Abstract
Macroautophagy is an evolutionarily conserved degradative process of eukaryotic cells. Double-membrane vesicles called autophagosomes sequester portions of cytoplasm and undergo fusion with the endolysosomal pathway in order to degrade their content. There is growing evidence that members of the small GTPase RAB protein family—the well-known regulators of membrane trafficking and fusion events—play key roles in the regulation of the autophagic process. Despite numerous studies focusing on the functions of RAB proteins in autophagy, the importance of their upstream regulators in this process emerged only in the past few years. In this review, we summarize recent advances on the effects of RABs and their upstream modulators in the regulation of autophagy. Moreover, we discuss how impairment of these proteins alters the autophagic process leading to several generally known human diseases.
Keywords: :
Introduction
The genetic regulation of macroautophagy (hereafter autophagy) was originally described in the yeast Saccharomyces cerevisiae, but it has also an important role in multicellular organisms as a cytoprotective response to stress and pathological conditions.Citation1 The process is characterized by the formation of double-membrane autophagosomes arising from a phagophore assembly site (PAS). The PAS is a well-defined cytoplasmic site marked by a subset of autophagy-related (ATG) proteins.Citation2 The formation of the phagophore from the PAS requires the subsequent activity of the ULK1/2 (Atg1 in yeast) kinase complex, which, together with the class III phosphatidylinositol 3-kinase (PIK3C3/VPS34) complex initiates the phagophore nucleation and expansion and 2 ubiquitin-like conjugation systems: the ATG12–ATG5-ATG16L1 complex and the phosphatidylethanolamine-conjugated LC3/Atg8. Completed autophagosomes undergo fusion events with various members of the endolysosomal system to form amphisomes or autolysosomes and in either case their content is degraded by lysosomal hydrolases.Citation3
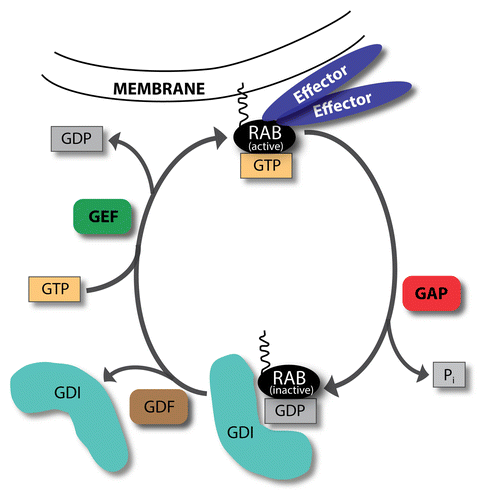
RAB proteins, members of the RAS GTPase superfamily, are key regulators of membrane trafficking and fusion events.Citation4 All of the RABs contain a conserved nucleotide binding domain which is able to bind both GTP and GDP. RABs generally cycle between GTP-bound active and GDP-bound inactive forms (). In their active state, RAB proteins recruit various effectors to the membrane, which they are attached to. RABs have a low intrinsic hydrolase activity; their hydrolysis rate depends on GTPase-activating proteins (GAPs). These proteins can complement the catalytic site of RABs, thereby promoting GTP hydrolysis. Inactive, GDP-bound RABs are removed from their target membrane and kept soluble in the cytoplasm by GDP dissociation inhibitors (GDIs). Upon RAB activation, GDIs are removed mainly by GDI displacement factors (GDFs). Thereafter, guanine nucleotide exchange factors (GEFs) activate RAB proteins by stimulating the exchange of GDP to GTP. Active RABs can stably attach to membrane surfaces via their C-terminal lipid (geranylgeranyl) anchor, provided by RAB escort proteins.
Figure 1. GTP-GDP exchange cycle of RAB proteins. RABs cycle between GTP-bound active and GDP-bound inactive forms. In their active, membrane-attached state they recruit various effectors. GTPase-activating proteins (GAPs) increase the GTP hydrolysis rate, thereby inactivating RABs. Inactive RABs are sequestered in the cytosol by GDP dissociation inhibitors (GDIs). Upon RAB activation, GDI displacement factors (GDFs) can displace GDIs. Afterwards, guanine nucleotide exchange factors (GEFs) activate RAB proteins by changing GDP to GTP.
