Abstract
Ribonucleotide reductase (RNR) plays a critical role in catalyzing the biosynthesis and maintaining the intracellular concentration of 4 deoxyribonucleoside triphosphates (dNTPs). Unbalanced or deficient dNTP pools cause serious genotoxic consequences. Autophagy is the process by which cytoplasmic constituents are degraded in lysosomes to maintain cellular homeostasis and bioenergetics. However, the role of autophagy in regulating dNTP pools is not well understood. Herein, we reported that starvation- or rapamycin-induced autophagy was accompanied by a decrease in RNR activity and dNTP pools in human cancer cells. Furthermore, downregulation of the small subunit of RNR (RRM2) by siRNA or treatment with the RNR inhibitor hydroxyurea substantially induced autophagy. Conversely, cancer cells with abundant endogenous intracellular dNTPs or treated with dNTP precursors were less responsive to autophagy induction by rapamycin, suggesting that autophagy and dNTP pool levels are regulated through a negative feedback loop. Lastly, treatment with si-RRM2 caused an increase in MAP1LC3B, ATG5, BECN1, and ATG12 transcript abundance in xenografted Tu212 tumors in vivo. Together, our results revealed a previously unrecognized reciprocal regulation between dNTP pools and autophagy in cancer cells.
Introduction
Autophagy is an evolutionarily conserved process that occurs in all eukaryotic cells. During autophagy, portions of the cytoplasm and intracellular organelles are sequestered in characteristic double-membrane autophagosomes that subsequently fuse with lysosomes, resulting in degradation and recycling of the intracellular components.Citation1 Mechanistically, there are 3 distinct forms of autophagy to deliver cytosolic cargo to the lysosomes: macroautophagy (referred to as autophagy in this report), chaperone-mediated autophagy, and microautophagy.Citation2 It is now becoming increasingly clear that autophagy exhibits significant diversity in its ability to deliver mitochondria (mitophagy), endoplasmic reticulum (reticulophagy), ribosomes (ribophagy), and peroxisomes (pexophagy) to lysosomes for degradation. In general, basal autophagy plays a critical role in cellular homeostasis by degrading excessive and/or damaged organelles and proteins, thereby performing “in-house” quality control of essential cellular components.Citation3,Citation4 In addition to its basal function, autophagy can be induced by diverse stimuli such as nutrient deprivation, serum starvation, metabolic stress, radiation, and anticancer drugs.Citation5-Citation9 Consequently, autophagic degradation provides a dynamic balance between biosynthetic and catabolic processes, thereby facilitating cell survival.
Basal autophagic activity and the upregulation of autophagy are relevant to many pathological and physiological conditions.Citation10 For example, autophagy plays a protective role against the progression of some human diseases such as cancer, muscular disorders, and neurodegeneration, which includes Huntington, Alzheimer, and Parkinson diseases. It also acts as a cellular defense mechanism to prevent infection by certain pathogenic bacteria and viruses.Citation11 However, the role of autophagy in tumorigenesis and treatment responsiveness is complicated and context-dependent.Citation12 Autophagy can suppress tumor growth during the early stages of tumorigenesis.Citation13 The allelic loss of BECN1, which encodes the protein involved in autophagy initiation, and reduced autophagy are found with high frequency in human prostate, breast, and ovarian cancers.Citation14,Citation15 As such, induction of autophagy would seem to be beneficial for inhibiting tumorigenesis. Conversely, autophagy can promote tumor growth in established tumors, and cancer cells can use enhanced autophagy to survive under metabolic and therapeutic stress.Citation12 For example, the pharmacological and/or genetic inhibition of autophagy reportedly sensitizes cancer cells to the lethal effects of various cancer therapies including chemotherapy, radiotherapy, and targeted therapies, suggesting that suppression of the autophagic pathway could represent a valuable sensitizing strategy for cancer treatments.Citation3,Citation16,Citation17 Lastly, the presence of autophagosomes in dying cells also raises the possibility that autophagy may also play an active role in cell death.Citation18 While autophagy is demonstrated to be essential for cancer cells to adapt to the metabolic demands of amino acids and fatty acids during tumorigenesis, the contribution of autophagy to the supply of the DNA building blocks for DNA synthesis and repair is largely unknown.
Human ribonucleotide reductase is an evolutionarily conserved and a well-studied enzyme composed of 2 identical large subunits (RRM1) and 2 identical small subunits (RRM2).Citation19 Functionally, RNR catalyzes the rate-limiting step for the de novo conversion of ribonucleoside triphosphates to deoxyribonucleoside triphosphates, which are the building blocks used in the synthesis of DNA during replication and repair. Unbalanced dNTP pools can cause genetic abnormalities, tumorigenesis, and cell death.Citation19,Citation20 RNR activity is regulated in a cell-cycle dependent manner to coordinate dNTP production and DNA replication. In general, RRM1 levels are constant throughout the cell cycle and always in excess of RRM2 levels, which fluctuate throughout the cell cycle and peak in the S phase.Citation21,Citation22 Furthermore, elevated RNR activity and overexpression of RRM2 have been found to increase cellular invasiveness, angiogenesis, and proliferation in human cancer cells.Citation23,Citation24 It would be of particular interest to investigate whether there is an additional signaling pathway to regulate dNTP pools.
Previous reports have indicated that nutrient starvation and glucose utilization inhibition induce autophagy.Citation16,Citation25 However, the connection between the intracellular dNTP pool and autophagy regulation is left unaddressed. As more evidence emerges to confirm the role of autophagy in recycling proteins and lipids, it also becomes critical to detect the role of autophagy in maintaining nucleic acid homeostasis. Our studies reported here fill in this gap and utilize combined biochemical, cell biological, and xenograft studies to demonstrate that regulation of the dNTP pool and autophagy are reciprocally coordinated.
Results
Autophagy decreases dNTP pools
To determine whether autophagy regulates dNTP pools, human hepatocellular carcinoma Huh-7 cells were treated with Earle's Balanced Salt Solution (EBSS) or rapamycin (10 μM) for 24 h, a known autophagy inducer. The intracellular dNTP pool concentration was then measured. As shown in , EBSS-treatment caused a marked decrease of intracellular dNTP levels. The relative (compared with control) whole cell dCTP, dGTP, dATP, and dTTP level in EBSS-treated cells was 14.79% (P = 0.00023), 46.73% (P = 0.00047), 28.64% (P = 0.00089), and 38.76% (P = 0.00044), respectively. In parallel, comparable results of decreased dNTP levels were obtained in rapamycin-treated cells (). The relative dCTP, dGTP, dATP, and dTTP level in rapamycin-treated cells was 20.49% (P = 0.00017), 62.31% (P = 0.00129), 63.27% (P = 0.00143), and 25.01% (P = 0.00175) respectively. We consistently observed similar effects using lower doses of rapamycin (Fig. S1). The relative whole cell dCTP, dGTP, dATP, and dTTP level in 0.1 μM rapamycin-treated cells was 72.48% (P = 0.00814), 85.31% (P = 0.00041), 56.21% (P = 0.00141), and 62.41% (P = 0.00002) respectively, and in 1 μM rapamycin-treated cells was 68.64% (P = 0.0046), 72.84% (P = 0.000002), 77.84% (P = 0.09697), and 55.69% (P = 0.00027) respectively. To monitor the short-term effect of rapamycin (10μM) on dNTP pools, a kinetic study was performed from 0.5 to 8 h (Fig. S2). The concentration of the dGTP pool was transiently increased to 2.7-fold (P = 8.11769E-7) at 0.5 h, likely due to stimulation of the salvage pathway, and then reduced to 36% after 8 h of treatment (P = 0.0002). The rest of the dNTP pools did not show profound changes within 8 h of treatment. Lastly, a significant decrease in RNR activity in rapamycin- or EBSS-treated cells was detected (P = 0.00086 and P = 0.00013 respectively) (). Increased MAP1LC3 lipidation coupled with decreased SQSTM1 levels, hallmarks of autophagy induction, were observed in EBSS- () or rapamycin- () treated Huh-7 cells to ascertain the effect of EBSS or rapamycin on autophagy induction. These findings demonstrated that RNR activity and the size of the dNTP pools correlate with autophagy induction.
