupted and monomers bind to PIK3C3, a class III phosphatidylinositol 3-kinase, forming the BECN1-PIK3C3 AIC.Citation5,Citation7 A regulatory protein, ATG14, subsequently associates with and directs the AIC to a phagophore assembly site, activating and initiating the biogenesis of autophagosomes.Citation5,Citation8,Citation9 The mechanisms that mediate dissociation of BECN1 from BCL2, disruption of BECN1 dimers, and formation of BECN1-PIK3C3 AIC have not been defined.
DIRAS3 is widely expressed in human epithelial cells from different organs and is downregulated in carcinomas of the ovary, breast, lung, prostate, thyroid, and pancreas.Citation10-Citation14 While DIRAS3 is highly expressed in normal ovarian surface epithelial cells,Citation13 it is downregulated in > 60% of ovarian cancers and is associated with decreased progression-free survival.Citation13 As an imprinted gene, only one DIRAS3 allele is expressed in normal adult cells. In ovarian and breast cancers, downregulation of the expressed DIRAS3 allele is mediated by multiple mechanisms including loss of heterozygosity, DNA methylation, transcriptional regulation, and shortened mRNA half-life.Citation15-Citation21
Ovarian cancer cell lines have been established with tet-inducible expression of DIRAS3 at physiological levels similar to those found in normal ovarian epithelial cells. Re-expression of DIRAS3 induces autophagy in cultured cells and in xenografts.Citation22 DIRAS3 downregulates the PtdIns3K-AKT-MTOR pathway, facilitating formation of the ULK1-ATG13-RB1CC1 complex. DIRAS3 also induces expression of the ATG4 cysteine protease that cleaves the microtubule-associated protein LC3-I (LC3-I) to form the autophagosome biomarker LC3-II. While re-expression of DIRAS3 at physiological levels in cell culture results in autophagic cell death within 3–4 d, re-expression of DIRAS3 in xenografts induces dormancy with persistent suppression of viable tumor growth. When DIRAS3 is downregulated after 6 wk of expression, dormant xenografts undergo angiogenesis and grow promptly to kill their murine hosts. Treatment of dormant avascular xenografts with chloroquine significantly delays outgrowth of tumors after downregulation of DIRAS3,Citation22 consistent with the possibility that DIRAS3-induced autophagy enhances survival of dormant cells in a nutrient-poor environment.
In this report, we show that DIRAS3 regulates assembly of the AIC in nutrient-deprived ovarian cancer cells in culture. DIRAS3 expression is increased upon nutrient deprivation and colocalizes with BECN1. Interaction of DIRAS3 and BECN1 is required for both starvation-induced and DIRAS3-induced autophagy. DIRAS3 promotes autophagosome biogenesis and triggers vesicle nucleation by binding to BECN1, inhibiting BECN1 dimerization, disrupting BECN1-BCL2 interaction, and promoting assembly of the BECN1-PIK3C3-ATG14 initiation complex.
The clinical relevance of these observations was evaluated by examining the coordinate expression of DIRAS3 and BECN1 with LC3 puncta, which served as a biomarker for autophagy in primary ovarian cancers. The level of autophagy was compared in primary ovarian cancers at initial cytoreductive surgery and in the small, dormant nodules of ovarian cancer removed from the peritoneal surface at “second-look” operations following primary chemotherapy, where nutrient deprivation is likely to occur. In primary cancers, punctate expression of DIRAS3, BECN1, and the autophagic biomarker LC3-II are highly correlated. Punctate expression of DIRAS3 and LC3-II was dramatically upregulated in small nodules of persistent cancer excised during second-look procedures performed in the same patients. These studies suggest that DIRAS3 not only regulates the AIC, but induces autophagy in dormant, nutrient-deprived ovarian cancer cells that remain after conventional chemotherapy and facilitates their survival.
Results
DIRAS3 expression is increased during amino acid starvation-induced autophagy
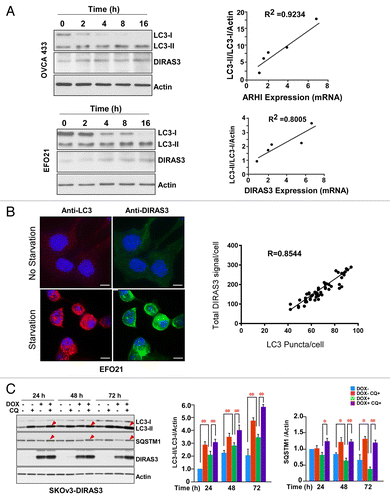
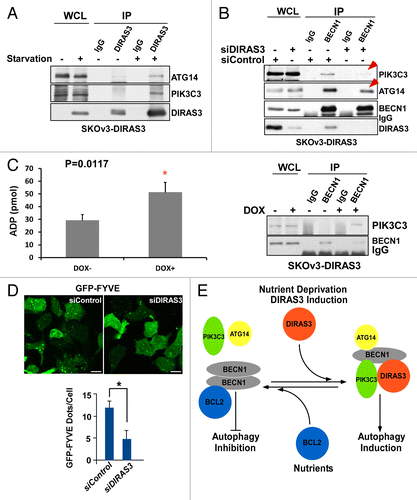
Our previous studies demonstrated that re-expression of DIRAS3 at physiological levels in ovarian cancer cell lines (SKOv3 and HEY) can induce autophagy, associated with an increase in the number of characteristic GFP-LC3 puncta in cultured cells and the formation of typical double-membrane vesicles both in cell culture and in xenografts.Citation22 Others have demonstrated that starvation of ovarian cancer cells can induce LC3 processing and autophagosome formation.Citation23 To examine the role of DIRAS3 in the regulation of amino acid starvation-induced autophagy, we first tested the relationship between endogenous DIRAS3 expression and nutrient deprivation in ovarian cancer cells. Both mRNA and endogenous protein levels of DIRAS3 were significantly increased upon nutrient withdrawal (). To measure the induction of autophagy during amino acid starvation, western blot analysis of cell lysates with antibody against LC3 demonstrated conversion of LC3-I to LC3-II in a time-dependent manner after nutrient withdrawal. Increasing DIRAS3 mRNA expression and protein level correlated with increased conversion of LC3-I to LC3-II in OVCA433 and EFO21 ovarian cancer cells following nutrient deprivation (). Consistent with these results, the amino acid starvation-induced increase in endogenous DIRAS3 protein also correlated with an increase in LC3 puncta in EFO21 cells (). We next examined changes in autophagic flux by comparing the levels of LC3-II in the presence and absence of the lysosome inhibitor chloroquine (CQ). Treatment with CQ significantly increased endogenous LC3-II accumulation after nutrient deprivation. Similarly, in tet-inducible SKOv3-DIRAS3 cells treated with doxycycline (DOX), induced re-expression of DIRAS3 at physiological levels increased LC3-II. Levels of LC3-II were further increased by CQ-mediated inhibition of autolysosome turnover (),Citation22 suggesting that re-expression of DIRAS3 had increased autophagic flux. To test the effect of DIRAS3 on autophagic flux, we measured changes in the levels of SQSTM1/p62, a selective substrate that is degraded in autolysosomes.Citation24 Consistent with the enhanced LC3 turnover, SQSTM1/p62 levels were significantly decreased after DIRAS3 induction (). Moreover, the DIRAS3-induced reduction of SQSTM1/p62 was prevented by CQ, consistent with autophagic degradation of SQSTM1/p62. Collectively, these results demonstrate that DIRAS3 expression is associated with a robust autophagic response upon nutrient deprivation and re-expression of DIRAS3 at physiological levels induces autophagic flux in ovarian cancer cells.
Figure 1. DIRAS3 expression is important for induction of autophagy. (A) Induction of DIRAS3 mRNA is correlated with increased conversion of LC3-I to LC3-II. OVCA433 and EFO21 ovarian cancer cells were incubated in growth medium or HBSS plus 0.3% glucose for 2, 4, 8, or 16 h. Cells were collected for western blotting with antibodies and RNA was extracted for analysis by real-time PCR analyses. Band intensities were quantified using ImageJ. (B) Increasing endogenous DIRAS3 protein is correlated with increasing LC3 puncta. EFO21 cells were incubated in growth medium or in HBSS plus 0.3% glucose for 4 h. Cells were stained for DIRAS3 or LC3 and imaged by immunofluorescence microscopy. The quantities of LC3 puncta and the intensities of endogenous DIRAS3 protein were quantified using ImageJ. Scale bars: 5 μm. (C) Re-expression of DIRAS3 at physiological levels increased the conversion of LC3-I to LC3-II and decreased the level of SQSTM1/p62. SKOv3-DIRAS3 cells were treated with DOX for the indicated intervals to induce DIRAS3 expression and then treated with CQ to block the function of autolysosomes. Band intensity was quantified using ImageJ. Data were obtained from 3 independent experiments. Values are the means ± SD (*P < 0.05; **P < 0.01).
DIRAS3 is required for induction of autophagy by nutrient depletion
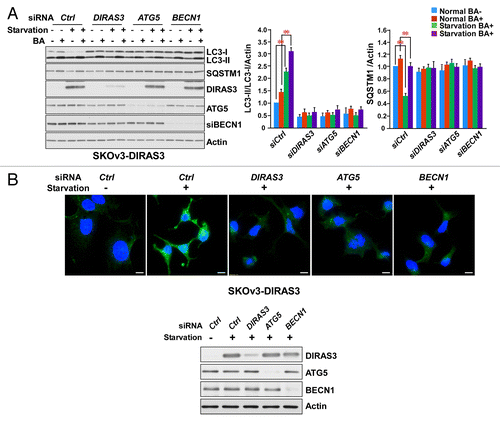
To determine whether DIRAS3 protein is required for the induction of autophagy, we measured the effect of siRNA-mediated DIRAS3 depletion on amino acid starvation-induced autophagy. Knockdown of DIRAS3 in SKOv3-DIRAS3 (), EFO21 (Fig. S1A) and OVCA433 cell lines (Fig. S1B) significantly impaired amino acid starvation-induced conversion of LC3-I to LC3-II (), autophagy-mediated SQSTM1/p62 degradation (), and formation of LC3 puncta (; Fig. S1C), consistent with inhibition of autophagosome formation. Inhibition of autophagy observed after DIRAS3 knockdown was equivalent to that seen upon depletion of 2 known components of the autophagic pathway, ATG5 and BECN1 (). Thus, DIRAS3 is required for the induction of amino acid starvation-induced autophagy.
Figure 2. DIRAS3 expression is required for induction of autophagy. (A) DIRAS3 depletion inhibits LC3 turnover. SKOv3-DIRAS3 cells in growth medium were transfected with siControl, siDIRAS3, siATG5 or siBECN1 for 48 h before incubation in growth medium or HBSS plus 0.3% glucose for 16 h. Cells were then treated with or without 400 nM bafilomycin A1 (BA) for 4 h. Cell lysates were subjected to western blotting. Band intensities were measured and analyzed for the relative accumulation of LC3-II/LC3-I. Data were obtained from 3 independent experiments. Values are the means ± SD (**P < 0.01). (B) DIRAS3 depletion inhibits accumulation of LC3 puncta. SKOv3-DIRAS3 cells were depleted of DIRAS3, ATG5 and BECN1 by siRNA transfection for 48 h before they were switched to growth medium or HBSS plus 0.3% glucose. Left panel, cells were stained for endogenous LC3 and imaged by immunofluorescence. Scale bars: 10 μm. Right panel, the expression of DIRAS3 and BECN1 knocked down with siRNA were analyzed by western blot.