In mammals, there are more than 70 known RAB proteins among which about 10 have a defined function in autophagy (). However, only a few of their upstream regulators were implicated in this process. Here we summarize recent advances on the role of RABs and their regulators in autophagy, showing their primary functions and highlighting many well-known GEFs and GAPs whose autophagic function needs further investigation. Furthermore, we discuss how impaired RAB cascades can cause alterations in the autophagic process leading to various human diseases.
Table 1. RAB proteins implicated in autophagy
Endosomal RABs: RAB4, RAB5, and RAB7
In parallel with autophagy, the endosomal system also provides macromolecules and energy sources for the cell. Cargo taken up by different ways of endocytosis is subsequently transported to early or sorting endosomes (EEs). RAB5 is a well-known marker of these structures: active RAB5 recruits effector proteins, which play a role in maintaining, trafficking, cargo recycling, and maturation of EEs. RAB5 is involved in vesicle fusion events as well: its yeast ortholog, Vps21, mediates homo- and heterotypic fusion of EEs with endocytic carriers through its effectors CORVET and EEA1/Vac1.Citation53 Furthermore, RAB5 participates in the endosomal recruitment of PIK3C3/VPS34, which in turn produces phosphatidylinositol 3-phosphate (PtdIns3P) on the EE membrane.Citation54
Lately, RAB5 was found to be a regulator of the early steps of autophagosome formation. Similarly to its endosomal functions, RAB5 may have a role in the recruitment of the autophagic BECN1-PIK3C3 complex and subsequent PtdIns3P production on the phagophore membrane. A previous work of Ravikumar and colleagues showed that RAB5 regulates the conjugation of ATG12 to ATG5 through its effector, PIK3C3, as a part of the autophagic BECN1 complex.Citation13 This study suggested a direct role for RAB5 in the process of autophagosome formation, since its effect on autophagy was not due to its endocytic roles or through the regulation of MTOR signaling. Moreover, both RAB5 and PIK3C3 were found to be required for the formation of autophagosomes during autophagy induced by the nonstructural protein 4B (NS4B) of hepatitis C virus.Citation14 Interestingly, recent studies showed that PIK3CB/p110-β, the catalytic subunit of the class IA phosphoinositide 3-kinase complex facilitates this RAB5 function.Citation15,Citation16 In these studies, Dou and colleagues showed that loss of PIK3CB leads to impaired autophagy. They found that following growth factor limitation, PIK3CB dissociates from the growth factor receptor complex and associates with RAB5, positively regulating its activity and enhances its interaction with PIK3C3, thereby promoting autophagosome formation.
RAB4 localizes to early endosomes and mediates cargo recycling toward the plasma membrane on the so-called fast recycling route.Citation55,Citation56 In addition, RAB4 was recently found to have a role in the early steps of autophagy.Citation18 This study showed an increased colocalization of RAB4 with LC3 due to autophagy induction by starvation or rapamycin treatment. Talaber and colleagues also showed that a C-terminally truncated native isoform of RAB4 shows an enhanced colocalization with LC3 and mitochondria and its expression leads to an increased formation of autophagic structures. Furthermore, overexpression of functional RAB4 protein results in the accumulation of mitochondria during autophagy induction, suggesting that it promotes the formation not only of autophagosomes, but also of the mitochondrial network, thereby preserving mitochondria upon autophagy induction. Whether RAB4 plays a direct role in the early stages of autophagy or acts indirectly is not understood yet, since the exact molecular mechanism through which RAB4 can orchestrate autophagy is still poorly characterized.