Figure 1. Autophagy causes a decrease in the dNTP pool. (A and B) EBSS and rapamycin decreased dNTP pool levels in Huh-7 cells. Huh-7 cells were treated with EBSS (A) or rapamycin (10 μM) (B) for 24 h. Cells were harvested and subjected to dNTP analysis. (C) EBSS and rapamycin decreased RNR activity in Huh-7 cells. Huh-7 cells were treated as described in (A) and analyzed for RNR activity. Mean ± SD from 3 independent experiments was shown. (*P < 0.05 vs. control). (D and E) EBSS and rapamycin induced autophagy in Huh-7 cells. Huh-7 cells were treated with EBSS (D) or rapamycin (10 μM) (E) for 24 h. The whole-cell lysates were subjected to western blotting to assess the expression of SQSTM1 and MAP1LC3. ACTB was used as a control to ensure equal loading in each lane. Ctrl, control; Rap, rapamycin.
Inhibition of RNR induces autophagy
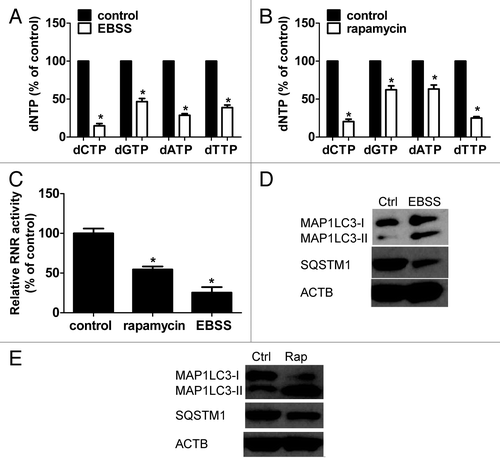
Next, we determined whether the reduction of intracellular dNTP pools upregulates autophagy. Two sets of experiments were performed. First, we have previously shown that downregulation of RRM2 by RRM2 siRNA depletes dNTP pools.Citation26 RRM2B, a homolog of RRM2, was not silenced, which demonstrates the specificity of RRM2 siRNA (data not shown). As predicted, depletion of RRM2 clearly resulted in an increase in the MAP1LC3-II level and a reduction in the SQSTM1 steady-state level in human hepatocellular carcinoma Huh-7 cells (). Similar results were also observed in human oropharyngeal carcinoma KB cells, human lung cancer cell line H460, and human head and neck squamous cell carcinoma Tu212 (). Biochemical analyses confirmed that the relative total dCTP, dGTP, dATP, and dTTP level in si-RRM2-treated Huh-7 cells was reduced to 53.24% (P = 0.00059), 70.25% (P = 0.00947), 44.29% (P = 0.00031), and 72.14% (P = 0.0052) respectively relative to si-control-treated cells (). RNR activity significantly decreased consistently after si-RRM2 treatment (P = 0.00012) ().
Figure 2. RNR inhibition by knockdown of RRM2 or hydroxyurea treatment induces autophagy in human cancer cells. (A) Knockdown of RRM2 induced autophagy in Huh-7, KB, H460, and Tu212 cells. Western blotting was performed to determine RRM2 expression, SQSTM1 degradation, and MAP1LC3-II accumulation in Huh-7, KB, H460, and Tu212 cells by depletion of RRM2 with specific siRNAs for 48 h. ACTB was used as an internal control to ensure that equal amounts of proteins were loaded in each lane. Ctrl, control. (B and C) Knockdown of RRM2 decreased dNTP pool levels and RNR activity in Huh-7 cells. Huh-7 cells were treated with control siRNA or RRM2 siRNA for 48 h. Cells were harvested for dNTPs (B) and RR activity (C) analysis. (D) Hydroxyurea treatment upregulated MAP1LC3-II and downregulated SQSTM1 in Huh-7 cells. Huh-7 cells were treated with hydroxyurea (0.5 mM, 24 h) and then whole-cell lysates were subjected to western blotting to assess the expression of MAP1LC3 and SQSTM1. ACTB was used as an internal control to ensure equal loading in each lane. (E and F) Hydroxyurea decreased dNTP pool levels and RNR activity in Huh-7 cells. Huh-7 cells were treated with hydroxyurea (0.5 mM, 24 h), and dNTP levels (E) and RNR activity (F) were analyzed. (G) Knockdown of RRM2 induced GFP-LC3 puncta in Huh7 cells. GFP-LC3 expressing cells derived from Huh-7 cell line were transfected with non-silencing control-siRNA or RRM2-siRNA and incubated at 37 °C for 48 h. Cells were fixed, stained with DAPI and examined by fluorescence microscopy. Representative merged photographs are shown. (H) TEM images verified autophagic vesicles or autophagosomes induced by si-RRM2 in Huh-7 cells. TEM images were collected by electron microscopy at 48 h post si-RRM2 transfection. The enlarged view shows the presence of typical autophagic vesicles (indicated by an arrow) in si-RRM2-transfected Huh-7 cells. (I) Detection of autophagic flux in RRM2-silenced Huh-7 cells. GFP-LC3 expressing cells derived from Huh-7 cell line were transfected with control-siRNA or RRM2-siRNA. Cells were treated with BafA1 24 h after transfection and incubated for additional 24 h. Fluorescent intensity of GFP-LC3 was measured by FACS analysis 48 h post-transfection.
Second, hydroxyurea (Hu), a chemical inhibitor of RNR, was added to Huh-7 cells prior to harvesting. As shown in , a reduction in the SQSTM1 level and an increase in the MAP1LC3-II level were observed in Hu (0.5 mM, 24 h)-treated Huh-7 cells. Biochemical analyses confirmed that the relative total dCTP, dGTP, dATP, and dTTP level in Hu-treated cells was decreased to 61.27% (P = 0.00117), 75.83% (P = 0.0063), 89.21% (P = 0.03216), and 57.63%: (P = 0.0009) respectively (). RNR activity markedly decreased consistently in Hu-treated cells (P = 0.00084) ().
To ascertain whether the knockdown of RRM2 induces autophagy, fluorescence microscopy and transmission electron microscopy (TEM) studies were employed to visualize autophagosome formation in cells transfected with si-RRM2. As shown in , RRM2 knockdown stimulated the redistribution of GFP-LC3 fusion protein from a diffuse pattern to distinct cytoplasmic puncta, symbolizing the formation of autophagosomes in Huh-7 cells. The percentage of cells containing cytoplasmic GFP-LC3 puncta was significantly increased in si-RRM2 transfected cells compared with the control (P = 0.00574) (Fig. S3). The accumulation of autophagosomes was visualized by TEM analyses (). The number of autophagosomes was significantly increased consistently in RRM2-silenced cells compared with the control (P = 0.00052) (Fig. S4). To further demonstrate that the reduction of dNTP pools elicits the complete process of autophagy, we knocked down RRM2 expression in a GFP-LC3-expressing stable cell line derived from Huh-7 cells (; Fig. S5). Autophagy flux was detected in si-RRM2-transfected cells as well as in EBSS-treated cells as indicated by the reduction in GFP-LC3 fluorescent intensity in the total cell population compared with the controls (; Fig. S6). Inhibition of autolysosomal activity by BafA1 increased the overall GFP-LC3 intensity in both control-siRNA and RRM2-siRNA treated cells () as well as in cells incubated with or without EBSS (Fig. S6). Altogether, these findings confirmed that the reduction of intracellular dNTP pools by knocking down RRM2 expression induced autophagy.