DIRAS3 colocalizes with ATG12 and LC3 during DIRAS3- and amino acid starvation-induced autophagy
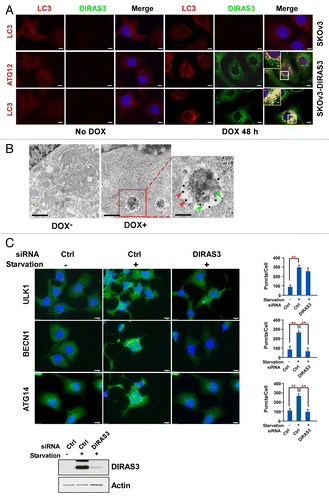
In our previous studies, colocalization of DIRAS3 and GFP-LC3 was observed using immunofluorescence confocal microscopy, and the association of DIRAS3 with LC3 in the autophagosome membrane was confirmed by UV cross-linking.Citation22 To further document the direct participation of DIRAS3 in autophagosome formation, we measured colocalization of DIRAS3, ATG12 and LC3 during DIRAS3-induced autophagy in SKOv3-DIRAS3 cells treated with DOX to induce DIRAS3. ATG12 is required for autophagy and forms a complex with ATG5 and ATG16L1 to form the ATG12–ATG5-ATG16L1 complex that is found within elongating phagophore membranes prior to LC3 lipidation and insertion into the maturing phagophore membrane.Citation25,Citation26 When autophagy was induced in SKOv3-DIRAS3 cells treated with DOX for 48 h, DIRAS3 formed punctate structures that were evenly distributed throughout the cytoplasm (; Fig. S2A). DIRAS3 colocalized with the elongation marker ATG12, and the maturation marker LC3, in cells by 24 h after the addition of DOX but stronger colocalization of DIRAS3 with ATG12 (Pearson r = 0.716988) and LC3 (Pearson r = 0.773758) was observed at 48 h post-induction (; Fig. S2A). Importantly, when autophagy was induced by nutrient deprivation in SKOv3-DIRAS3 cells, colocalization of DIRAS3 puncta with ATG12 puncta was observed within 8 h after nutrient withdrawal (Fig. S2B). By 16 h, DIRAS3 puncta further accumulated, many of which colocalized with LC3 (Fig. S2B). Finally, immuno-gold electron microscopy was performed to verify the colocalization of DIRAS3 with endogenous LC3. After induction of DIRAS3 in SKOv3-DIRAS3 cells for 48 h with DOX, small gold particles conjugated with anti-DIRAS3 immunoglobulin were clustered with large gold particles conjugated with anti-LC3 immunoglobulin on the membrane of autophagosomes (), suggesting direct participation of DIRAS3 with LC3 at the autophagosome. Taken together, our results demonstrate that DIRAS3 participates directly in the formation of autophagosomes.
Figure 3. Expression of DIRAS3 induces colocalization of DIRAS3 with markers of autophagosomes. (A) DIRAS3 colocalizes with ATG12 and LC3 in SKOv3-DIRAS3 cells. SKOv3-DIRAS3 cells and SKOv3 cells were treated with DOX for 48 h. Immunofluorescence staining of DIRAS3 and endogenous ATG12 and LC3 was analyzed by confocal microscopy. Scale bars: 5 μm. Higher magnifications of indicated regions are shown. Scale bars: 1 µm. (B) DIRAS3 colocalizes with LC3. Representative immuno-gold electron microscopy images of induced and uninduced SKOv3-DIRAS3 cells are shown. The green arrows indicate DIRAS3 labeled with small gold particles in the autophagosome vesicles, and the red arrows indicate LC3 labeled with large gold particles. Scale bars: 0.2 μm. Higher magnification of an indicated region is shown. Scale bar: 1 µm. (C) DIRAS3 depletion inhibits accumulation of BECN1 and LC3 puncta, but not ULK1 puncta. SKOv3-DIRAS3 cells in growth medium were transfected with DIRAS3 siRNA 48 h before incubation in growth medium or amino acid starvation medium (HBSS + 3% glucose) for 16 h. Cells were stained for endogenous ULK1, BECN1, and ATG14 and imaged by immunofluorescence microscopy. Fluorescence intensity was quantified using ImageJ. Data were obtained from 3 independent experiments. Values are the means ± SD (**P < 0.01). Scale bars: 5 μm.
DIRAS3 is required for the punctate formation involving ATG14 and BECN1, but not ULK1, during amino acid starvation-induced autophagy
To determine whether DIRAS3 participates in earlier stages of autophagosome formation, we studied the effect of siRNA-mediated DIRAS3 depletion on early stages of amino acid starvation-induced autophagy using immunofluorescent staining with antibodies against ULK1, ATG14, and BECN1. ULK1 is a mammalian ortholog of Atg1, which forms a complex with ATG13 and RB1CC1, and is required for the initial step of autophagosome biogenesis after downregulation of MTOR activity.Citation6,Citation27,Citation28 Activation of ULK1 leads to formation of the BECN1-ATG14-PIK3C3-PIK3R4 complex.Citation29-Citation31 Depletion of DIRAS3, in SKOv3-DIRAS3 cells, did not affect puncta accumulation of ULK1 after amino acid starvation for 16 h (). In marked contrast, DIRAS3 depletion significantly impaired amino acid starvation-induced accumulation of BECN1 and ATG14 puncta (), suggesting that DIRAS3 might regulate autophagy during formation of the BECN1-ATG14-PIK3C3-PIK3R4 complex.
DIRAS3 colocalizes with BECN1 and binds directly to BECN1 protein
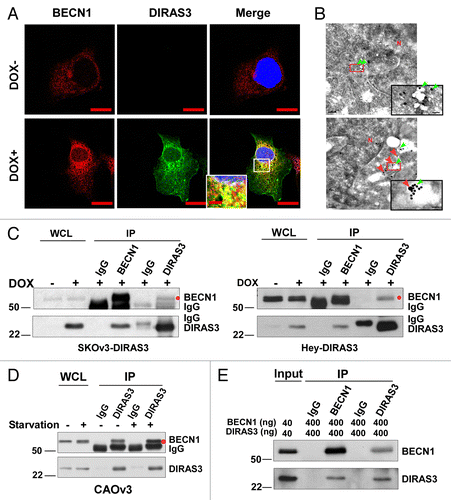
BECN1 is an important component of an autophagy-specific class III PtdIns3K complex (BECN1-ATG14-PIK3C3-PIK3R4 complex) and the activity of this complex coats a cup-shaped phagophore membrane with PtdIns3P, which serves as a signal to recruit additional membrane to form the autophagosome.Citation32 To test whether DIRAS3 is involved in regulation of the BECN1 autophagy initiation complex, we first asked whether DIRAS3 colocalized with endogenous BECN1 by immunofluorescent staining. BECN1 colocalized with DIRAS3 after DIRAS3 induction with DOX (Pearson r = 0.762936) (; Fig. S3A) or amino acid starvation (Pearson r = 0.702286) (Fig. S3B). Furthermore, immunogold electron microscopy analysis of SKOv3-DIRAS3 cells showed that anti-BECN1 immunoglobulin labeled with large gold particles, clustered near anti-DIRAS3 immunoglobulin labeled with small gold particles (). We next examined whether DIRAS3 could interact directly with endogenous BECN1 in ovarian cancer cells. When DIRAS3 was re-expressed at physiological levels in SKOv3-DIRAS3 or Hey-DIRAS3 ovarian cancer cells, DIRAS3 could be coprecipitated with anti-BECN1 antibodies and BECN1 could be coprecipitated with anti-DIRAS3 antibodies in both cell lines (). Interaction of DIRAS3 with BECN1 was also demonstrated in CAOv3 ovarian cancer cells that have high endogenous DIRAS3 levels and high basal autophagy (Fig. S3D and S3E). Thus, robust interaction of DIRAS3 with BECN1 was verified with endogenous levels of these proteins () and, more importantly, this interaction was further enhanced by amino acid starvation (), indicating that DIRAS3-BECN1 interactions are strengthened during nutrient deprivation of ovarian cancer cells. Co-immunoprecipitation of DIRAS3 and BECN1 might not be mediated by direct protein-protein interactions, but rather through BECN1-interacting proteins. To clarify whether DIRAS3 and BECN1 interact directly, co-immunoprecipitation was performed ex vivo with purified recombinant DIRAS3 and using BECN1 proteins from 2 different sources. Purified DIRAS3 could be co-precipitated with purified BECN1 using DIRAS3 antibody and purified BECN1 could be co-precipitated with purified DIRAS3 using anti-BECN1 antibody (; Fig. S3C), indicating direct protein-protein interaction between DIRAS3 and BECN1.
Figure 4. DIRAS3 colocalizes with BECN1 and co-immunoprecipitates with BECN1. (A) DIRAS3 colocalizes with BECN1. SKOv3-DIRAS3 cells were treated with DOX for 24 h. Immunofluorescent staining of expressed DIRAS3 and endogenous BECN1 was analyzed by confocal microscopy. Scale bars: 10 μm. (B) DIRAS3 colocalizes with BECN1. TEM images were obtained after staining with immunogold conjugates of anti-DIRAS3 (red arrow) and anti-BECN1 (green arrow) antibodies in SKOv3-DIRAS3 cells. Scale bars: 0.2 μm. Higher magnifications of indicated regions are shown. Scale bars: 1 µm. (C) DIRAS3 interacts with BECN1. SKOv3-DIRAS3 and Hey-DIRAS3 cells were treated with DOX. Endogenous DIRAS3-BECN1 complexes were immunoprecipitated with anti-BECN1 or anti-DIRAS3 and analyzed for co-immunoprecipitation of DIRAS3-BECN1 conjugates (IP). Host species-matched nonspecific IgG served as negative controls. Whole-cell lysates (WCL) are included for comparison. (D) Amino acid starvation enhances association of DIRAS3 with BECN1. CAOv3 cells were incubated in growth medium or amino-acid starvation medium for 16 h. BECN1-DIRAS3 complexes were immunoprecipitated with anti-DIRAS3 and analyzed for co-immunoprecipitated BECN1. (E) DIRAS3 interacts with BECN1 in vitro. Purified recombinant DIRAS3 and BECN1 proteins were incubated together to allow complex formation. The DIRAS3-BECN1 complexes that formed were immunoprecipitated with anti-BECN1 or anti-DIRAS3 antibodies and analyzed by western blotting.
DIRAS3 inhibits the BECN1-BCL2 interaction
Having demonstrated that DIRAS3 interacts directly with BECN1, we sought to examine the effect of DIRAS3 on the ability of BECN1 to bind BCL2. It is known that BCL2 inhibits autophagy by binding to BECN1, preventing its interaction with PIK3C3 to form the active ‘core complex’.Citation7,Citation33,Citation34 SKOv3-DIRAS3 cells were transfected with a Flag-BCL2 plasmid and treated with DOX to induce DIRAS3 at physiological levels. Induction of DIRAS3 increased formation of the DIRAS3-BECN1 complex while dramatically reducing BECN1-BCL2 interaction, as indicated by the dramatically reduced BCL2 co-immunoprecipitated with anti-BECN1 antibody (). DIRAS3 inhibition of the BECN1-BCL2 interaction was further validated in Hey-DIRAS3 cells (). Therefore, induction of autophagy by DIRAS3 closely correlates with a concomitant decrease in BECN1-BCL2 interaction. To verify that disruption of the BECN1-BCL2 complex is mediated by DIRAS3, we knocked down DIRAS3 and examined the BECN1-BCL2 interaction. As shown in , inhibition of BECN1-BCL2 interaction was completely reversed by siDIRAS3, demonstrating that dissociation between BECN1 and BCL2 is a DIRAS3-dependent event.