After cargo recycling, EEs mature into late endosomes (LEs).Citation57 During the maturation process, inactivated RAB5 is substituted by RAB7 on the endosomal membrane. Subsequently, RAB7 recruits its effectors which are required for LE trafficking and fusion with lysosomes.Citation58 Two well-known RAB7 effectors, RILP (Rab-interacting lysosomal protein) and OSBPL1A/ORP1L (oxysterol-binding protein-like 1A) are necessary for the minus end-directed transport of late endosomes along microtubules.Citation59-Citation61 These proteins bind to active RAB7 simultaneously; their interaction facilitates the association of SPTBN2/βIII spectrin and the dynein-dynactin complex with the endosomal membrane, which in turn mediates the microtubule-attached vesicle movement. Furthermore, RAB7 through the homotypic fusion and vacuole protein sorting (HOPS) complex and its associated SNAREsCitation62,Citation63 and through RNF115/Rabring7 (ring finger protein 115/RAB7-interacting RING finger protein),Citation64 plays a key role in the late endosome-lysosome fusion. In addition, the BECN1-binding UVRAG (UV radiation resistance associated), while binding class C VPS members of the HOPS complex, also enhances RAB7 activity.Citation65 Accordingly, KIAA0226/Rubicon acts as a negative regulator of endosome maturation by sequestering UVRAG from the class C VPS complex and preventing it from RAB7 activation.Citation22,Citation23,Citation66,Citation67
During autophagosome maturation RAB7 acts similarly as in endosome maturation ().Citation24 However, while OSBPL1A has not yet been implicated in autophagy, RAB7-RILP interaction was recently found to be required for neuronal autophagy.Citation19 This study showed that during neuronal stress, the presence of IGF1 enhances the interaction of RAB7 with RILP, thereby facilitating autophagic flux. A further study showed that RAB7 together with its LC3-interacting effector FYCO1 (FYVE and coiled-coil domain containing 1) participates in the microtubular transport of autophagosomes.Citation20 FYCO1 is observed on endosomes and autophagosomes as well, where it forms a complex and colocalizes with RAB7. Loss of FYCO1 results in the accumulation of autophagosomes in the perinuclear region of the cells, suggesting that it plays a key role in plus end-directed microtubule transport of autophagosomes. These findings suggest that RAB7 and its effectors can directly mediate autophagosome trafficking on microtubule tracks in both directions. Supporting this, a recent study showed that autophagosomes at the early stage of their maturation process can move in neurons using either kinesin or dynein motors. After a while, however, they show unidirectional movement toward the cell soma using only dynein motor proteins.Citation21 In HeLa cells only plus-end directed transport of autophagosomes is observed, and all evidence of minus-end directed autophagosomal transport and bidirectional movement are derived from neuron models; it is possible that these latter types of autophagosome trafficking is a specific features of neurons.
Figure 2. Various roles of RAB7 in autophagy. Through its interaction partners, RAB7 is implicated in the bidirectional transport of autophagosomes. RILP participates in the recruitment of dynein-dynactin complex, whereas FYCO1 interacts with kinesins. Other RAB7 effectors, UVRAG, KIAA0226/Rubicon, and RNF115 have a role in autophagosome maturation. While the HOPS complex associated SNAREs, VTI1B, and VAMP8, are involved in autophagosome-lysosome fusion, the HOPS itself, although a well-known RAB7 effector, is not implicated in this process. Furthermore, RAB7 is implicated in the regulation of TORC1 activity, and, together with its effector PLEKHF1/Phafin1, is also described in autophagosome (AP) formation.
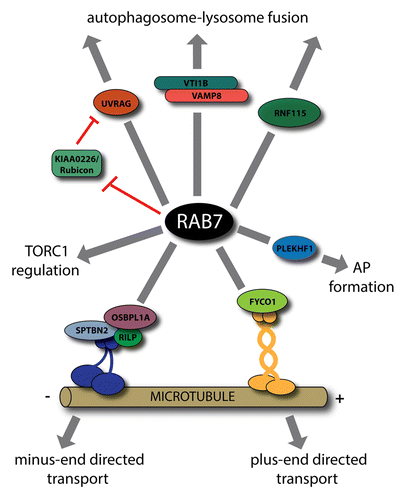
Similarly to its endosomal functions, RAB7 is required for fusion of autophagosomes with late endosomes or lysosomes.Citation25-Citation28 These studies show that RAB7 colocalizes with LC3 and its silencing or overexpression of the GDP-locked, dominant negative RAB7 lead to an increase in size and number of autophagosomes due to their impaired fusion with lysosomes. Supporting these findings, a recent study revealed a similar role for RAB7 in selective autophagy as well.Citation29 Although some results show that the HOPS complex-associated SNAREs VTI1B and VAMP8 play a role in this fusion process,Citation30 the HOPS member VPS16 was not found to be necessary for it.Citation31 Furuta and colleagues found that during antimicrobial autophagy induced by Streptococcus pyrogenes (also known as Group A Streptococcus, GAS) infection, VTI1B, and VAMP8 colocalize with LC3 on autophagic structures and these SNAREs are required for elimination of bacteria through mediating the fusion of GAS-containing xenophagosomes with lysosomes.Citation30 Furthermore, several studies indicated that UVRAG and KIAA0226/Rubicon also have an effect on autophagic maturation. These studies indicate that UVRAG physically interacts with the HOPS complex, thereby facilitating RAB7 activity and the maturation process of both endosomes and autophagosomes.Citation65,Citation66 In contrast, KIAA0226 inhibits PIK3C3 activity and downregulates autophagy, probably through its interaction with RAB7.Citation22,Citation67