Increased intracellular dNTPs desensitize cells to autophagy induction
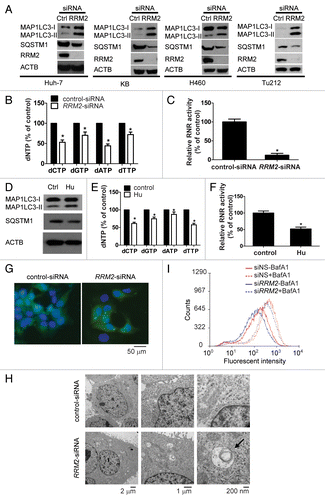
To examine whether autophagy induction is affected by intracellular dNTP pools, a pair of RRM2 overexpression KB cells, KB-M2D (overexpressing exogenous RRM2) and KB-Hur (hydroxyurea resistant cells), were employed. First, RRM2 overexpression was confirmed in KB-M2D and KB-Hur cells by western blot analysis (). As predicted, the relative dNTP levels and RNR activity were markedly increased in KB-M2D and KB-Hur cells (KB-M2D: dATP: P = 0.01489; dCTP: P = 0.00589; dGTP: P = 0.00646; dTTP: P = 0.00652; KB-Hur: dATP: P = 0.0021; dCTP: P = 0.006; dGTP: P = 0.00633; dTTP: P = 0.01002; RNR activity: KB-M2D: P = 0.00684; KB-Hur: P = 0.00967) (). Next, we determined the responsiveness, i.e., MAP1LC3-II accumulation and decrease of SQSTM1, of RRM2-overexpressing cells to rapamycin treatment. While KB-M2D and KB-Hur cells exhibited a very modest increase in MAP1LC3-II levels and a slight decrease in SQSTM1 levels in comparison to KB cells prior to rapamycin treatment, overexpression of RRM2 clearly suppressed rapamycin-induced MAP1LC3 lipidation and SQSTM1 reduction (). Collectively, these results suggest that a greater amount of intracellular dNTPs or overexpression of RRM2 in RRM2-overexpressing cells is likely to desensitize KB cells to rapamycin-induced autophagy. Notably, the RRM2 level was reduced in rapamycin-treated KB cells but not in KB-M2D or KB-Hur cells. To further determine whether the reduction of RRM2 by rapamycin was dependent on the cell cycle, cell cycle analyses were performed in KB cells. As shown in , there was no significant difference in cell cycle distribution between the control and rapamycin-treated KB cells. These results indicate that the decrease in RRM2 levels by rapamycin is not caused by the accumulation of cells in the G1/G0 phase. Consistently, we observed profound RRM2 downregulation following EBSS treatment in Huh-7 cells (, lane 1 vs. lane 5). S phase population was only modestly decreased following EBSS treatment (from 31% to 18%) suggesting that cell cycle arrest only contributed partially to the significant reduction of RRM2 protein level. To determine whether RRM2 was degraded during autophagy by autolysosome- or proteasome-dependent manner, we induced autophagy by EBSS treatment and cells were either treated with BafA1 or MG132 to inhibit autolysosome or proteasome activity respectively. BafA1 was unable to rescue degradation of RRM2 in the presence of EBSS (, lane 2 vs. lane 1). Strikingly, MG132 completely restored RRM2 protein level in the presence of EBSS to that in untreated control (Fig. H, lane 3 and 4 vs. lane 5). In contrast, no proteasomal-dependent degradation of RRM2 was detected in cells without induction of autophagy (, lane 7 and 8 vs. lane 5). Our data suggest that RRM2 undergoes proteasome-dependent degradation upon induction of autophagy.
Figure 3. Cancer cells with higher levels of the intracellular dNTPs pool are less responsive to autophagy inducers. (A) Overexpression of RRM2 in KB-M2D and KB-Hur cells. Western blot analysis was performed to determine RRM2 expression in KB, KB-M2D, and KB-Hur cells. (B and C) Increased dNTP pool levels in KB-M2 cells (B) and KB-Hur cells (C). (D) Increased RNR activity in KB-M2 cells and KB-Hur cells. (E and F) KB-M2D (E) and KB-Hur (F) were relatively resistant to rapamycin-treatment. Western blot analysis was performed to determine RRM2 expression, SQSTM1 degradation, and MAP1LC3-II accumulation in KB, KB-M2D, and KB-Hur cells with or without rapamycin-treatment (10 μM, 24 h). (G) Cell cycle analysis in KB cells with or without rapamycin-treatment (10 μM, 24 h) (n = 3, Mean ± SD). (H) The effect of autolysosome or proteasome inhibitors on RRM2 protein level upon autophagy induction by EBSS. Huh-7 cells were treated with BafA1 (10 nM) or MG132 (2 or 10 μM) in the cells that were treated or untreated with EBSS. Whole-cell lysates were subjected to western blotting to assess the expression of RRM2 and MAP1LC3. ACTB was used as an internal control to ensure equal loading in each lane.
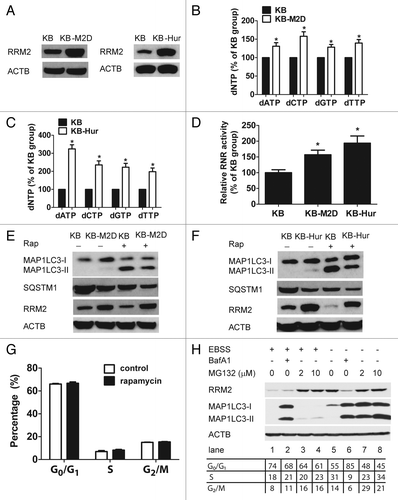
Lastly, we investigated whether dNTP levels influence rapamycin-mediated autophagy induction. As dNTPs penetrate cell membranes poorly, we used their precursor dNMPs to pretreat cells for 30 min and then treated them with rapamycin for 24 h. Pretreatment with 0.2 μM dNMPs resulted in a marked reduction of MAP1LC3 lipidation in rapamycin-treated KB cells (). Moreover, preaddition of 0.2 μM dNMPs effectively reversed the decrease in rapamycin-induced RRM2 () and dNTP levels (dCTP: P = 0.0145; dGTP: P = 0.00569; dATP: P = 0.00586; dTTP: P = 0.00199) (), suggesting that dNMP supplements could, at least in part, antagonize rapamycin-induced autophagy. We also assessed the effect of dNMP supplements on rapamycin-inhibited phosphorylation of Thr70, as well as Thr37 and Thr46 of EIF4EBP1, a known downstream target of MTOR. As shown in , downregulation of the phosphorylation of EIF4EBP1 by rapamycin was only slightly reversed by dNMP pretreatment, suggesting that dNTPs modulate downstream of MTOR. Taken together, our results suggest that an increase in intracellular dNTP pool concentrations attenuates the ability of rapamycin to induce autophagy.
Figure 4. Exogenous supplementation of dNTPs attenuates autophagy induced by rapamycin. (A) Elevated levels of dNTPs rescued the RRM2 level in response to rapamycin. KB cells were treated with rapamycin (Rap 10 μM) for 24 h, with or without dNMPs (0.02–0.2 μM) pretreatment for 30 min, and then whole-cell lysates were subjected to western blot analysis to assess the expression of MAP1LC3 and RRM2. ACTB was used as an internal control to ensure that equal amounts of proteins were loaded in each lane. (B) Incubation with dNMP increased dNTP pool levels in KB cells in the presence of rapamycin. KB cells were treated with rapamycin (10 μM) for 24 h, with or without dNMPs (0.2 μM) pretreatment for 30 min. Cells were then harvested for the dNTP pool assay. Data represents means ± SD of 3 independent experiments, P represents significant differences between conditions where P < 0.05. (C) dNMP supplements partially reversed phosphorylation of EIF4EBP1 by rapamycin. KB cells were treated with rapamycin (rap 10 μM) for 24 h, with or without dNMPs (0.2 μM) pretreatment for 30 min, and whole-cell lysates were subjected to western blot analysis to assess the expression of Phospho-EIF4EBP1, Thr70 (p-EIF4EBP1, Thr70), Phospho-EIF4EBP1, Thr37 and Thr46 (p-EIF4EBP1, Thr37 and Thr46), and total EIF4EBP1. ACTB was used as an internal control to ensure that equal amounts of proteins were loaded in each lane.