Figure 5. DIRAS3 disrupts the BECN1-BCL2 complex by competing for BECN1 binding. (A and B) Expression of DIRAS3 inhibits BECN1 and BCL2 interaction in vivo. SKOv3-DIRAS3 cells and Hey-DIRAS3 cells were transfected with Flag-BCL2 followed by DOX treatment. The BECN1-BCL2 complex was immunoprecipitated with anti-BECN1 and analyzed for co-immunoprecipitation of BECN1-BCL2 interaction. (C) DIRAS3 depletion increases BECN1-BCL2 interaction. SKOv3-DIRAS3 cells were transfected with siDIRAS3 for 48 h to knock down DIRAS3 expression. The BECN1-BCL2 complex was immunoprecipitated with anti-BECN1 and analyzed with the indicated antibodies. (D) Expression of DIRAS3 inhibits BECN1 and BCL2 interaction in a time-dependent manner. SKOv3-DIRAS3 cells were treated with DOX for the indicated intervals. The BECN1-BCL2 complex was immunoprecipitated with anti-BECN1 and analyzed by western blot. (E) DIRAS3 protein competes with BCL2 for BECN1 binding. Increasing amounts of Flag-DIRAS3 (Coomassie Blue staining) was added to BECN1-BCL2 complex and BECN1-interacting proteins were immunoprecipitated with anti-BECN1 and analyzed by western blotting. (F) DIRAS3 is required for amino acid starvation-induced dissociation of BECN1 and BCL2. SKOv3-DIRAS3 cells were depleted of DIRAS3 by incubation with siRNA for 48 h before they were incubated in growth medium or in HBSS plus 0.3% glucose for 16 h. The BECN1-BCL2 and BECN1-DIRAS3 complexes were immunoprecipitated with anti-BECN1 and analyzed by western blotting. (C, siControl; A, siDIRAS3.)
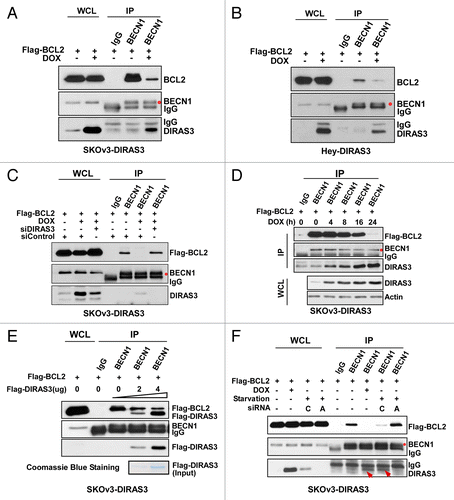
Inhibition of BECN1-BCL2 interaction following DIRAS3 induction is also time-dependent () and correlates with increased DIRAS3 expression. To examine whether this inhibition may be due to competition of DIRAS3 with BCL2 for BECN1 binding, purified DIRAS3 was added to cell lysates from SKOv3-DIRAS3 in the absence of DOX. As increasing amounts of purified DIRAS3 were added, BCL2 was displaced from BECN1 in a dose-dependent manner ().
Under nutrient-rich conditions, BECN1 is complexed with BCL2 in SKOv3-DIRAS3 and Hey-DIRAS3 ovarian cancer cells (; Fig. S4A). Nutrient depletion could, however, produce nearly complete dissociation of this complex, which could be blocked by depletion of DIRAS3 (; Fig. S4A). Taken together, our results indicate that disruption of the BECN1-BCL2 complexes may be one mechanism by which cells promote autophagy and survival during nutrient deprivation. Moreover, the amino acid starvation-induced disruption of the BECN1-BCL2 complex depends upon DIRAS3 expression.
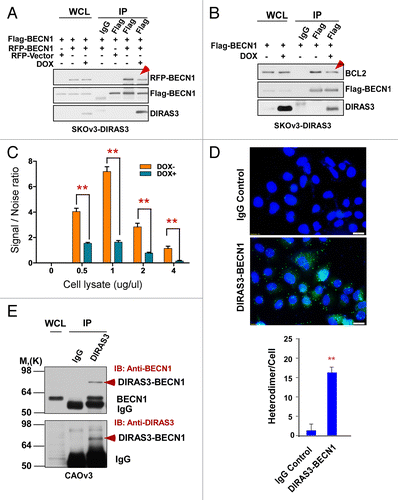
DIRAS3 inhibits BECN1 homodimerization
BECN1 regulates the induction of autophagy by switching from a BCL2-bound BECN1 homodimer to a BECN1 monomer. This monomer binds PIK3C3 and ATG14, upregulating PIK3C3 kinase activity and increasing PtdIns3P levels to induce autophagy.Citation35 In agreement with our previous observation that induction of autophagy by DIRAS3 requires its N-terminal 34 amino acids, expression of NTD in SKOv3-NTD cells did not alter LC3-II.Citation22 To examine whether DIRAS3 mediates dissociation of BCL2 from BECN1 and whether DIRAS3 facilitates disruption of BECN1 homodimers to monomers, we treated SKOv3-DIRAS3, SKOv3-NTD, and Hey-DIRAS3 cells with DOX or diluent to modulate physiological DIRAS3 expression and then transfected treated cells with both Flag-BECN1 and RFP-BECN1. In the absence of DIRAS3 expression, substantial amounts of RFP-BECN1 were detected in the anti-Flag immunoprecipitates, indicating the presence of Flag-BECN1-RFP-BECN1 heterodimers (). Induction of DIRAS3, however, reduced RFP-BECN1 levels in the immunoprecipitates, while not affecting Flag-BECN1 protein levels, indicating that re-expression of DIRAS3 reduced Flag-BECN1-RFP-BECN1 interaction and reduced Flag-BECN1-RFP-BECN1 dimer formation (; Fig. S4B). Concomitant with decreased BECN1 dimer formation, DIRAS3 also reduced the Flag-BECN1-endogenous BCL2 interaction while increasing DIRAS3-BECN1 complex formation (). However, expression of NTD had no inhibitory effects on Flag-BECN1-RFP-BECN1 dimer formation (Fig. S4C), and did not disrupt Flag-BECN1-BCL2 interaction (Fig. S4D) confirming that the DIRAS3 N-terminal 34 amino acids are required for DIRAS3 to induce autophagy. To further document a direct role of DIRAS3 in disrupting BECN1 dimers, the association of BECN1 dimers was measured with LANCE TR-FRET with and without induction of DIRAS3. The LANCE TR-FRET assay measures protein-protein interaction using an assembly of donor and acceptor dye pairs to detect protein binding. SKOv3-DIRAS3 cells were treated with DOX to induce DIRAS3 expression and then transfected with both Flag-BECN1 and HA-BECN1. In the absence of DIRAS3 expression, interactive fluorescence was observed (), whereas in the presence of DIRAS3, fluorescence was markedly decreased (). To demonstrate formation of BECN1-DIRAS3 heterodimers in intact cells, we used the in situ PLA system, which is a proximity ligation assay that enables detection and quantification of protein interactions in fixed cells. Ligation of antibody-conjugated oligonucleotides with a bridging probe in a proximity-dependent manner permits rolling-circle amplification of signals. The product is then detected by complementary fluorescent probes. As shown in , the green dots indicate that BECN1 and DIRAS3 formed heterodimers in CAOv3 cells that have high basal autophagy. Finally, a protein cross-linking experiment was performed with CAOv3 cells and further confirmed the heterodimer formation of DIRAS3 with BECN1 in the anti-DIRAS3 immunoprecipitates (). Taken together, these results indicate that DIRAS3 heterodimerizes with BECN1 and prevents the formation of BECN1 homodimers.
Figure 6. DIRAS3 inhibits BECN1 homo-dimerization. (A) DIRAS3 inhibits Flag-BECN1 and RFP-BECN1 dimerization. SKOv3-DIRAS3 cells were treated with DOX for 24 h and then transfected with Flag-BECN1 and RFP-BECN1 or RFP-Vector. Flag-BECN1-RFP-BECN1 or Flag-BECN1-RFP-Vector was immunoprecipitated with anti-Flag and analyzed by western blotting. (B) DIRAS3 inhibits Flag-BECN1 and endogenous BCL2 interaction. SKOv3-DIRAS3 cells were treated with DOX for 24 h, then transfected with Flag-BECN1. Flag-BECN1-BCL2 complexes were immunoprecipitated with anti-Flag and analyzed by western blotting. (C) DIRAS3 inhibits Flag-BECN1 and HA-BECN1 dimerization. SKOv3-DIRAS3 cells were treated with DOX for 24 h and then transfected with Flag-BECN1 and HA-BECN1. Cell lysates were subjected to LANCE TR-FRET protein-protein interaction analysis. Data were obtained from 2 independent experiments performed in triplicate. Values are the means ± SD (**P < 0.01). (D) BECN1 and DIRAS3 formed heterodimers in CAOv3 cells. BECN1 and DIRAS3 complexes were analyzed with an in situ PLA assay. Data were obtained from 2 independent experiments performed in triplicate. Values are the means ± SD (**P < 0.01). (E) BECN1 and DIRAS3 formed heterodimers in CAOv3 cells. BECN1 and DIRAS3 complexes were immunoprecipitated with anti-DIRAS3 and analyzed by western blotting.
The central region (amino acids/aa 84–200) of GTP-bound DIRAS3 binds to the N terminus (aa 1–144) of BECN1
To delineate the interaction domains between DIRAS3 and BECN1, a series of vectors that express deletions of DIRAS3 or BECN1 were constructed;Citation36-Citation39 Each Myc-tagged DIRAS3 mutant was tested for binding with Flag-BECN1. The DIRAS3 C-terminal deleted DIRAS3 (Myc-CTD), N-terminal deleted DIRAS3 (Myc-NTD), and Myc-DIRAS3 (84-229) were able to bind BECN1, albeit with reduced affinity when compared with wild-type DIRAS3 (; Fig. S5A). However, Myc-DIRAS3(1-84) and Myc-DIRAS3(1-64), were unable to bind BECN1. Thus, the central region of DIRAS3 is critical for interaction with BECN1. Since DIRAS3 exists in an active (GTP-bound) or an inactive (GDP-bound) state, we asked whether its interaction with BECN1 would be affected by binding to GTP. Unlike wild-type Ras, DIRAS3 has very low intrinsic GTPase activityCitation38 and exists predominantly in a constitutively activated state. Previously, we have reported that point mutations that increase DIRAS3′s GTPase activity reduce its tumor suppressor function.Citation38 Here we validated the interaction of DIRAS3 mutant, Myc-M2 (L93A G95Q) (; Fig. S5B) with BECN1 using expression co-immunoprecipitation. Myc-M2 showed significantly reduced co-immunoprecipitation with BECN1 (; Fig. S5B). In addition, the GST-affinity isolation assay confirmed that BECN1 predominately co-immunoprecipitated with the GTP-bound DIRAS3 (Fig. S5D) suggesting that BECN1 interacts preferentially with the active GTP-bound DIRAS3.