However, our growing knowledge on the convergence of endosomal and autophagic routes raises the possibility that these RAB7 effector proteins have only an indirect impact on the late stages of autophagy.Citation68 There are several lines of evidence that closed autophagosomes fuse not only with lysosomes, but also with various populations of endosomesCitation69-Citation71 to form hybrid organelles called amphisomes.Citation72 In many cell types amphisome formation seems to be essential for subsequent lysosomal fusion and degradation. Thus, in these cells the presence of mature endosomes is required for the completion of the autophagic process. Consequently, it is possible that impairment of autophagic maturation due to depletion of RAB7 or UVRAG is caused in fact by the lack of mature endosomes. Supporting this, recent studies showed that direct autophagosome-lysosome fusion is mediated independently of RAB7 and its effectors, by the SNARE protein STX17 (syntaxin 17).Citation73,Citation74 All of these studies suggest that RAB7 effectors facilitating endosome maturation have a cell type-dependent role in autophagy.
There are only a few studies revealing a role for RAB7 in other stages of autophagy. A former paper indicated that MTOR (mechanistic target of rapamycin) regulates the process of autophagic lysosome reformation likely through RAB7.Citation32 Yu and colleagues found that upon prolonged autophagy, reactivation of MTOR is responsible for the generation of protolysosomal tubules, which subsequently mature into functional lysosomes. They showed that GTP hydrolysis by RAB7 is required for this process, since treatment of cells with a nonhydrolizable GTP analog results in the presence of enlarged, long-lasting autolysosomes. In addition, through its effector PLEKHF1/Phafin1,Citation33 RAB7 is also implicated at an early step, at the formation of autophagosomes. This study reported that PLEKHF1 is recruited to lysosomes by RAB7, based on its colocalization with the lysosomal marker LysoTracker Red. It was also found that overexpression of PLEKHF1 results in the accumulation of LC3-positive autophagosome-like structures, suggesting that PLEKHF1 has a role in the induction of autophagosome formation, but the underlying mechanism is poorly understood yet. Similarly, 2 recent studies showed that RAB7 is required at the initial steps during the antibacterial autophagic response against GAS, since depletion of RAB7 leads to a decrease in formation of GAS-containing autophagic vacuoles.Citation34,Citation35
Further studies indicated that RAB5 and RAB7 play a role even in the activation of the main nutrient and energy sensor of eukaryotic cells, MTORC1, which controls the balance of anabolic and catabolic processes and is one of the most important regulators of autophagy. The core member of this complex, MTOR, blocks autophagy induction through the inhibitory phosphorylation of several autophagic proteins.Citation75 Recently, MTORC1 was shown to be activated at the surface of lysosomes by an amino acid-sensing protein cascade (reviewed in ref. Citation76). In agreement with this, another study found that intact late endosomes are essential for MTORC1 signaling, revealing a novel function of RAB7 as a required regulator of MTOR activity.Citation36 Two studies proposed a similar role for RAB5 in regulation of TORC1 activity and localization. Li and colleagues showed that either knockdown or uncontrolled expression of wild-type, constitutively active or dominant negative forms of Rab5 significantly inhibits TOR activity in Drosophila S2 cells, potentially through the regulation of the amino acid sensing RAG GTPase complex.Citation12 Meanwhile Bridges and others found that disruption of RAB5 impairs TOR activity and localization in yeast and mammalian cells.Citation17 Their study suggests that RAB5 carries out this role through acting on PIK3C3 kinase and its PtdIns3P production. All of these results indicate that fine-tuning of RAB cascades is important for their proper autophagic function.