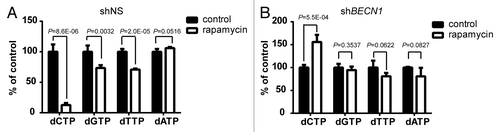
The reduction in dNTP pools following rapamycin treatment depends upon an intact autophagy pathway
To determine whether autophagy has a direct role in regulating the levels of dNTPs, we knocked down BECN1/BECLIN1 by expressing shBECN1 (Fig. S7) to block initiation of the autophagy pathway. Concentrations of dNTP were measured in stable cell lines that were treated with DMSO control or rapamycin for 24 h (). Huh-7 cells expressing nonsilencing shRNA (shNS) displayed similar but slightly different sensitivity to rapamycin in reducing dNTP levels compared with the parental cells (). The relative dCTP, dGTP, dTTP, and dATP level in rapamycin-treated cells was 12.66% (P = 8.6E-06), 73.45% (P = 0.0032), 70.78% (P = 2.0E-06), and 105.96% (P = 0.0516) respectively (). Profoundly, treatment of rapamycin in BECN1-silenced Huh-7 cells did not trigger significant reduction in dCTP, dGTP, dTTP, and dATP pools (). In fact, the dCTP pool was slightly increased after rapamycin treatment (1.6-fold increase, P = 5.5E-04) (). The data suggest that autophagy pathway mediates downregulation of dNTPs following rapamycin treatment.
Figure 5. The rapamycin-induced reduction of dNTP levels was alleviated by inhibition of autophagy. Huh-7 cells were transduced with lentiviruses expressing non-silencing shRNA (shNS) or shBECN1. Stable cell lines were established after puromycin selection. Cells were treated with 10 μM rapamycin or DMSO (control) for 24 h. Concentration of each dNTP in (A) shNS or (B) shBECN1 cells was measured and plotted as % of DMSO control. (n = 4, mean ± SD).
The decrease in dNTPs by RRM2 siRNA administration induces autophagy in vivo.
We have previously tested si-RNA-nanoparticles targeting RRM2 in treating xenografted Tu212 tumors.Citation27 RRM2 siRNA is successfully delivered to the tumor cells and significantly reduced tumor progression by suppressing cell proliferation and inducing apoptosis.Citation27 Since Tu212 cells responded to RRM2 siRNA treatment in vivo and induction of autophagy marker was observed following silencing of RRM2 in cell culture (), we further evaluated the role of dNTP levels in modulating autophagy in Tu212 in vivo. Tu212 cells were inoculated subcutaneously into the hind flank of NOD-SCID mice. These mice then received a single high-pressure (hydrodynamic) tail vein injection of RRM2 siRNA or scrambled siRNA 26 d after inoculation. To confirm that RRM2 protein levels were reduced in the tumor by si-RRM2 treatment, immunohistochemical staining (IHC) with an anti-RRM2 antibody was performed in pre- and post-treated samples. A significant reduction of the RRM2 signal was observed in a si-RRM2-treated group compared with a si-control-treated group (). Tumor RRM2 mRNA levels were measured by quantitative real-time polymerase chain reaction (qRT-PCR). A decrease in RRM2 mRNA levels was observed in si-RRM2-treated samples (P = 0.0009) (). In addition, the MKI67 level, as assayed by IHC, was lower in si-RRM2-treated samples than in the scrambled-siRNA-treated group (), confirming the antiproliferative activity of RRM2 siRNA.Citation28
Figure 6. Treatment of RRM2 siRNA induced autophagy in human cancers grafted in NOD-SCID mice. (A) RRM2 siRNA induces autophagy in vivo. RRM2, MKI67, and MAP1LC3B levels in xenografted tumor sections of vehicle-, scrambled siRNA-, and RRM2 siRNA-treated mice were visualized by immunohistochemical analyses. (B) Knockdown of RRM2 upregulates selected ATG gene transcript abundance in xenograft tumors. Selected mRNA expression levels of RRM2, MAP1LC3B, SQSTM1, ATG5, BECN1, ATG7, and ATG12 in xenografted tumor tissues of vehicle-, scrambled siRNA-, and RRM2 siRNA-treated mice were assayed by qRT-PCR analyses. (C) Decreased RNR activity in tumor xenografts treated with RRM2 siRNA. RNR activity in respective xenografted tumor tissues of vehicle-, scrambled siRNA-, and siRNA-treated mouse was assayed. (D) Decreased dNTP pool levels in tumor xenografts treated with RRM2 siRNA. Levels of dNTPs in tumor tissues of vehicle-, scrambled siRNA-, and RRM2 siRNA-treated mice were analyzed and compared. Data of column represents means ± SD of 3 independent experiments (*P < 0.05 vs. vehicle-treated group).
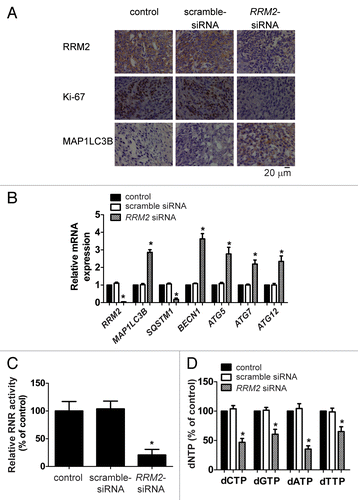
Since si-RRM2-treatment upregulated autophagy in cultured human cancer cells, we next determined the function of si-RRM2 in vivo. As shown in , tumors from si-RRM2-treated mice exhibited an increase in MAP1LC3B staining compared with the control group. Furthermore, an increase in the transcript levels of MAP1LC3B (P = 0.00043) () ATG5 (P = 0.01444), BECN1 (P = 0.00428), ATG7 (P = 0.00961), and ATG12 (P = 0.01204), accompanied with a decrease in the SQSTM1 mRNA level (P = 0.00027), were detected in si-RRM2-treated samples by qRT-PCR analyses (). These results implicated that an increase in transcript levels encoding for members of the autophagy machinery could account for the increased autophagy activity by si-RRM2 in the xenografted tumor tissues.
Given that a decrease in RNR activity and dNTPs levels was found in si-RRM2-treated cultured cells, we then investigated the efficacy of this siRNA on RNR activity and dNTPs in vivo. As shown in , there was a significant decrease in RNR activity (P = 0.00622) in si-RRM2-treated xenografted tumor samples compared with the control siRNA-treated group. Correspondingly, all dNTPs decreased dramatically. The relative dCTP, dGTP, dATP, and dTTP level in RRM2 siRNA-treated tumors was 46.84% (P = 0.00037), 60.55% (P = 0.00426), 35.29% (P = 0.00057), and 65.14% (P = 0.00621) respectively (). Altogether, we concluded that the decrease in dNTPs resulting from downregulation of RRM2 stimulates autophagy in vitro and in vivo.