Figure 7. The GTP-binding motif of DIRAS3 and N terminus of BECN1 are required for their interaction. (A) The central region of DIRAS3 is critical for interaction with BECN1. Myc-tagged wild type and mutant DIRAS3 were cotransfected with Flag-BECN1 into 293T cells. Interaction between Flag-BECN1 and each DIRAS3 mutant was analyzed by western blotting following immunoprecipitation with anti-Flag antibodies (BECN1), anti-Myc antibodies (DIRAS3) or control IgG and followed by western blotting with anti-Myc or anti-Flag as indicated. (B) BECN1 interacts preferentially with the active, GTP-bound DIRAS3. Wild type and DIRAS3 mutant Myc-M2(K93A G95Q) with point mutations that increased DIRAS3′s GTPase activity were cotransfected with Flag-BECN1 and their interaction analyzed by immunoprecipitation and western blotting. (C) The N-terminal region of BECN1 interacts with DIRAS3. Flag-tagged full-length and deletion mutants of BECN1 were cotransfected with Myc-DIRAS3. Interaction between Myc-DIRAS3 and each BECN1 mutant was analyzed by co-immunoprecipitation. The semiquantitative analysis of the DIRAS3-BECN1 interaction presented in was based on the ratio of anti-Myc (DIRAS3) relative to the amount of Flag-BECN1 that was affinity isolated with the anti-Flag antibody, and the anti-Flag (BECN1) ratio relative to the amount of Myc-DIRAS3 that was affinity isolated with the anti-Myc antibody.
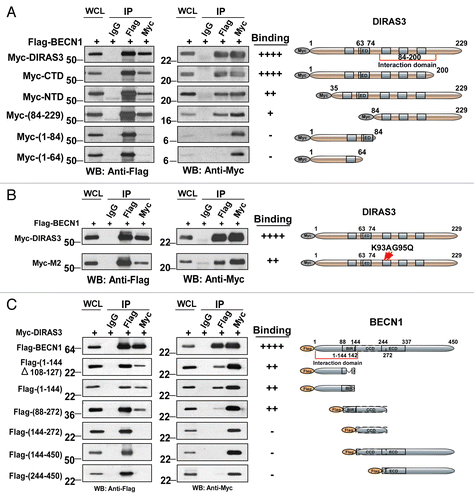
We also generated a series of Flag-tagged BECN1 deletion mutants and examined their interaction with Myc-DIRAS3. The N terminus of BECN1 (aa 1–144), including the BIR domain (aa 88–144) was required for interaction with DIRAS3. However, the N terminus of BECN1 with a deleted BCL2–interacting domain (aa 1–144∆108–127), which did not bind BCL2, did associate with DIRAS3 (; Fig. S5C). These results further suggest that the disruption of BCL2 interaction with the BECN1 BIR domain by DIRAS3 may not be due to direct competition for its physical binding site, but rather through inhibition of BECN1-BECN1 homodimerization by DIRAS3. Inhibition of this homodimerization reduces the BECN1 interaction with its negative regulator BCL2. BECN1’s coiled-coil domain (aa 144–244) and the evolutionarily conserved domain (aa 244–337) were not required for interaction with DIRAS3 (; Fig. S5C). Thus, interaction between DIRAS3 and BECN1 is mediated through the central domain of DIRAS3 with the N-terminal region of BECN1.
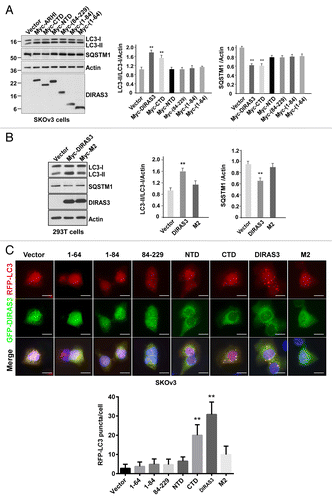
DIRAS3 mutants that fail to bind BECN1 cannot induce autophagy
To examine whether DIRAS3′s ability to induce autophagy is dependent on its interaction with BECN1, wild-type DIRAS3 and DIRAS3 mutants were transfected into SKOv3 () and 293T (Fig. S6) cells and autophagy was assessed by changes in the levels of LC3-II. Increased LC3-II was clearly evident in cells transfected with DIRAS3 and a C-terminal deletion mutant (CTD), but not with other DIRAS3 mutants, including NTD, M2, 84-229, 1-84, and 1-64 (; Fig. S6A). In line with the enhanced levels of LC3-II, SQSTM1/p62 expression levels were significantly decreased in the cells transfected with DIRAS3 and C-terminal deletion mutant (; Fig. S6A). Similar results were observed using RFP-LC3 as a readout for autophagy (). Taken together, our data indicate that the N-terminal 34 amino acids of DIRAS3 and its active GTP-bound state are required for interaction with BECN1 and are critical for DIRAS3′s ability to induce autophagy.
Figure 8. DIRAS3 induces autophagy dependent on its interaction with BECN1. (A and B) DIRAS3 mutants that fail to bind BECN1 cannot induce autophagy in SKOv3 and 293T cells. DIRAS3 and DIRAS3 deletion mutants were transfected into SKOv3 and 293T cells for 24 h. Induction of autophagy was examined by western blotting of LC3 and SQSTM1/p62. Band intensity was quantified using ImageJ. Data were obtained from 3 independent experiments. Values are the means ± SD (**P < 0.01). (C) DIRAS3 and CTD mutants can induce autophagy in SKOv3 cells. GFP-DIRAS3 and GFP-DIRAS3-mutants were cotransfected into SKOv3 cells with RFP-LC3 for 24 h. Cells were analyzed using confocal microscopy and RFP-LC3 puncta were counted. The puncta were quantified using ImageJ. Data were obtained from 3 independent experiments. Values are the means ± SD (**P < 0.01). Scale bars: 5 μm.
DIRAS3 is an integral component of the BECN1-PIK3C3-ATG14 complex
BECN1 interacts with PIK3C3 and ATG14 to induce autophagosome formation and interacts with PIK3C3 and UVRAG to regulate autophagosome maturation and endocytic trafficking.Citation5,Citation8,Citation40 PIK3C3 in association with BECN1 is a biochemical trigger for the initiation of phagophore assembly, elongation, and maturation.Citation41,Citation42 ATG14 in complex with BECN1 and PIK3C3 is required to initiate autophagy.Citation5,Citation8,Citation9 ATG14 and UVRAG bind to BECN1 in a mutually exclusive manner.Citation43,Citation44 To investigate the presence of DIRAS3 in the BECN1-PIK3C3-ATG14 complex, we performed sequential co-immunoprecipitation using cell lysates from 293T cells that were transfected with Flag-BECN1 and Myc-DIRAS3 together with HA-PIK3C3, HA-ATG14 or HA-UVRAG. Cell lysates were first immunoprecipitated with anti-Flag antibodies (BECN1) or control IgG. Proteins eluted with the Flag peptide were then immunoprecipitated with either anti-HA (PIK3C3, ATG14 or UVRAG) or anti-Myc (DIRAS3) antibodies. DIRAS3 was found together with BECN1, PIK3C3, and ATG14 in both sequential immunoprecipitations (), but not in a complex with BECN1 and UVRAG. However, BECN1 was found together with UVRAG and with DIRAS3 independently (), suggesting that DIRAS3 participates in the same complexes with BECN1 and PIK3C3 or ATG14 and is therefore an integral component of the BECN1 initiation “core complex.”
Figure 9. DIRAS3 is an integral component of the BECN1-PIK3C3-ATG14 multiprotein complex. (A) DIRAS3 is in the same complex with BECN1 and PIK3C3. 293T cells were transfected with Flag-BECN1, HA-PIK3C3, and Myc-DIRAS3 or Flag-Vector, HA-Vector and Myc-Vector for 24 h. Cell lysates were immunoprecipitated first with anti-Flag antibodies (BECN1), and the Flag peptide eluates were then immunoprecipitated with anti-HA or anti-Myc. The immunoprecipitates from the first and the second immunoprecipitations were analyzed by western blotting. (B) DIRAS3 is in the same complex with BECN1 and ATG14. Experiments were done as in (A), except HA-ATG14 was used in the co-transfection instead of HA-PIK3C3. (C) DIRAS3 is not in the same complex with BECN1 and UVRAG. Experiments were done as in (A), except HA-UVRAG was used in the co-transfection instead of HA-ATG4. (D) DIRAS3 interacts directly with PIK3C3. SKOv3-DIRAS3 cells were transfected by siBECN1 for 24 h followed by DOX treatment. The DIRAS3-PIK3C3 complex was immunoprecipitated with anti-DIRAS3 and analyzed by western blotting. (E) DIRAS3 interacts with ATG14 through BECN1. SKOv3-DIRAS3 cells were transfected with siBECN1 for 24 h followed by DOX treatment. The DIRAS3-ATG14 complex was immunoprecipitated with anti-DIRAS3 and analyzed by western blotting. (F and G) DIRAS3 increases interaction of BECN1 with ATG14 and PIK3C3. SKOv3-DIRAS3 cells were treated with or without DOX for 24 h. The BECN1-ATG14 or BECN1-PIK3C3 complexes were immunoprecipitated with anti-BECN1 and analyzed by western blotting.
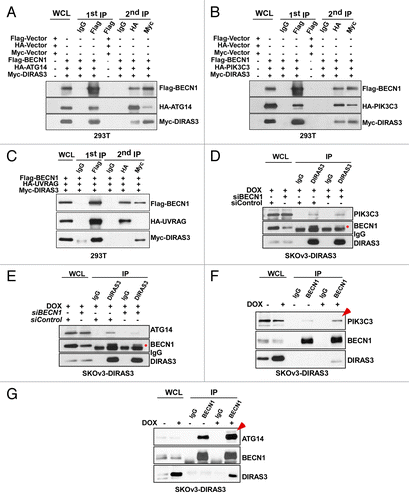
The presence of DIRAS3 in the same immune complex with PIK3C3 and ATG14 may be through direct protein-protein interactions or through association mediated by their mutual interaction with BECN1. Knockdown of BECN1 had no effect on the interaction between DIRAS3 and PIK3C3 (), but significantly reduced DIRAS3 and ATG14 interaction (), suggesting that DIRAS3 might interact directly with PIK3C3, but associates indirectly with ATG14 through BECN1. As knockdown of BECN1 was not complete we cannot, however, exclude the possibility that BECN1 can also mediate DIRAS3 and PIK3C3 interaction.
DIRAS3 enhances BECN1 binding to ATG14-PIK3C3 and increases PIK3C3 kinase activity
We next examined the role of DIRAS3 in forming or stabilizing the BECN1-ATG14-PIK3C3 AIC and the ability of DIRAS3 to regulate PIK3C3 kinase activity. When SKOv3-DIRAS3 cells were treated with DOX to induce DIRAS3 expression, the re-expression of DIRAS3 at physiological levels increased the amount of endogenous PIK3C3 and ATG14 that could be co-immunoprecipitated with anti-BECN1 (). Moreover, amino acid starvation for 16 h substantially increased the association of DIRAS3 with PIK3C3 and ATG14 (). Conversely, knockdown of DIRAS3 reduced the interaction between BECN1, PIK3C3, and ATG14 (). The BECN1-PIK3C3 interaction is associated with increased PIK3C3 kinase activity. Re-expression of DIRAS3 increased PIK3C3 kinase activity (), and knockdown of DIRAS3 reduced the accumulation of GFP-2X-FYVE puncta in SKOv3-DIRAS3 cells treated with DOX to induce DIRAS3 (), or in cells incubated with amino acid starvation medium (Fig. S6B). These results suggest that binding of DIRAS3 to BECN1 can enhance or stabilize the interaction with the BECN1, PIK3C3, and ATG14 core complex, contributing to increased PIK3C3 kinase activity, which is required for the initiation of the autophagosome and the induction of autophagy.