Although RAB4, RAB5, and RAB7 were implicated in various autophagic roles and several of their GEFs and GAPs are known, only 2 of these proteins were studied in the context of autophagy and implicated in the regulation of this process (). A recent study showed that in the nematode Caenorhabditis elegans the siRNA-mediated silencing of a well-known RAB5 GEF, RABGEF1/RABEX5 results in an enhanced autophagy.Citation77 Disruption of the ubiquitin-binding domain of RABGEF1 is responsible for autophagy induction, suggesting that this motif can be responsible for switching the involvement of RABGEF1 between autophagy and the endocytic pathway. Another RAB5 GEF protein, ALS2/Alsin, was observed as a key participant of late stages of autophagy.Citation78 Missense mutation of ALS2 leads to decreased amphisome formation and impaired autophagic degradation. However, it is possible that ALS2 has only an indirect impact on autophagy through its role in endosome maturation. As endosomal RABs have important functions in autophagy regulation, clarifying and further investigating their GAP and GEF proteins in this process can be essential for understanding the distinct roles of RAB5 and RAB7 cascades in orchestrating autophagy.
Table 2. Rab GEFs and GAPs and their role in autophagy
The Golgi Gatekeepers: RAB1 and RAB11
Yeast Ypt1 and its mammalian homolog RAB1 mediate the vesicular transport between the endoplasmic reticulum (ER) and Golgi apparatus. RAB1 participates in vesicle trafficking and—through its interaction partners—facilitates tethering and subsequent fusion of ER-derived vesicles with the cis-Golgi.Citation79 Recently, RAB1 was described as an important factor of autophagosome formation as well, since depletion of functional RAB1 protein leads to an impairment in this process.Citation5
Several studies revealed the existence of special ER sites where the phagophore is formed from a potentially ER-derived membrane source.Citation80-Citation82 Upon autophagy induction, ATG proteins are recruited to these PAS sites in a well-defined order and mediate the subsequent formation and completion of autophagosomes.Citation83 First, the members of the ULK1/2-RB1CC1 (Atg1-Atg17 in yeast) complex gather to the ER-associated autophagic sites. This is followed by the recruitment of the ATG14-containing BECN1-complex and PtdIns3P production due to PIK3C3 activity. In yeast, Atg17 functions as a scaffold protein during autophagosome biogenesis.Citation84 Upon autophagy induction it binds to Atg29 and Atg31. This interaction is required for directing Atg9-decorated vesicles to the PAS and for phagophore organization. ATG9/Atg9, the only known transmembrane protein required for autophagosome formation, primarily localizes to recycling endosomes and small Golgi-derived vesicles, shuttling between peripheral cytoplasmic sites and the PAS, and also participates in providing a membrane source for autophagosomes.Citation85,Citation86
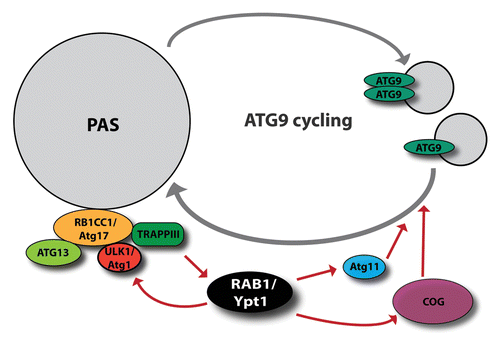
Recently, Wang and colleagues found that Atg17 directly binds to the well-known RAB1/Ypt1 GEF, TRAPPIII (transport protein particle III) thereby recruiting it to the PAS.Citation6 Once TRAPPIII is bound to Atg17, it activates Ypt1, which subsequently recruits Atg1 to the PAS. The specificity of Atg1 recruitment is indicated by the observation that overexpression of Ypt1 results in a significant increase in the presence of Atg1 at the PAS and in enhanced autophagy, while there is no detectable change in the amount of other Atg proteins on the phagophore. However, while in yeast there are 3 TRAPP complexes in existence, only TRAPPIII and its specific component, Trs85 were found to be involved in the early stages of both nonselective autophagy and pexophagy, the selective engulfment of peroxisomes.Citation7 In line with these findings, Meiling-Wesse and colleagues showed that lack of Trs85 lead to defects both in autophagosome formation and the cytoplasm to vacuole targeting (Cvt) pathway, since recruitment of Atg8 is impaired in these mutant yeast cells.Citation8 In addition, a further study reported that Trs85 is responsible for the PAS targeting of TRAPPIII and the subsequent recruitment and activation of Ypt1.Citation9