Discussion
There are 2 major findings in this report. First, we demonstrated that the knockdown of RRM2 or decrease in intracellular dNTP pool levels results in autophagy induction in human cancer cells, suggesting that autophagy initiators monitor the availability of intracellular dNTPs in addition to amino acid and nutrient levels. Several experimental paradigms, the inhibition of RNR activity by pharmacological inhibitor Hu or the depletion of RRM2, were utilized to reach a similar conclusion (; ). However, the exact mechanism(s) underlying the induction of autophagy by inhibition of the de novo synthesis of dNTPs remains to be elucidated. Posttranslational modification of autophagy machinery by several kinases stimulates initiation of autophagy.Citation29 One of the regulating kinases is MAPK8/JNK1. Phosphorylation of BCL2 by MAPK8/JNK1 stimulates dissociation of the BECN1-BCL2 complex and allows BECN1 to associate with the PtdIns3K complex I, thereby inducing autophagy.Citation30 However, no activation of MAPK/JNK activity was observed following silencing of RRM2 expression in Huh-7 cells (Fig. S8). Our data suggests that upregulation of autophagy transcripts such as MAP1LC3B, ATG5, BECN1, and ATG12 by silencing RRM2 might be involved in the initiation and formation of the autophagosome.Citation31 Nevertheless, such regulation seems to be exclusively limited to the animal studies since silencing RRM2 in cultured Huh-7 cells did not alter autophagy transcripts (Fig. S9). Additional factors within the tumor microenvironment are likely to be involved in the transcriptional regulation of autophagy genes.
The MTOR complex 1 (MTORC1) is a multiprotein signaling complex regulated by oncogenes and tumor suppressors. It integrates environmental cues to control cell growth (increase in cell mass) and cell division.Citation32 Outputs downstream of MTORC1 include RPS6KB1 (ribosomal protein S6 kinase, 70 kDa, polypeptide 1), EIF4EBP1 (eukaryotic translation initiation factor 4E binding protein 1), and autophagy.Citation33 Our second major finding showed that rapamycin treatment resulted in the downregulation of RRM2, a decrease in RNR activity, and a decrease in intracellular dNTP pool levels. Interestingly, an increase in intracellular dNTP levels by overexpression of RRM2 or supplementation of exogenous dNMPs effectively attenuated rapamycin-induced autophagy, which coincides with activation of the EIF4EBP1 pathway, suggesting a reciprocal regulation of autophagy and dNTPs, a previously unrecognized regulatory mechanism. Although cell growth and cell cycle division are often controlled by distinct regulators, these 2 biological processes need to be coordinated to ensure the maintenance of cell size after each division. Our observations reinforce the possibility that MTOR not only regulates cell growth by promoting protein translation through RPS6KB1 and EIF4EBP1, but also accelerates cell cycle progression by accumulating dNTPs via inhibition of autophagy.
It is not entirely clear how autophagy induces a decrease in dNTP levels. A recent report has suggested that autophagy targets micronuclei, which arise as a result of deficient bipolar chromosome segregation in cells exposed to cell cycle perturbations.Citation34 These authors show that treatment with Hu increases the frequency of micronuclei surrounded by autophagic puncta, which is consistent with our observations. Autophagy is the process by which intracellular organelles and proteins are degraded in lysosomes.Citation35 However, our results seem counterintuitive because dNTP pool levels actually decreased, which is contradictory to the recycling purpose. We propose that the nucleases in and acidic pH of the lysosomes would cause the breakage of nucleic acids and glycosidic bonds between the deoxyriboses and bases, thereby decreasing dNTP levels. This hypothesis is supported by our observations that silencing of BECN1 to block initiation of autophagy desensitized the cells to rapamycin-induced downregulation of dNTPs. Additionally, the decrease in dNTP pool levels could be enhanced by a decrease in RRM2 levels following induction of autophagy. Dyavaiah et al. report that a large fraction of the yeast ortholog of RRM1 (yeast Rnr1) protein is packaged into a membrane-bound structure and transported to the vacuole for autophagic degradation following treatment with the alkylating agent methyl methanesulfonate.Citation36 Unlike yeast Rnr1, our data suggest that human RRM2 is targeted for proteasome-dependent but autolysosome-independent degradation upon induction of autophagy.
The biological significance of autophagy in sensing and regulating the dNTP pool is still unclear. RNR is the rate-limiting enzyme in the de novo synthesis of dNTP, which plays an important role in mammalian DNA replication and DNA repair.Citation19 Hence, the activity of RNR is dynamically regulated at various levels to ensure sufficient dNTP levels for biological processes. Besides allosteric regulation on the RNR enzyme, RRM2 can be upregulated transcriptionally in the S phase.Citation37 and targeted for ubiquitin-dependent proteasome degradation in the G1 and G2/M phases,Citation38 when dNTPs are no longer needed. It is likely that autophagy is triggered to rapidly deplete the dNTP pool by lysosomal degradation of dNTPs and indirectly through the downregulation of RRM2 in metabolically stressed cells.
It is well characterized that rapamycin inhibits cancer cell growth at least in part by suppressing protein translation via the MTOR-EIF4EBP1 pathway. In this present study, we have demonstrated that rapamycin triggers autophagy to turnover nucleic acids in cancer cells. Both dNTP depletion and retardation of protein biosynthesis are likely to coordinate an antiproliferation effect in rapamycin treatment via suppression of the MTOR pathway. However, autophagy has been proposed to be a prosurvival mechanism in established cancer cells that are exposed to metabolic stress, since metabolites that are generated by the autophagic degradation of cellular components can be used as bioenergetic and anabolic substrates to support cancer growth.Citation5,Citation39 The opposing effects of autophagy in depleting dNTPs and promoting survival might account for the minimal efficacy of rapamycin in limiting cancer cell growth as a single therapeutic agent.Citation40 The combination of rapamycin with inhibitors to interfere with cancer metabolism might be beneficial to enhance clinical outcomes in cancer patients.
In summary, we have demonstrated for the first time that autophagy leads to a decrease in dNTP pool levels and conversely, high levels of dNTP pools desensitize cells to the induction of autophagy, suggesting a direct linkage between nucleotide pools and autophagy in various human cancer cells. Importantly, our findings suggest that increased RRM2 expression or dNTP pool levels in a subset of human tumors could likely modulate the clinical response to a rapamycin-based therapy.
Materials and Methods
Cell culture
Human lung cancer cell line H460 (American Type Culture Collection, HTB-177), human oropharyngeal carcinoma KB cells (American Type Culture Collection, CCL-17), human liver cancer cell line Huh-7 (Japanese Collection of Research Bioresources Cell Bank, JCRB0403) were cultured in DMEM medium containing 10% fetal bovine serum (Atlanta Biologicals, S12450H), 100 units/ml penicillin, and 100 units/ml streptomycin in a humidified cell incubator with an atmosphere of 5% CO2 at 37 °C. RRM2-overexpressing KB-M2D cells were cultured in the same medium as KB cells with 300 μg/ml G-418.Citation41 KB-Hur, a hydroxyurea-resistant clone induced by hydroxyurea, was incubated and maintained in the presence of 1 mM hydroxyurea (Sigma, H8627).Citation41 Human head and neck squamous cell carcinoma Tu212 was cultured in RPMI 1640 medium (Corning Cellgro, 10-040-CV) containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 units/ml streptomycin in a humidified cell incubator with an atmosphere of 5% CO2 at 37 °C.
dNTP pool assay
An optimized dNTP pool assay has been described in our previous study.Citation42 About 1 × 106 cell pellets were harvested and added to 100 μl of 15% trichloroacetic acid. Supernatant fractions were saved and extracted with 2 50-μl aliquots of 1,1,2-trichlorotrifluoroethane/trioctylamine (55:45; Sigma-Aldrich, 270369, and T81000). The assay reaction mixture (50 μl) contained 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 5 mM DTT, 0.25 mM [3H]-dATP (for dCTP, dGTP, and dTTP pool detection) or [3H]-dTTP (for dATP pool detection), 0.2 units sequenase (Affymetrix, 70775Y), and a diluted sample. After 20 min of incubation, 40 μl of aliquots were applied to circular Whatman DE81 ion exchange paper (GE Healthcare Life Sciences, 3658-325). After 3 washes, radioactivity was counted in a liquid scintillation counter and compared with a standard sample prepared in the presence of 0, 0.25, 0.50, 0.75 and 1.0 pmol/ml of dATP, dTTP, dGTP, and dCTP each.