Figure 10. DIRAS3 enhances BECN1-ATG14 and BECN1-PIK3C3 interaction and increases PIK3C3 kinase activity. (A) Amino acid starvation increases interaction of DIRAS3 with ATG14 and PIK3C3. SKOv3-DIRAS3 cells were incubated in growth medium and amino acid starvation medium for 16 h. The DIRAS3-ATG14 or DIRAS3-PIK3C3 complexes were immunoprecipitated with anti-DIRAS3 antibody and analyzed by western blotting. (B) Knockdown of DIRAS3 reduced interaction between BECN1 and ATG14 but not interaction between BECN1 and PIK3C3. SKOv3-DIRAS3 cells were transfected by siDIRAS3 for 24 h. The BECN1-ATG14 and BECN1-PIK3C3 complexes were immunoprecipitated with anti-BECN1 and analyzed by western blotting. (C) Expression of DIRAS3 increases PIK3C3 kinase activity. SKOV3-DIRAS3 cells were treated with DOX to induce DIRAS3 for 24 h. Left panel shows PIK3C3 protein was immunoprecipitated with anti-BECN1 antibody. Right panel shows kinase activity assay. The ADP-GloTM Kinase assay kit was used to measure the kinase activity of the PIK3C3 protein immunoprecipitated by anti-BECN1 antibody. Data were obtained from 2 independent experiments in triplicate. Values are the means ± SD (*P < 0.05). (D) Depletion of DIRAS3 reduced the accumulation of GFP-2X-FYVE puncta. SKOV3-DIRAS3 cells were transfected with siDIRAS3 for 24 h and then transfected with GFP-2x-FYVE for 24 h. GFP-2x-FYVE puncta were quantified using ImageJ. Scale bars: 10 μm. Data were obtained from 2 independent experiments in triplicate. Values are the means ± SD (*P < 0.05). (E) A working model of DIRAS3-dependent regulation of the autophagosome initiation complex.
Expression of DIRAS3 correlates positively with BECN1 and punctate LC3 staining in surgical specimens of ovarian cancer
Our studies in cell culture indicate that nutrient deprivation increases levels of DIRAS3 and that DIRAS3 regulates the induction of autophagy by binding to the BECN1-PIK3C3 AIC. To assess the clinical relevance of these observations, immunohistochemical methods have been developed to measure association of DIRAS3 and BECN1 in formalin-fixed, paraffin-embedded tissues. Staining conditions for the 3 antibodies were optimized on sections from formalin-fixed, paraffin-embedded pellets of DIRAS3-induced and uninduced SKOv3 human ovarian cancer cells that had been treated with or without DOX. Tissue microarray (TMA) samples of primary ovarian cancers were obtained from the Yale Human Investigation Committee (Table S1) and the “second-look” ovarian cancers were from the gynecological oncology tumor bank of Memorial Sloan Kettering Cancer Center (Table S2). Expression of DIRAS3, BECN1, and LC3 was measured by immunohistochemical staining of tissue microarray sections that contained cores from more than 200 primary ovarian cancers. Different staining patterns, reflecting punctate vs. diffuse staining, between DIRAS3, LC3, and BECN1 were recorded for each specimen in the tissue array () and were compared with staining of induced and uninduced SKOv3-DIRAS3 controls (Fig. S7A). Immunohistochemical staining was measured on a scale of 0 to 3 for both total staining intensity and for puncta or diffuse staining, which represents the presence or absence of autophagosome formation. A dot plot shows the distribution of cases with total or punctate staining for DIRAS3, LC3, and BECN1 at different levels of intensity (Fig. S8).
Figure 11. DIRAS3 expression associates with punctate LC3 and BECN1 staining in autophagosomes in ovarian cancers. (A) LC3 punctate staining is associated with robust DIRAS3 and BECN1 punctate staining in an ovarian cancer tissue microarray. Chi squared analysis was used to confirm the positive association between DIRAS3 and LC3 and BECN1 for punctate staining. (B) Representative tissue microarray images show different combinations of DIRAS3, BECN1 and LC3 expression in different ovarian cancers. Patient A: Positive for DIRAS3, BECN1, and LC3 with punctate staining. Lower magnification, scale bars: 25 μm. Higher magnification (inset), scale bars: 10 μm. The red arrows indicate the autophagosome vesicles. Patient B: Negative for DIRAS3, BECN1, and LC3. Patient C: Low DIRAS3 with only diffuse LC3 and BECN1 staining. Patient D: Low BECN1 with diffuse DIRAS3 and LC3 staining.
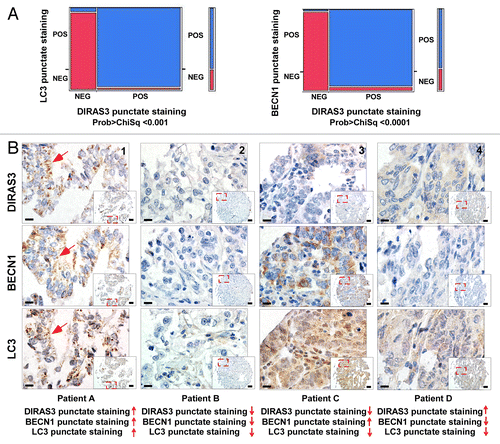
We previously reported that DIRAS3 is downregulated in > 60% of ovarian and breast cancers.Citation13 DIRAS3 expression was measured on a scale of 0 to 3, with 0 to 1 being negative and 2 to 3 being positive. In the current study, in comparison with staining observed in normal ovarian surface epithelial cells (Fig. S7B), total (diffuse and puncta) DIRAS3 staining was observed in 41% of ovarian cancers (n = 223). When LC3 and BECN1 were measured using the same scale, total (diffuse and puncta) LC3 staining was observed in 60% (n = 200) and total (diffuse and puncta) BECN1 staining in 71% (n = 232) of cases. Punctate staining was also measured for DIRAS3, LC3, and BECN1 using the same scale system. Punctate staining for DIRAS3 was observed in 30% (n = 176), LC3 in 41% (n = 163) and BECN1 in 31% (n = 170) of cases.
When the entire scale from 0 to 3 was considered, DIRAS3 correlated positively with LC3 and BECN1 for both total staining (Spearman rho, 0.322, P = 0.0001; 0.442, P = 0.0001, respectively) and for the presence of punctate staining (Spearman rho, 0.797, P = 0.0001; 0.847, P = 0.0001, respectively) (Table S3). Chi squared analysis further confirmed the positive association of DIRAS3 with LC3 and BECN1 for punctate and total staining (; Figs. S9 and S10). When LC3 punctate staining was positive, only 2 cases (1.4%; n = 139) were observed in which DIRAS3 showed only diffuse staining without puncta (Fig. S11). Similarly, we observed one case (0.7%; n = 137) in which DIRAS3 showed only diffuse staining while BECN1 punctate staining was present (Fig. S12). In triple-positive cases (represented by patient A), all 3 markers were detected and punctate staining was prominent, consistent with detection of autophagosome structures (). In triple negative cases (represented by patient B), none of the 3 proteins stained immunohistochemically (). When little or no DIRAS3 (represented by patient C) or BECN1 (represented by patient D) could be detected, only diffuse staining for LC3 was observed, consistent with an absence of autophagosome structure(s) (). The remarkable correlation of punctate staining for LC3 with DIRAS3 and BECN1 suggests that these molecules are colocalized in human ovarian cancers undergoing autophagy and that DIRAS3 and BECN1 are important for inducing autophagy in ovarian cancer.
DIRAS3 expression is increased in second-look specimens of ovarian cancer
Nutrient deprivation is likely to occur in poorly vascularized portions of primary ovarian cancers as well as in small collagen-dense nodules of dormant ovarian cancer that persist on the peritoneal surface after primary treatment. In the past, “second-look” operations have been performed after conventional surgery and chemotherapy to detect persistent disease. Nests of ovarian cancer cells have been found within small, avascular collagenous nodules on the surface of the peritoneal cavity. Even when persistent disease has been detected, a prolonged period of many months to years may elapse before clinical recurrence, consistent with the presence of dormant disease.
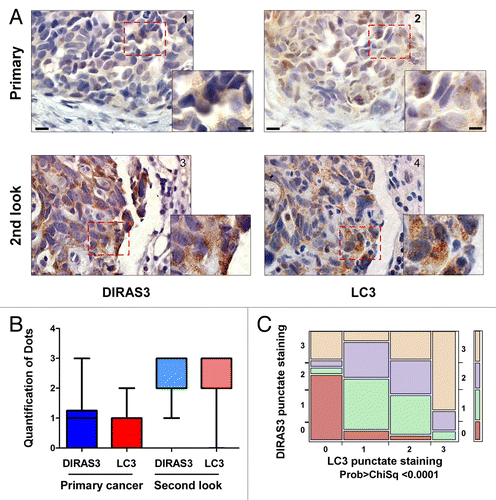
If DIRAS3 were required to induce autophagy, and autophagy facilitates the survival of nutrient-deprived dormant cancer cells, then DIRAS3 expression and punctate LC3 expression should be enhanced in the small deposits of dormant ovarian cancer cells that survive in poorly vascularized scars on the peritoneal cavity. To test this hypothesis, we prepared serial sections from primary ovarian cancers and second-look surgical specimens from the same 34 patients (Table S2). All patients were in a complete clinical remission at the completion of adjuvant chemotherapy defined by normal CA-125 level, normal CT scans of the abdomen and pelvis, and a normal physical exam. When immunohistochemical analysis was performed, DIRAS3-positive staining was observed in 41% of primary cancers and 97% of second-look cases. LC3-positive staining was observed in 35% of primary cancers and 85% of second-look cases. Punctate DIRAS3 staining was observed in 23% of primary cancers and 84% of second-look cases, whereas punctate LC3 staining was observed in 21% of primary cancers and 81% of second-look cases. Positive staining of both total and punctate DIRAS3 and LC3 was significantly higher in second-look specimens than in primary cases (; Figs. S13 and S14). DIRAS3 and LC3 punctate staining correlated positively at a highly significant level with Pearson Chi-square analysis (). Taken together, these data suggest that DIRAS3 is associated with punctate LC3 expression in autophagosomes in human ovarian cancers undergoing autophagy and that DIRAS3 and autophagy are upregulated in dormant ovarian cancer cells that survive in poorly vascularized scars on the peritoneal cavity after conventional surgery and chemotherapy.
Figure 12. DIRAS3 expression associates with punctate LC3 and BECN1 staining in autophagosomes in ovarian cancer second-look samples. (A) Representative primary and second-look images stained with DIRAS3 and LC3. Lower magnification, scale bars: 10 μm. Higher magnification, scale bars: 5 μm. (B) Positive staining of the punctate DIRAS3 and LC3 is significantly higher in second-look than in primary cases. A bar graph of staining was generated by analysis of 34 pairs of primary and second-look ovarian cancer patient tissue samples. (C) DIRAS3 punctate staining correlates significantly with LC3 punctate staining. Contingency analysis of DIRAS3 puncta by LC3 puncta with Pearson Chi-square test (0: negative; 1: +; 2: ++; 3: +++).