In a recent study, Atg11 was identified as an effector of the yeast Ypt1.Citation10 Atg11, a scaffold protein at the PAS, is involved in different types of selective autophagy, for instance in the Cvt pathway.Citation87 Moreover, Atg11 interacts with Atg9 and regulates its cycling through the PAS during selective autophagy.Citation88 In addition to these results, the conserved oligomeric Golgi (COG) complex, another known effector of Ypt1,Citation89 is also implicated in autophagy.Citation11 This study showed that COG localizes to the PAS where it interacts with Atg proteins. Furthermore, COG is required for the proper subcellular localization of Atg9. Supporting these results, RAB1 is required for selective autophagy in higher eukaryotic cells as well: it was described as a key regulator of Salmonella clearance in mammalian cells.Citation90 A further study indicated a role for RAB1A in homeostasis of SNCA/α-synuclein the major component of Lewy bodies (protein aggregates specific for Parkinson disease).Citation91
Taken together, these studies suggest a key role for RAB1/Ypt1 in the organization of the very early steps of autophagy (). Once Atg17—the most upstream factor in the regulation of autophagosome formation—is attached to the PAS, besides other ATG proteins it recruits the TRAPPIII complex, which subsequently activates Ypt1. Through its effector proteins, Ypt1 orchestrates autophagosome biogenesis: it recruits Atg11 and COG, both of which participate in the regulation of Atg9-dependent membrane transport to the PAS, thereby providing a membrane source for the forming autophagosomes. Parallel with these, Ypt1 recruits to the PAS Atg1 kinase, a core component of autophagy induction complex, which is required for the regulation of phagophore formation and expansion.
Figure 3. RAB1/Ypt1 has a key role in autophagosome formation. Through its interactors, the conserved oligomeric Golgi (COG) complex and Atg11, it facilitates ATG9/Atg9 cycling. Moreover, activated by the RAB1 GEF, TRAPPIII, RAB1 participates in the recruitment of ULK1/Atg1 to the PAS.
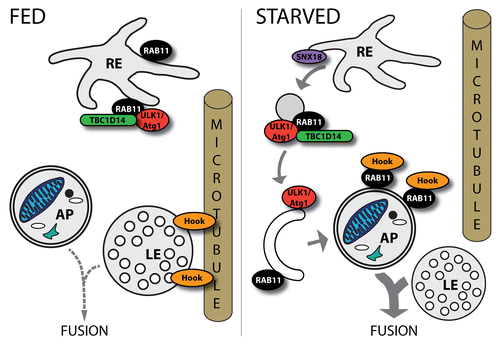
Ypt31/32, the yeast homologs of RAB11 have a key role in trafficking of secretory vesicles from the Golgi to the plasma membrane,Citation92 whereas RAB11 is a widely used marker of recycling endosomes (REs) in mammals and is required for cargo transport from REs toward the plasma membrane.Citation93 As was mentioned above, ATG9-marked vesicles can derive not only from Golgi, but also from REs: Longatti and colleagues found that ATG9 localizes to RAB11-positive REs.Citation41 This study revealed a role for the RAB GAP TBC1D14 as an effector of RAB11 in the early steps of autophagosome formation (). The study showed that TBC1D14 binds ULK1 kinase and this interaction is responsible for the recycling endosomal localization of ULK1. Furthermore, loss of RAB11 results in impairment of TBC1D14-driven RE tubulation and autophagosome formation. All of these data suggest that REs are a possible membrane source of autophagy and RAB11 is required for the transport of RE-derived membrane to the autophagosome precursors. A similar role for RAB11 was suggested in a recent work in Drosophila melanogaster showing that RAB11, together with the Drosophila homolog of sorting nexin 18 (SNX18), participates in membrane traffic from REs to the autophagosome formation sites, providing a RE-derived membrane source for phagophore expansion.Citation42 Moreover, Puri and colleagues demonstrated that several autophagic membrane sources converge at the level of REs and the fusion of different populations of plasma membrane-derived vesicles is essential for autophagosome formation.Citation43 They showed that ATG9 participates in membrane transport from ATG16L1-decorated autophagic precursor vesicles to the REs, where these carriers fuse with endosomes in a SNARE-dependent manner. They observed the correlation of these fusion events with the rate of autophagosome formation and upon autophagy induction they found an increased fusion of vesicles serving as a membrane source of autophagosomes and a decreased recycling, suggesting that this mechanism has a role in the regulation of early autophagic steps. Taken together these results suggest that RAB11 may directly contribute to autophagosome formation through providing various membrane sources for the phagophore.