RNR activity assay
The RNR activity assay was performed as described.Citation42 Cell extracts were passed through a ZebaTM Spin Desalting column (Thermo Scientific, 87768) to remove endogenous nucleotides. The reaction mixture contained 0.15 mM [3H]-CDP, 50 mM HEPES (Calbiochem, 391338; pH 7.2), 6 mM DTT, 4 mM magnesium acetate, 2 mM ATP (Roche Applied Science, 12906450), 0.05 mM CDP, and a specific amount of cell lysate. Mixtures were incubated at 37 °C for 20 min. The formed dCDP and remaining CDP were dephosphorylated by phosphodiesterase. Cytidine and deoxycytidine were separated by HPLC using a C18 reversed-phase column (Phenomenex, 00F-4375-E0) connected to a Model 2b-RAM Radio Flow-Through detector (IN/US Systems/Lablogic, Sheffield, UK).
Western blot analysis
Western blot analysis was conducted as previously described.Citation26 About 30 μg total protein was separated by electrophoresis on 10% or 15% standard SDS-polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Bio-Rad, 162-0177). After blocking with 5% nonfat dry milk in Tris-buffered saline (10 mM Tris-Cl, 150 mM NaCl) containing 0.05% Tween-20 (TJ Baker, X251-07), the membrane was incubated with the primary antibody (1:1,000 dilution) overnight at 4 °C. After 3 to 4 washes, the membrane was incubated with horseradish peroxidase-conjugated secondary antibodies (1:2,000 dilutions; Bio-Rad, 170-6515 and 170-6516) for 1 h. After sequential washes, the immunoreactive protein bands were visualized by Enhanced Chemiluminescence (Perkin Elmer, NEL102001EA) according to the manufacturer’s instructions. The following antibodies were used: MKI67/Ki67 (Santa Cruz Biotechnology, sc-15402), RRM2 (Santa Cruz Biotechnology, sc-10844), RRM1 (Santa Cruz Biotechnology, sc-11733), SQSTM1 (Santa Cruz Biotechnology, sc-28359), MAP1LC3A (Cell Signaling Technology, 4599), MAP1LC3B (Cell Signaling Technology, 3868), EIF4EBP1/4E-BP1 (Cell Signaling Technology, 6944), pEIF4EBP1 (Thr70, as well as Thr37 and Thr46; Cell Signaling Technology, 6455 and 2855), MPAK/JNK (Cell Signaling Technology, 9258), pMAPK/pJNK (Thr183 and Thr185; Cell Signaling Technology, 4668), ACTB/β-ACTIN (Sigma, A5441) and BECN1 (BD Biosciences, 612113).
Silencing of RRM2 by siRNA
The human RRM2 siRNA (sc-36338) and control siRNA (sc-36338) were purchased from Santa Cruz Biotechnology. A total of 2 × 105 cells per well were briefly cultured in 6-well plates with 2 mL antibiotic-free growth medium at 37 °C in a CO2 incubator for 24 h. SiRNA duplex-lipofectamine was prepared by 6.0 μL of 10 μmol/L RRM2 siRNA or control siRNA and 3.0 μl lipofectamine RNAiMAX (Invitrogen, 1378-150) in 500 μl siRNA Transfection Medium (Santa Cruz Biotechnology, sc-36868) according to the manufacturer's instructions and directly added into the media. Cells were collected at time points as indicated in each experiment. The knockdown of RRM2 was measured by quantitative reverse transcription-PCR (q-RT-PCR) or western blot analysis.
GFP-LC3 fluorescence
Huh-7 cells were transiently transfected with a GFP-LC3 lentiviral construct using Lipofectamine 2000 (Invitrogen, 11668-019) according to the manufacturer’s instructions. Then, cells were siRNA-treated 24 h after transfection and then returned to the incubator for various times up to an additional 48 h. Alternatively, Huh-7 cells were transduced with GFP-LC3 lentivirus and GFP-positive cells were sorted. Sorted GFP-LC3-expressing cells were transfected with control siRNA or RRM2 siRNA and incubated for 48 h. After incubation, cells were examined by fluorescence microscopy and photographed. For detection of autophagic flux, sorted GFP-LC3-expressing Huh-7 cells were transfected with control siRNA or RRM2 siRNA. Twenty-four h post-transfection, cells were treated either with DMSO as control or bafilomycin A1 (BafA1; 10 nM) and incubated for an additional 24 h. In addition, sorted GFP-LC3-expressing Huh-7 cells were treated with DMSO (control) or BafA1 (10 nM) in the presence of Earle's Balanced Salt Solution (EBSS) or complete media. Fluorescent intensity of GFP-LC3 was measured by FACS analysis 48 h post-transfection.
Transmission electron microscopy
The cell pellets were fixed with 2% glutaraldehyde (Electron Microscopy Science, 16220) in 0.1M cacodylate buffer [Na(CH3)2AsO2 · 3 H2O; (Sigma, C0250)], pH 7.2, at 4 °C overnight. The cell pellets were washed 3 times with 0.1 M cacodylate buffer, pH 7.2, fixed with 1% OsO4 in 0.1 M cacodylate buffer for 30 min and 3 more times with 0.1 M cacodylate buffer. The samples were then dehydrated through 60%, 70%, 80%, 95%, and 100% ethanol, absolute ethanol (twice), propylene oxide (twice), and left in propylene oxide/Eponate (Electron Microscopy Science, 20401; Ted Pella Inc., 18028) (1:1) overnight at room temperature in sealed vials. The next day, the vials were left open for 2 to 3 h to evaporate the propylene oxide. The samples were infiltrated with 100% Eponate and polymerized at ~64 °C for 48 h. Ultra-thin sections (~70-nm thick) were cut using a Leica Ultra cut UCT ultramicrotome (Leica Microsystems, Vienna, Austria) with a diamond knife and then picked up on 200 mesh copper EM grids (Ted Pella Inc., 1GC200). Grids were stained with 2% uranyl acetate for 10 min followed by Reynold’s lead citrate staining for 1 min. Electron microscopy was done on an FEI Tecnai 12 transmission electron microscope equipped with a CCD camera (Gatan Inc., Pleasanton, CA).
Cell cycle analysis
Cell cycle distribution was analyzed by flow cytometry. Cells incubated with or without rapamycin for 24 h were trypsinized, washed twice with PBS (0.2 g/L KCl, 0.24 g/L KH2PO4, 8 g/L NaCl and 1.44 g/L Na2HPO4), and fixed in 70% ethanol for 24 h at 4 °C. Fixed cells were washed with PBS and incubated with propidium iodide (PI; Sigma, P4170) staining solution (50 μg/ml of PI, 1% Triton X-100 [TJ Baker, X198-07], 100 μg/ml of ribonuclease A [Roche, 109169]) on ice for 30 min in the dark. The stained cells were analyzed by FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Data were acquired with Cell Quest software, and the percentages of G1/G0, S, and G2/M phase cells were calculated with MODFIT software.