Discussion
Our findings document for the first time that DIRAS3 is a member of the BECN1 interactome and can act as a regulatory switch to promote initiation of autophagosome formation (). Re-expression of DIRAS3 at physiological levels induces autophagy, whereas DIRAS3 depletion blocks amino acid starvation-induced autophagy. Under nutrient-poor conditions, DIRAS3 is upregulated, displaces BCL2 from BECN1, disrupts BECN1 dimers, forms DIRAS3-BECN1 heterodimers and promotes interaction of BECN1 monomers with PIK3C3 and ATG14, increasing PIK3C3 kinase activity and ultimately inducing autophagy.
Binding or dissociation of BECN1 and BCL2 is an important mechanism by which autophagy can be regulated in response to diverse stimuli.Citation7 Molecular mechanisms that govern such interaction include posttranslational modifications of BECN1 or BCL2,Citation45 disruption of BECN1-BCL2 interaction by BH3-only proteins,Citation46 and binding of BECN1 by different receptors or their adaptor proteins.Citation47,Citation48 BECN1 forms homodimers via its coiled-coil domain and BCL2 preferentially interacts with the BECN1 dimer.Citation49 Binding of UVRAG disrupts this homodimerization, freeing BECN1 to bind other core complex proteins.Citation49 One member of the initiation core complex is ATG14,Citation5,Citation8,Citation9 and ATG14 and UVRAG are found with the BECN1-PIK3C3 complexes in a mutually exclusive manner.Citation40 Similar to UVRAG, DIRAS3 can also disrupt BECN1 dimers and facilitate BECN1 dissociation from BCL2. In addition, DIRAS3 appears to participate directly in the same complex with BECN1 and PIK3C3 as well as with BECN1 and ATG14. Consequently, our data suggest that there are 2 populations of BECN1 complexes, one involving ATG14 driven by DIRAS3 and a second driven by UVRAG.
Analysis of DIRAS3 mutants identified 3 regions of the DIRAS3 protein that are important for inducing autophagy. First, the N-terminal extension of DIRAS3 is required to induce autophagy.Citation22 By contrast, deletion of the C-terminal domain of DIRAS3 (CTD) did not prevent interaction with BECN1 to induce autophagy, despite the lack of a membrane anchor. The role of this N-terminal extension in inducing autophagy is not fully elucidated. It is, however, intriguing to speculate that the leucine-rich domain within the DIRAS3 N-terminal extension is necessary for intercalation and structural distortion of BECN1’s coiled-coil dimerization domain, which contains the metastable leucine zipper sequences. Because recent structural studies indicate that the BECN1 coiled-coil dimerization domain is anti-parallel,Citation50 the DIRAS3 molecule may associate with the N terminaus of BECN1 on opposite sides of the BECN1 dimer, intercalating into the weak coiled-coil of BECN1 from opposite sides by its N-terminal leucine-rich domain, thus helping to destabilize the BECN1 dimer. Second, the central domain of DIRAS3 is involved in direct protein-protein interaction with BECN1 and is also critical for induction of autophagy. Third, the GTP-bound form of DIRAS3 preferentially interacts with BECN1, suggesting that DIRAS3 induces autophagy in a GTP-dependent manner. A deeper understanding of DIRAS3-BECN1 interaction awaits crystallization and structural analysis of the complex.
TP53 regulates autophagy in a dual fashion whereby the pool of cytoplasmic TP53 protein represses autophagy in a transcription-independent fashion and the pool of nuclear TP53 stimulates autophagy in a transcription-dependent fashion. However, we performed all protein-protein interaction experiments with both SKOv3-DIRAS3 cells (TP53 null) and Hey-DIRAS3 cells (TP53 wild type), suggesting that DIRAS3 had very comparable results in both SKOv3 cells and Hey cells. It is without question that TP53 plays an important role in regulating autophagy in other systems.
A growing body of evidence shows that cancer cells exhibit significant metabolic alterations that promote nutrient uptake, enabling cancer cell proliferation, dysregulation, and autonomy. In nutrient-poor environments within primary cancers or dormant metastases, induction of autophagy may be critical to maintain the viability of metabolically dysfunctional cancer cells. DIRAS3 expression is increased during amino acid starvation and is required for starvation-mediated autophagy in cancer cells, but it is unaltered in normal ovarian epithelial cells (Fig. S15); cancer cells that lack DIRAS3 expression may be incapable of utilizing this mechanism to maintain their viability in nutrient-poor conditions and may be less prone to dormancy. DIRAS3 expression and punctate LC3 expression were, however, enhanced in the small deposits of dormant ovarian cancer cells that survive in poorly vascularized scars on the peritoneal cavity and that are found at second-look surgical surveillance procedures. Our previous studies showed that functional inhibition of autophagy with CQ markedly inhibits growth of dormant, autophagic ovarian cancer cells that express DIRAS3.Citation22 As DIRAS3-associated autophagic cells have been found in 84% of positive second-look specimens, trials of adjuvant therapy with CQ or hydroxychloroquine might be considered as maintenance therapy.
Survival factors provided by the tumor or microenvironment may also be an important factor in determining the fate of dormant autophagic cancer cells. In our previous study, induction of DIRAS3 in SKOv3-DIRAS3 cells in culture led to autophagic death within 4 d, judged by clonogenic assays. Autophagic cells could be rescued in culture by the addition of survival factors found within the xenograft, including VEGFA (vascular endothelial growth factor A), IL8 (interleukin 8) and IGF1 (insulin-like growth factor 1 [somatomedin C]). Treatment with antibodies against these survival factors or inhibition of their receptors might eliminate dormant cancer cells with or without functional inhibition of autophagy using CQ or hydroxychloroquine. Of interest is that maintenance therapy with the anti-VEGFA antibody bevacizumab has prolonged progression-free survival in multiple clinical trials.Citation51-Citation54
While our studies of DIRAS3 have been performed with ovarian cancer, DIRAS3 is downregulated in a fraction of cancers from several other sites, including breast, lung, prostate, thyroid, and pancreas. Consequently, DIRAS3-associated autophagy could contribute to maintaining dormant cancer cells from a significant fraction of cancer patients.
Materials and Methods
Antibodies and recombinant proteins
Antibodies against LC3 (2775), ATG5 (2630), BECN1 (3738), Actin (2367), Flag-tag (2368), Myc-tag (2278), and HA-tag (3724, 2367) were purchased from Cell Signaling Technology, and anti-ATG14 (PD045) was obtained from MBL. Antibodies against Flag-tag (F1804), HA-tag (H9658) and BECN1 (B6061) were purchased from Sigma. Anti-ULK1 (NB110-74844) was obtained from Novus Biologicals. Anti-ATG14 was purchased from Acris antibodies (AP55472PU-IV). Murine monoclonal antibodies against DIRAS3 were generated in our laboratory. Purified recombinant human BECN1 (TP301627) and DIRAS3 (DIRAS3; TP302787) proteins were purchased from OriGene Technologies. Purified GST-BECN1 (human, H00009077-P01) was purchased from Abnova, and BECN1-purified recombinant human GST-DIRAS3 and BECN1 (aa 1–144) were obtained through a collaboration with Dr Choel Kim.
Cell culture
Tet-on inducible SKOv3-DIRAS3 ovarian cancer cells (TP53 null) were grown in McCoy’s medium supplemented with 10% FBS, 200 µg/mL G418 (Fisher Scientific, 61-234-RG) and 0.12 µg/mL puromycin (Sigma, P7266).Citation22 Tet-on inducible Hey-DIRAS3 ovarian cancer cells (TP53 wild type) were cultured in RPMI-1640 medium supplemented with 10% FBS 25 µg/mL blasticidin (InvivoGen, Ant-bl-1) and 1 µg/mL puromycin.Citation22 DIRAS3 expression was induced by adding 1 µg/mL DOX (Sigma, D9891) to the culture medium. In addition to 2 inducible cell lines, we also used Caov3, OVCa433 and EFO21 cells, which have relative higher endogenous DIRAS3 expression by comparison with other ovarian cancer cell lines, to help to demonstrate the physiological relevance of the study. HEK 293T cells and EFO21 ovarian cancer cells were grown in RPMI-1640 medium supplemented with 10% FBS, while OVCA433 and CAOv3 ovarian cancer cells were grown in DMEM medium supplemented with 10% FBS. In previous studies, we have shown that induced levels of DIRAS3 expression closely resemble expression levels found in normal ovarian surface epithelial cells.Citation22,Citation55 To further demonstrate this point, we have compared DIRAS3 expression at the protein level in doxycycline-induced SKOv3-DIRAS3 cells with normal ovarian epithelium tissue by immunohistochemical staining with anti-DIRAS3 antibody (Fig. S1D). During amino acid starvation, cells were incubated with Hanks’ balanced salt solution (HBSS) (Sigma H9269) supplemented with glucose with a final concentration of 3 g/L for the indicated time periods.
Plasmids
Flag-BCL2, HA-PIK3C3, HA-ATG14, Flag-BECN1, RFP- Myc-tagged wild-type, mutant DIRAS3 constructs (Myc-DIRAS3, Myc-CTD, Myc-NTD, Myc-DIRAS3[84-229], Myc-DIRAS3[1-84], Myc-DIRAS3[1-64], Myc-M2[K93A G95Q], and Myc-M3[A46V K93A G95Q]) as well as Flag-tagged wild-type and mutant BECN1 constructs (Flag-BECN1[1-450], Flag-BECN1[1-144∆108-127], Flag-BECN1[1-144], Flag-BECN1[88-272], Flag-BECN1[142-272], Flag-BECN1[144-450], and Flag-BECN1[273-450]) were custom made by Origene.
Transient transfection
Ovarian cancer cells were transfected with Lipofectamine 2000 (Invitrogen, 11668-019) for plasmid DNA or DharmaFECT4 (ThermoScientific, T-2004-02) for siRNA, using manufacturer’s protocols. The predesigned siRNAs were from Dharmacon (ThermoScientific).
RT-PCR
Ovarian cancer cell lines were starved by incubation in Hank's solution supplemented with glucose with a final concentration of 3 g/L. Total RNA was extracted using Trizol reagent (Invitrogen, 15596-026) and reversely transcribed into cDNA from 2 ug total RNA with the SuperScript III kit (Invitrogen, 11904-018). SYBR Green-based real-time PCR reactions were conducted in triplicate in 7900HT Fast Real-Time PCR system (Applied Biosystems) with 20 ng cDNA using iTaq SYBR green supermix with Rox (Bio-Rad, 172-5853). Human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as an endogenous control. Data were analyzed using the delta Ct method. Primers for DIRAS3 are forward (NY2P2) 5′-TCTCTCCGAG CAGCGCA and reverse (3Y2SP2) 5′-CGTCGCCACT CTTGCTGTCG; primers for GAPDH are forward (F45) 5′- TCGACAGTCA GCCGCATCTT CTTT and reverse (R138) 5′- ACCAAATCCG TTGACTCCGA CCTT.