In vivo animal studies
The animal protocol for the tumorigenicity assay in vivo was approved by the Institutional Animal Care and Use Committee of Zhejiang University. A total of 24 6- to 8-wk-old NOD-SCID female mice were subcutaneously inoculated in the right flank with 5 × 106 Tu212 cells resuspended in 100 μL serum-free RPMI-1640 medium. The mice were divided into 3 groups with 8 mice in each group. Tumor xenografts appeared approximately 3 to 6 d following cell injection. The mice received a single high-pressure tail vein injection. SiRNA was prepared in D5W (5% wt/vol glucose in water) such that a 10% (vol/wt) injection provided doses of 5 mg/kg for siRNA. Tumor xenograft diameters were measured with digital calipers twice a wk. Mice were sacrificed 30 d after inoculation. Tumor xenografts were dissected and the final volume and wet weight were determined. The xenografts were subsequently cut through the median, with 1 part fixed in formalin and embedded in paraffin and the other parts embedded in Tissue-Tek (Sakura, 4583) and snap frozen in liquid nitrogen. Tumor tissues from representative mice were sectioned, embedded in paraffin, and stained with H&E (Mayer’s Hematoxylin from American Master Tech, JXMMHPT and EOSIN Y from American Master Tech, STE0157) for histopathological evaluation. The rest of the tumors were harvested for RNR activity and dNTP pools.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from xenograft tumor tissue or Huh-7 cells transfected with control-siRNA and RRM2-siRNA with the Qiagen RNeasy Mini Kit (74104). Residual genomic DNA was removed by incubating the RNA with DNase (Qiagen, 79254). cDNA was synthesized from 1.0 μg of total RNA using the Superscript III first-strand cDNA synthesis kit (Invitrogen, 18080-051) in a final volume of 20 μl with 0.25 μg of random hexamer and 200 units of Superscript reverse transcriptase (Invitrogen, 18080-051). The reaction mixture was first incubated at 25 °C for 5 min and then at 50 °C for 50 min. Quantitative real-time PCR was performed in the ABI Prism 7900 HT Sequence Detection System (Applied Biosystems, Warrington, UK). The 20 μl reaction mixture consisted of 1 × ABI SYBR Green PCR Master Mix (4309155), 0.25 μl of cDNA, and 0.2 μM of each primer. The following protocol was used for PCR: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The following primers were used: RRM2, 5′-ACAGAAGCCC GCTGTTTCTA-3′ (forward) and 5′-CCCAGTCTGC CTTCTTCTTG-3′ (reverse); MAP1LC3B (LC3B), 5′-AGCAGCATCC AACCAAAATC-3′ (forward) and 5′-CTGTGTCCGT TCACCAACAG-3′ (reverse); SQSTM1, 5′-CACCTGTCTG AGGGCTTCTC-3′ (forward) and 5′-CACACTCTCC CCAACGTTCT-3′ (reverse); ATG5, 5′-TGGGATTGCA AAATGACAGA-3′ (forward) and 5′-TTCCCCATCT TCAGGATCAA-3′ (reverse); BECN1, 5′-CTCAGAATTG GAGGGCTCTG-3′ (forward) and 5′-TGGGCAGATT AATTCGGAAG-3′ (reverse); ATG7, 5′-ACCCAGAAGA AGCTGAACGA-3′ (forward) and 5′-CTCATTTGCT GCTTGTTCCA-3′ (reverse); ATG12, 5′-CTTACGGATG TCTCCCCAGA-3′ (forward) and 5′-ATGAGTCCTT GGATGGTTCG-3′ (reverse); ACTB, 5′-GGACTTCGAG CAAGAGATGG-3′ (forward) and 5′-AGCACTGTGT TGGCGTACAG-3′ (reverse).
A relative gene-expression quantification method was used to calculate the fold change of mRNA expression according to the comparative Ct method using ACTB for normalization. Final results were determined as follows: 2-(ΔCt sample-ΔCt control), where ΔCt values of the control and sample were determined by subtracting the Ct value of the target gene from the value of the housekeeping gene: ACTB. Data was represented as a ratio of the treated sample value to the control sample value.
Immunohistochemical analysis
Immunohistochemical staining was performed on 5-μm thick sections prepared from formalin-fixed, paraffin-embedded tissues. Tissue sections were deparaffinized in xylene followed by 100% ethanol. Samples were then quenched in 3% hydrogen peroxide (Macron Fine Chemicals, 5240-02) and pretreated with the steam of DIVA solution (Biocare Medical, DV2004MX) to promote antigen retrieval. After antigen retrieval, slides were incubated in Protein Block (Dako, X0909) for 5 min. Slides were then incubated with primary antibody recognizing RRM2 [1:5 dilution 1 one h at room temperature (RT)], MAP1LC3B (1:100 dilution for 30 min at RT), or MKI67/Ki67 (1:50 dilution for 30 min at RT), washed in Dako buffer (Dako Corporation, K8000), and incubated with DAKO Envision + System-HRP (Dako Corporation, K4001/K4003) with mouse or rabbit polymer antibody for 30 min. After washing with Dako buffer, slides were incubated with the chromogen diaminobenzidine tetrahydrochloride (Dako, K8002), counterstained with hematoxylin, and mounted.
Statistical analysis
The Student t test was used to evaluate the statistical differences between the experimental values of 2 samples being compared. P < 0.05 is considered statistically significant.
| Abbreviations: | ||
| ACTB | = | actin, beta |
| ATG | = | autophagy-related |
| BafA1 | = | bafilomycin A1 |
| BCL2 | = | B-cell CLL/lymphoma 2 |
| BECN1 | = | Beclin 1, autophagy-related |
| CCD | = | charge-coupled device |
| CDP | = | cytidine diphosphate |
| DAPI | = | 4′,6-diamidino-2-phenylindole |
| DMSO | = | dimethyl sulfoxide |
| dNMPs | = | deoxyribonucleoside monophosphates |
| dNTPs | = | deoxyribonucleoside triphosphates |
| DTT | = | dithiothreitol |
| EBSS | = | Earle's Balanced Salt Solution |
| EIF4EBP1 | = | eukaryotic translation initiation factor 4E binding protein 1 |
| GFP | = | green fluorescent protein |
| Hu | = | hydroxyurea |
| MAP1LC3 | = | microtubule-associated protein 1 light chain 3 |
| MKI67 | = | marker of proliferation Ki-67 |
| MPAK8/9/10 | = | mitogen-activated protein kinase 8/9/10 |
| MTOR | = | mechanistic target of rapamycin (serine/threonine kinase) |
| MTORC1 | = | MTOR complex 1 |
| NOD-SCID | = | nonobese diabetic/severe combined immunodeficiency |
| PBS | = | phosphate-buffered saline |
| PI | = | propidium iodide |
| Rap | = | rapamycin |
| shNS | = | nonsilencing short hairpin RNA |
| shRNA | = | short hairpin RNA |
| siRNA | = | short interfering RNA |
| RNR | = | ribonucleotide reductase |
| Rnr1 | = | yeast ribonucleotide reductase large subunit 1 |
| RPS6KB1 ribosomal protein S6 kinase | = | 70 kDa, polypeptide 1 |
| RRM1 | = | ribonucleotide reductase M1 |
| RRM2 | = | ribonucleotide reductase M2 |
| RRM2B | = | ribonucleotide reductase M2 B (TP53 inducible) |
| SQSTM1 | = | sequestosome 1 |
| TEM | = | transmission electron microscopy |
Additional material
Download Zip (710.7 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Sofia Loera and her colleagues from her core facility for assisting with the immunohistochemical analysis, Dr Brian Armstrong, Mariko Lee, and Tina Patel for helping with the digital image capturing in the Microscope Core Lab, Dr Zhuo Li for helping with the digital image capturing in the EM Core Lab, and Dr Frank Hong for his critical review of the manuscript. This work was supported by the National Natural Science Foundation of China grant No. 31000775 (WC), Scientific Research Foundation for Returned Overseas Chinese Scholars grant (Human Resources and Social Security Department, 2012) (WC), National Key Technology R&D Program of China grant No. 2012BAD33B08 (WC), Research Fund for the Doctoral Program of Higher Education of China grant No. 20103326120006 (WC), City of Hope’s Women’s Cancers Program Award (Y-RC) and NIH grant 5R01DE10742, R01DE14183 (DKA) and 5R01CA127541 (LZ, KZ, BZ, M-LK, SH, and YY).