Immunoprecipitation and immunoblotting
SKOv3-DIRAS3 cells were incubated in lysis buffer (50 mM HEPES, pH 7.0, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10 mM NaF, 10 mM sodium pyrophosphate, 10% glycerol, 1% Triton X-100) plus protease and phosphatase inhibitors (1 mM PMSF (Sigma, P7626), 10 µg/mL leupeptin (Sigma, L2884), 10 µg/mL aprotinin (Sigma, A6103) and 1 mM Na3VO4 (Sigma, 450243)). Cells were lysed for 30 min on ice, and then centrifuged at 17,000 × g for 30 min at 4 °C. The protein concentration was assessed using a bicinchoninic acid protein assay (Thermo, 23223/23224). Lysates (0.8–1 mg protein) were diluted with lysis buffer to 1 mL. Immune complexes were incubated overnight with 2 µg of the antibody and precipitated with protein G magnetic beads (Thermo Scientific, 88803) for 60 min. Complexes were washed in lysis buffer (3 × 5 min) and in PBS (phosphate-buffered saline, 3 × 5 min). Immunoprecipitated proteins were separated by SDS-PAGE and transferred to PVDF membranes. Immunoblot analysis was performed with the indicated antibodies and visualized with an ECL-enhanced chemiluminescence detection kit (Perkin Elmer, NEL104001EA). Band intensity from western blots was quantified using the ImageJ program.
Cross-linking assay
To examine heterodimer formation of DIRAS3 with BECN1, dilute formaldehyde solution (0.4%) was used as a cross-linker.Citation56 Cells were incubated with formaldehyde solution with mild agitation for 10 min and quenched with ice-cold 1.25 M glycine in PBS, and then cells were washed with glycine in PBS and solubilized with lysis buffer. Co-immunoprecipitation and western blotting were performed as describe in the figure legends.
GST affinity isolation assay
200 µg of purified GST-DIRAS3 and 200 µg of purified BECN1 (1–144 aa) were added into binding buffer (4.3 mM Na2HPO4, 1.47 mM NaCl, 2.7 mM KCl, pH 7.3 1 mM PMSF) with 1 mM GTPγS or GDP for 4 °C overnight incubation, then exactly 40 μL of protein G (Clontech, 635607) was added for 1 h incubation at 4 °C and the precipitate was washed 3 times with wash buffer (4.3 mM Na2HPO4, 1.47 mM NaCl, 2.7 mM KCl, pH 7.3). The bound proteins were eluted after boiling with a 4× SDS sample buffer and eluates were analyzed by co-immunoprecipitation followed by western blotting.
Immunofluorescence
Cells on coverslips were fixed in 4% formaldehyde in PBS for 10 min, rinsed twice in PBS, and then permeabilized in 100% ethanol for 1 h at -20 °C. Coverslips were rinsed twice in PBS and blocked with 5% BSA (Sigma, A7906) in PBS for 1 h at room temperature followed by incubation with primary antibodies diluted in 1.5% BSA in PBS overnight at 4 °C. Cells were then washed 4 × 5 min in PBS and incubated with secondary antibodies diluted in 1.5% BSA for 1 h at room temperature. Coverslips were then washed 4 × 5 min in PBS and mounted on glass slides with Vectashield fluorescent mounting medium (Vector Lab, H-1500). Goat anti-mouse or goat anti-rabbit secondary antibodies conjugated to Alexa Fluor 488 (A-11017), or 594 (A-11072) were purchased from Invitrogen. Puncta were quantified by ImageJ. Briefly, cells in a region of interest were encircled using the program’s freehand selection tool. Puncta from the images were measured and counted. Dots per cell were calculated by dividing the total number of dots by the total number of cells (at least 30–50 cells) within each sample. Colocalizations of 2 proteins were quantified by ImageJ “Colocalization_Finder” plugin (Christophe Laummonerie; 2006/08/29: Version 1.2).
LANCE time-resolved fluorescence resonance energy transfer (LANCE TR-FRET)
LANCE Eu-labeled anti-HA antibody (donor) and Surelight APC anti-Flag antibody (acceptor) were purchased from PerkinElmer (AD0059F). SKOv3-DIRAS3 cells were treated with DOX to induce DIRAS3 expression, and induced cells were transfected with Flag-BECN1 and HA-BECN1 plasmids. Cells were rinsed with cold PBS and solubilized with lysis buffer after 24 h. The protein concentration was assessed using a bicinchoninic acid protein assay. Cell lysate was incubated with LANCE Eu-labeled anti-HA antibody (2 nM) and Surelight APC anti-Flag antibody (50 nM) in a 96-well plate at room temperature for 1 h. Energy transfer signal was measured with PHERstar FS plate reader (BMG Lab TECH) at both donor (Eu at 615 nm) and acceptor (APC at 665 nm) emissions.
DUOLINK in situ assay
Duolink in situ PLA probes and Duolink in situ detection reagents were purchased from Sigma (DUO92014). BECN1-DIRAS3 heterodimer formation was studied in CAOv3 ovarian cancer cells. Procedures were performed following the manufacturer’s instructions. Briefly, cells were seeded on chamber slides, fixed, blocked, incubated with primary antibodies, and then with secondary antibodies, conjugated with oligonucleotides (PLA probe MINUS and PLA probe PLUS). Finally, cells were incubated sequentially with ligation solution and amplification solution followed by analysis with fluorescence microscopy.
Kinase activity assay
The PIK3C3 assay was performed using the ADP-GloTM Kinase assay kit (Promega, V9101) which quantifies the amount of ADP produced by the PIK3C3 reaction. Briefly, 10 µl of PIK3C3 protein immunoprecipitated by anti-BECN1 antibody from SKOv3-DIRAS3 lysates, 5 µl of PtdIns3K lipid substrate (Life technologies, PV5371) and 5 µl of kinase dilution buffer were added to a pre-cooled 96-well white opaque plate, followed by the addition of 5 µl of 250 µM ATP assay solution. The reaction mixture was sonicated in the 96-well plate for 10 s and the reaction mixture incubated at 30 °C for 15 min. The reaction was terminated by adding 25 µl of ADP-Glo reagent. The plate was then incubated with agitation for 40 min at room temperature. Finally, 50 µl of kinase detection reagent was added to the plate, which was incubated for an additional 30 min, and intensity measured on a plate reader (BioTek Synergy 2). The specific activity (ADP) was calculated based on an ADP standard curve.
Transmission electron microscopy (TEM)
SKOv3-DIRAS3 cells were fixed with a mixture of 3% paraformaldehyde (Electron Microscopy Science, 15720) and 0.15% glutaraldehyde (Electron Microscopy Science, 15720) in PBS for 20 min at room temperature. Cells were then treated with 0.1% sodium borohydride (Sigmam, 213462) in PBS for 15 min and washed with PBS (4 × 10 min) followed by permeabilization with 2.5% Triton X-100 in PBS for 15 min at room temperature. Specimens were then blocked with 5% BSA for 20 min at room temperature. Cells were incubated with 2 different species of antibodies for 24 h at 4 °C, followed by washing with PBS (4 × 10 min). Cells were then incubated with 2 secondary antibodies labeled with 2 different sizes of gold particles that recognize IgG from different species and washed with PBS (4 × 10 min) followed by post fixation with 2.5% glutaraldehyde in PBS for 20 min. Finally, cells were stained with aqueous uranyl acetate and lead citrate before imaging with a Jeol-100 (JEOL) TEM at 80 kV.
Immunohistochemistry
Formalin-fixed, paraffin-embedded ovarian tumors were resected from 429 patients at Yale University/New Haven hospitals between 1981 and 2001. Tissues were obtained from the archives of the Pathology Department, Yale University (New Haven, CT). Specimens and associated clinical information were collected under informed consent under the ethics guidelines and approval of the Yale Human Investigation Committee. Cohort detail characteristics are described in Table S1. Paired samples of paraffin-embedded, formalin-fixed ovarian cancer from patients who had undergone primary and “second-look” operations were obtained from the gynecological oncology tumor bank of Memorial Sloan Kettering Cancer Center with the approval of their Institutional Review Board. All patients underwent a second-look procedure which is a surgical re-evaluation of the peritoneal cavity preformed at the completion of adjuvant chemotherapy for patients in clinical remission defined by normal CA-125 level, normal CT scan of the abdomen and pelvis, and a normal physical exam. Clinical information regarding the patients donating tissues are described in Table S2. To provide positive and negative controls, SKOv3-DIRAS3 cells were grown for 24 h in McCoy’s medium supplemented with 10% FBS, 200 µg/mL G418 and 0.12 µg/mL puromycin as described above, with or without 1 ug/mL DOX. Cells were harvested in 0.25% trypsin (Fisher Scientific, MT-25-052-CI) washed 2 times in PBS, fixed in 10% neutralized formalin, and embedded in paraffin.
Oven incubation at 60 °C for 20 min was used to restore antigenic reactivity followed by 2 20 min incubations in xylene. After slides were rehydrated, antigen retrieval was performed in 6.5 mM sodium citrate buffer (pH 6.0) for 10 min. 3% bovine serum albumin in 0.1 M Tris-buffered saline was used for blocking. The primary antibody (anti-DIRAS3 mouse monoclonal, 1:1000, R.C. Bast Laboratory; anti-LC3B rabbit monoclonal, 1:250, Cell Signaling Technology, 2775; or anti-BECN1 rabbit monoclonal 1:800, Epitomics, 2026) were incubated for 4 °C overnight incubation. Mouse (GM601H) or rabbit (GR602H) secondary antibody was then applied for 1 h at room temperature (Biocare Medical) followed by washing 3 times in PBS for 10 min. DAB chromagen (Biocare Medical, BDB 2004L) was added for 1 min per slide followed by 3 additional washes in PBS for 10 min and then hematoxylin staining was performed for 1 min per slide followed by 3 additional washes in PBS for 10 min. Serial sections of cell line test arrays and IgG staining served as positive and negative controls and were stained alongside TMAs to confirm assay reproducibility. Omission of the primary antibody served as a negative control for each immunostaining event.
Immunohistochemical staining was evaluated for a) total staining intensity and b) punctate staining (indicating the presence of an autophagosome structure) vs. diffuse staining (indicating staining but with absence of autophagosome structure(s) and/or autophagosome formation) by Zhen Lu (ZL) and Maria T Baquero (MTB). A TMA serial section stained for mouse IgG1 served as an additional negative control. Total staining intensity was determined as 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). Punctate staining was also evaluated and scored on the scale described above. All slides were evaluated independently by 2 investigators (ZL and MTB) without knowledge of the identity of the patient or clinical outcome.
Statistics
All experiments were repeated independently at least 2 times and the data (bar graphs) expressed as mean ± s.e. Statistical analysis was performed using the Student t test (2-sample assuming unequal variances). The criterion for statistical significance was taken as P < 0.05 (2-sided). Staining expression values for DIRAS3, LC3B, and BECN1 were obtained from each TMA or tissue slide and treated as ordinal variables. Spearman rho was used to assess the direction and strength of association between expression scores for DIRAS3, LC3B, and BECN1. Chi-square analysis was used to compare positive and negative expression values between the 3 markers (JMP Statistical Discovery Software, Version 7.0.1 (SAS Institute, Inc., Cary, NC).
| Abbreviations: | ||
| aa | = | amino acids |
| AIC | = | autophagosome initiation complex |
| BA | = | bafilomycin A1 |
| CQ | = | chloroquine |
| CTD | = | C-terminal deleted DIRAS3 |
| DIRAS3 | = | DIRAS family, GTP-binding RAS-like 3 |
| DOX | = | doxycycline |
| MAP1LC3 | = | microtubule-associated protein 1 light chain 3 |
| NTD | = | N-terminal deleted DIRAS3 |
| TEM | = | transmission electron microscopy |
| TMA | = | tissue microarray |
Additional material
Download Zip (1.4 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
These studies were supported in part by a grant from the National Cancer Institute R01 CA135354, by the MD Anderson SPORE in Ovarian Cancer NCI P50 CA83639, the Shared Resources of the MD Anderson CCSG NCI P30 CA16672, the Ovarian Cancer Research Fund, the National Foundation for Cancer Research, philanthropic support from the Zarrow Foundation and Stuart and Gaye Lynn Zarrow and Chia Family Foundation.