References
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8:931 - 7; http://dx.doi.org/10.1038/nrm2245; PMID: 17712358
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67 - 93; http://dx.doi.org/10.1146/annurev-genet-102808-114910; PMID: 19653858
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069 - 75; http://dx.doi.org/10.1038/nature06639; PMID: 18305538
- Eskelinen E-L, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta 2009; 1793:664 - 73; http://dx.doi.org/10.1016/j.bbamcr.2008.07.014; PMID: 18706940
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005; 120:237 - 48; http://dx.doi.org/10.1016/j.cell.2004.11.046; PMID: 15680329
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 2005; 280:31582 - 6; http://dx.doi.org/10.1074/jbc.M506736200; PMID: 16027116
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21:1621 - 35; http://dx.doi.org/10.1101/gad.1565707; PMID: 17606641
- Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol 2005; 26:1401 - 10; PMID: 15809734
- Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ 2007; 14:548 - 58; http://dx.doi.org/10.1038/sj.cdd.4402030; PMID: 16946731
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069 - 75; http://dx.doi.org/10.1038/nature06639; PMID: 18305538
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ 2005; 12:Suppl 2 1542 - 52; http://dx.doi.org/10.1038/sj.cdd.4401765; PMID: 16247502
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12:401 - 10; http://dx.doi.org/10.1038/nrc3262; PMID: 22534666
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27 - 42; http://dx.doi.org/10.1016/j.cell.2007.12.018; PMID: 18191218
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999; 59:59 - 65; http://dx.doi.org/10.1006/geno.1999.5851; PMID: 10395800
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672 - 6; http://dx.doi.org/10.1038/45257; PMID: 10604474
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21:1621 - 35; http://dx.doi.org/10.1101/gad.1565707; PMID: 17606641
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 2007; 21:1367 - 81; http://dx.doi.org/10.1101/gad.1545107; PMID: 17510285
- Baehrecke EH. Autophagy: dual roles in life and death?. Nat Rev Mol Cell Biol 2005; 6:505 - 10; http://dx.doi.org/10.1038/nrm1666; PMID: 15928714
- Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem 2006; 75:681 - 706; http://dx.doi.org/10.1146/annurev.biochem.75.103004.142443; PMID: 16756507
- López LC, Akman HO, García-Cazorla A, Dorado B, Martí R, Nishino I, Tadesse S, Pizzorno G, Shungu D, Bonilla E, et al. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum Mol Genet 2009; 18:714 - 22; http://dx.doi.org/10.1093/hmg/ddn401; PMID: 19028666
- Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem 2000; 275:17747 - 53; http://dx.doi.org/10.1074/jbc.M000799200; PMID: 10747958
- D’Angiolella V, Donato V, Forrester FM, Jeong Y-T, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 2012; 149:1023 - 34; http://dx.doi.org/10.1016/j.cell.2012.03.043; PMID: 22632967
- Duxbury MS, Whang EE. RRM2 induces NF-kappaB-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun 2007; 354:190 - 6; http://dx.doi.org/10.1016/j.bbrc.2006.12.177; PMID: 17222798
- Zhang K, Hu S, Wu J, Chen L, Lu J, Wang X, Liu X, Zhou B, Yen Y. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: implication of RRM2 in angiogenesis. Mol Cancer 2009; 8:11; http://dx.doi.org/10.1186/1476-4598-8-11; PMID: 19250552
- Aki T, Yamaguchi K, Fujimiya T, Mizukami Y. Phosphoinositide 3-kinase accelerates autophagic cell death during glucose deprivation in the rat cardiomyocyte-derived cell line H9c2. Oncogene 2003; 22:8529 - 35; http://dx.doi.org/10.1038/sj.onc.1207197; PMID: 14627994
- Liu X, Zhou B, Xue L, Yen F, Chu P, Un F, Yen Y. Ribonucleotide reductase subunits M2 and p53R2 are potential biomarkers for metastasis of colon cancer. Clin Colorectal Cancer 2007; 6:374 - 81; http://dx.doi.org/10.3816/CCC.2007.n.007; PMID: 17311703
- Rahman MA, Amin AR, Wang X, Zuckerman JE, Choi CH, Zhou B, Wang D, Nannapaneni S, Koenig L, Chen Z, et al. Systemic delivery of siRNA nanoparticles targeting RRM2 suppresses head and neck tumor growth. J Control Release 2012; 159:384 - 92; http://dx.doi.org/10.1016/j.jconrel.2012.01.045; PMID: 22342644
- Heidel JD, Liu JY, Yen Y, Zhou B, Heale BS, Rossi JJ, Bartlett DW, Davis ME. Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin Cancer Res 2007; 13:2207 - 15; http://dx.doi.org/10.1158/1078-0432.CCR-06-2218; PMID: 17404105
- Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713 - 20; http://dx.doi.org/10.1038/ncb2788; PMID: 23817233
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008; 30:678 - 88; http://dx.doi.org/10.1016/j.molcel.2008.06.001; PMID: 18570871
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22:124 - 31; http://dx.doi.org/10.1016/j.ceb.2009.11.014; PMID: 20034776
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274 - 93; http://dx.doi.org/10.1016/j.cell.2012.03.017; PMID: 22500797
- Nyfeler B, Bergman P, Triantafellow E, Wilson CJ, Zhu Y, Radetich B, Finan PM, Klionsky DJ, Murphy LO. Relieving autophagy and 4EBP1 from rapamycin resistance. Mol Cell Biol 2011; 31:2867 - 76; http://dx.doi.org/10.1128/MCB.05430-11; PMID: 21576371
- Rello-Varona S, Lissa D, Shen S, Niso-Santano M, Senovilla L, Mariño G, Vitale I, Jemaá M, Harper F, Pierron G, et al. Autophagic removal of micronuclei. Cell Cycle 2012; 11:170 - 6; http://dx.doi.org/10.4161/cc.11.1.18564; PMID: 22185757
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ 2005; 12:Suppl 2 1542 - 52; http://dx.doi.org/10.1038/sj.cdd.4401765; PMID: 16247502
- Dyavaiah M, Rooney JP, Chittur SV, Lin Q, Begley TJ. Autophagy-dependent regulation of the DNA damage response protein ribonucleotide reductase 1. Mol Cancer Res 2011; 9:462 - 75; http://dx.doi.org/10.1158/1541-7786.MCR-10-0473; PMID: 21343333
- Eriksson S, Martin DW Jr.. Ribonucleotide reductase in cultured mouse lymphoma cells. Cell cycle-dependent variation in the activity of subunit protein M2. J Biol Chem 1981; 256:9436 - 40; PMID: 6270086
- Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci U S A 2003; 100:3925 - 9; http://dx.doi.org/10.1073/pnas.0330774100; PMID: 12655059
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10:51 - 64; http://dx.doi.org/10.1016/j.ccr.2006.06.001; PMID: 16843265
- Gaur S, Chen L, Yang L, Wu X, Un F, Yen Y. Inhibitors of mTOR overcome drug resistance from topoisomerase II inhibitors in solid tumors. Cancer Lett 2011; 311:20 - 8; http://dx.doi.org/10.1016/j.canlet.2011.06.005; PMID: 21764510
- Zhou BS, Tsai P, Ker R, Tsai J, Ho R, Yu J, Shih J, Yen Y. Overexpression of transfected human ribonucleotide reductase M2 subunit in human cancer cells enhances their invasive potential. Clin Exp Metastasis 1998; 16:43 - 9; http://dx.doi.org/10.1023/A:1006559901771; PMID: 9502076
- Zhou BS, Ker R, Ho R, Yu J, Zhao YR, Shih J, Yen Y. Determination of deoxyribonucleoside triphosphate pool sizes in ribonucleotide reductase cDNA transfected human KB cells. Biochem Pharmacol 1998; 55:1657 - 65; http://dx.doi.org/10.1016/S0006-2952(98)00042-2; PMID: 9634002