References
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6:463 - 77; http://dx.doi.org/10.1016/S1534-5807(04)00099-1; PMID: 15068787
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci 2005; 118:7 - 18; http://dx.doi.org/10.1242/jcs.01620; PMID: 15615779
- Bray K, Mathew R, Lau A, Kamphorst JJ, Fan J, Chen J, Chen HY, Ghavami A, Stein M, DiPaola RS, et al. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS One 2012; 7:e41831; http://dx.doi.org/10.1371/journal.pone.0041831; PMID: 22848625
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ 2005; 12:Suppl 2 1542 - 52; http://dx.doi.org/10.1038/sj.cdd.4401765; PMID: 16247502
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11:385 - 96; http://dx.doi.org/10.1038/ncb1846; PMID: 19270696
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20:1981 - 91; http://dx.doi.org/10.1091/mbc.E08-12-1248; PMID: 19211835
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22:140 - 9; http://dx.doi.org/10.1016/j.ceb.2010.01.001; PMID: 20097051
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A 2008; 105:19211 - 6; http://dx.doi.org/10.1073/pnas.0810452105; PMID: 19050071
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360 - 72; http://dx.doi.org/10.1091/mbc.E08-01-0080; PMID: 18843052
- Wu X, Liang L, Dong L, Yu Z, Fu X. Effect of ARHI on lung cancer cell proliferation, apoptosis and invasion in vitro. Mol Biol Rep 2013; 40:2671 - 8; http://dx.doi.org/10.1007/s11033-012-2353-x; PMID: 23247805
- Yu Y, Xu F, Peng H, Fang X, Zhao S, Li Y, Cuevas B, Kuo WL, Gray JW, Siciliano M, et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci U S A 1999; 96:214 - 9; http://dx.doi.org/10.1073/pnas.96.1.214; PMID: 9874798
- Dalai I, Missiaglia E, Barbi S, Butturini G, Doglioni C, Falconi M, Scarpa A. Low expression of ARHI is associated with shorter progression-free survival in pancreatic endocrine tumors. Neoplasia 2007; 9:181 - 3; http://dx.doi.org/10.1593/neo.06838; PMID: 17401457
- Rosen DG, Wang L, Jain AN, Lu KH, Luo RZ, Yu Y, Liu J, Bast RC Jr.. Expression of the tumor suppressor gene ARHI in epithelial ovarian cancer is associated with increased expression of p21WAF1/CIP1 and prolonged progression-free survival. Clin Cancer Res 2004; 10:6559 - 66; http://dx.doi.org/10.1158/1078-0432.CCR-04-0698; PMID: 15475444
- Wang L, Hoque A, Luo RZ, Yuan J, Lu Z, Nishimoto A, Liu J, Sahin AA, Lippman SM, Bast RC Jr., et al. Loss of the expression of the tumor suppressor gene ARHI is associated with progression of breast cancer. Clin Cancer Res 2003; 9:3660 - 6; PMID: 14506155
- Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, Fishman DM, Yu Y, Bast RC Jr.. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer 2008; 112:1489 - 502; http://dx.doi.org/10.1002/cncr.23323; PMID: 18286529
- Feng W, Shen L, Wen S, Rosen DG, Jelinek J, Hu X, Huan S, Huang M, Liu J, Sahin AA, et al. Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res 2007; 9:R57; http://dx.doi.org/10.1186/bcr1762; PMID: 17764565
- Fujii S, Luo RZ, Yuan J, Kadota M, Oshimura M, Dent SR, Kondo Y, Issa JP, Bast RC Jr., Yu Y. Reactivation of the silenced and imprinted alleles of ARHI is associated with increased histone H3 acetylation and decreased histone H3 lysine 9 methylation. Hum Mol Genet 2003; 12:1791 - 800; http://dx.doi.org/10.1093/hmg/ddg204; PMID: 12874100
- Hisatomi H, Nagao K, Wakita K, Kohno N. ARHI/NOEY2 inactivation may be important in breast tumor pathogenesis. Oncology 2002; 62:136 - 40; http://dx.doi.org/10.1159/000048259; PMID: 11914599
- Yu Y, Fujii S, Yuan J, Luo RZ, Wang L, Bao J, Kadota M, Oshimura M, Dent SR, Issa JP, et al. Epigenetic regulation of ARHI in breast and ovarian cancer cells. Ann N Y Acad Sci 2003; 983:268 - 77; http://dx.doi.org/10.1111/j.1749-6632.2003.tb05981.x; PMID: 12724231
- Yuan J, Luo RZ, Fujii S, Wang L, Hu W, Andreeff M, Pan Y, Kadota M, Oshimura M, Sahin AA, et al. Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers. Cancer Res 2003; 63:4174 - 80; PMID: 12874023
- Lu Z, Luo RZ, Peng H, Rosen DG, Atkinson EN, Warneke C, Huang M, Nishmoto A, Liu J, Liao WS, et al. Transcriptional and posttranscriptional down-regulation of the imprinted tumor suppressor gene ARHI (DRAS3) in ovarian cancer. Clin Cancer Res 2006; 12:2404 - 13; http://dx.doi.org/10.1158/1078-0432.CCR-05-1036; PMID: 16638845
- Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, Kondo S, Kondo Y, Yu Y, Mills GB, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest 2008; 118:3917 - 29; PMID: 19033662
- Kueck A, Opipari AW Jr., Griffith KA, Tan L, Choi M, Huang J, Wahl H, Liu JR. Resveratrol inhibits glucose metabolism in human ovarian cancer cells. Gynecol Oncol 2007; 107:450 - 7; http://dx.doi.org/10.1016/j.ygyno.2007.07.065; PMID: 17825886
- Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 2009; 452:181 - 97; http://dx.doi.org/10.1016/S0076-6879(08)03612-4; PMID: 19200883
- Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy 2013; 9:424 - 5; http://dx.doi.org/10.4161/auto.22931; PMID: 23321721
- Matsushita M, Suzuki NN, Obara K, Fujioka Y, Ohsumi Y, Inagaki F. Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem 2007; 282:6763 - 72; http://dx.doi.org/10.1074/jbc.M609876200; PMID: 17192262
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20:1992 - 2003; http://dx.doi.org/10.1091/mbc.E08-12-1249; PMID: 19225151
- Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009; 284:12297 - 305; http://dx.doi.org/10.1074/jbc.M900573200; PMID: 19258318
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 2011; 144:253 - 67; http://dx.doi.org/10.1016/j.cell.2010.12.018; PMID: 21241894
- Tanida I. Autophagy basics. Microbiol Immunol 2011; 55:1 - 11; http://dx.doi.org/10.1111/j.1348-0421.2010.00271.x; PMID: 21175768
- Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal 2011; 14:2201 - 14; http://dx.doi.org/10.1089/ars.2010.3482; PMID: 20712405
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20:5971 - 81; http://dx.doi.org/10.1093/emboj/20.21.5971; PMID: 11689437
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447:1121 - 5; PMID: 17589504
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 2008; 15:344 - 57; http://dx.doi.org/10.1016/j.devcel.2008.08.012; PMID: 18804433
- Ku B, Woo JS, Liang C, Lee KH, Jung JU, Oh BH. An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy 2008; 4:519 - 20; PMID: 18334862
- Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res 2007; 17:839 - 49; http://dx.doi.org/10.1038/cr.2007.78; PMID: 17893711
- Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 2005; 1:46 - 52; http://dx.doi.org/10.4161/auto.1.1.1542; PMID: 16874027
- Luo RZ, Fang X, Marquez R, Liu SY, Mills GB, Liao WS, Yu Y, Bast RC. ARHI is a Ras-related small G-protein with a novel N-terminal extension that inhibits growth of ovarian and breast cancers. Oncogene 2003; 22:2897 - 909; http://dx.doi.org/10.1038/sj.onc.1206380; PMID: 12771940
- Luo RZ, Peng H, Xu F, Bao J, Pang Y, Pershad R, Issa JP, Liao WS, Bast RC Jr., Yu Y. Genomic structure and promoter characterization of an imprinted tumor suppressor gene ARHI. Biochim Biophys Acta 2001; 1519:216 - 22; http://dx.doi.org/10.1016/S0167-4781(01)00226-3; PMID: 11418188
- Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy 2009; 5:534 - 6; http://dx.doi.org/10.4161/auto.5.4.8062; PMID: 19223761
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J 2009; 28:2244 - 58; http://dx.doi.org/10.1038/emboj.2009.159; PMID: 19590496
- Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci 2006; 119:259 - 70; http://dx.doi.org/10.1242/jcs.02735; PMID: 16390869
- Matsunaga K, Noda T, Yoshimori T. Binding Rubicon to cross the Rubicon. Autophagy 2009; 5:876 - 7; PMID: 19550146
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11:385 - 96; http://dx.doi.org/10.1038/ncb1846; PMID: 19270696
- Wei Y, Sinha S, Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 2008; 4:949 - 51; PMID: 18769111
- Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy 2007; 3:374 - 6; PMID: 17438366
- Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ 2009; 16:1006 - 17; http://dx.doi.org/10.1038/cdd.2009.34; PMID: 19325567
- Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem 2008; 283:33175 - 82; http://dx.doi.org/10.1074/jbc.M804478200; PMID: 18772134
- Noble CG, Dong JM, Manser E, Song H. Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin1. J Biol Chem 2008; 283:26274 - 82; http://dx.doi.org/10.1074/jbc.M804723200; PMID: 18641390
- Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, Zhang M, Yue Z, Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun 2012; 3:662; http://dx.doi.org/10.1038/ncomms1648; PMID: 22314358
- Kroep JR, Nortier JW. The role of bevacizumab in advanced epithelial ovarian cancer. Curr Pharm Des 2012; 18:3775 - 83; http://dx.doi.org/10.2174/138161212802002689; PMID: 22591420
- Aggarwal C, Somaiah N, Simon G. Antiangiogenic agents in the management of non-small cell lung cancer: where do we stand now and where are we headed?. Cancer Biol Ther 2012; 13:247 - 63; http://dx.doi.org/10.4161/cbt.19594; PMID: 22481432
- Sato S, Itamochi H. Bevacizumab and ovarian cancer. Curr Opin Obstet Gynecol 2012; 24:8 - 13; http://dx.doi.org/10.1097/GCO.0b013e32834daeed; PMID: 22123222
- Tol J, Punt CJ. Monoclonal antibodies in the treatment of metastatic colorectal cancer: a review. Clin Ther 2010; 32:437 - 53; http://dx.doi.org/10.1016/j.clinthera.2010.03.012; PMID: 20399983
- Badgwell DB, Lu Z, Le K, Gao F, Yang M, Suh GK, Bao JJ, Das P, Andreeff M, Chen W, et al. The tumor-suppressor gene ARHI (DIRAS3) suppresses ovarian cancer cell migration through inhibition of the Stat3 and FAK/Rho signaling pathways. . Oncogene . 2012; ; . 31::. 68 - -. 79; http://dx.doi.org/10.1038/onc.2011.213; PMID: 21643014
- Klockenbusch C, Kast J. Optimization of formaldehyde cross-linking for protein interaction analysis of non-tagged integrin beta1. J Biomed Biotechnol 2010; 2010:927585; http://dx.doi.org/10.1155/2010/927585; PMID: 20634879