Figure 4. Roles of RAB11 in the regulation of autophagy. RAB11 seems to have different roles in distinct steps of autophagy. Together with TBC1D14 it plays a role in membrane trafficking from the recycling endosome (RE) to the PAS. SNX18 is also implicated in this process. Furthermore, upon autophagy induction by starvation, RAB11 removes Hook—a negative regulator of endosome maturation—from LEs allowing subsequent fusion of a LE with an autophagosome (AP).
In addition to these, both Rab1 and Rab11 were described as required factors for TORC1 activity in Drosophila, since knockdown of these proteins decreases significantly the phosphorylation of RPS6KB/S6K, a well-known TOR target.Citation12
Interestingly, besides autophagosome formation, RAB11 seems to play an important role in another stage of autophagy. It was previously shown that RAB11 is required for the fusion of autophagosomes with multivesicular bodies (MVBs).Citation44-Citation46 These studies reported that depletion of RAB11 results in the decreased convergence of RAB11-decorated MVBs with LC3-labeled autophagosomes. Furthermore, this role of RAB11 may be affected by the toxic aggregates of the HTT/huntingtin protein during Huntington disease (HD), which can lead to neurodegeneration. In line with these studies, our latest work revealed a role for Rab11 in amphisome formation in Drosophila, since loss of functional Rab11 protein resulted in the accumulation of autophagosomes and enlarged, acidic late endosomes.Citation47 Our results suggest that upon autophagy induction Rab11 translocates from REs to autophagosomes labeled with Atg8a, where it physically interacts with the microtubule binding protein Hook, a negative regulator of endosome maturation. This interaction presumably prevents Hook from anchoring late endosomes to microtubules thereby allowing subsequent endosome-autophagosome fusion. These data suggest that upon autophagy induction, when the maturation of autophagosomes requires an increased input from the endolysosomal system, Rab11 can facilitate endosome maturation by regulating the subcellular localization of Hook ().
Although, both RAB1 and RAB11 have several known upstream regulators, only the RAB1 GEF TRAPPIII was described as a player in autophagy (). TRAPP complexes are common GEFs of the yeast homologs of RAB1 and RAB11, Ypt1 and Ypt31/32, respectively.Citation94,Citation95 Moreover, these small GTPases were found in the same Rab cascade in yeast: the GEF of Ypt32 is a putative effector of Ypt1.Citation96 In addition to this, Ypt32 facilitates the proper localization and activity of the Ypt1 GAP, and Gyp1.Citation97 Taken together, these results suggest the existence of a Rab conversion mechanism in the yeast secretory pathway, during which Ypt1 is substituted with Ypt32. The close relationship of these 2 proteins and their involvement at the same stage of autophagy raises the possibility that more of their upstream regulators may have a role in this process, but this question requires further investigation.
The Other Golgi RABs: RAB9 and RAB33
RAB9 functions at the level of late endosomes in cargo recycling toward the trans-Golgi network (TGN).Citation98 Similarly to RAB7, RAB9 plays a role in the autophagic clearance of GAS.Citation39 RAB9A is recruited to matured GAS-containing autophagosomes and is crucial for homotypic and lysosomal fusion of these structures. Supporting these findings, depletion of RAB9A lead to the impaired degradation of GAS. A former study found that RAB9, facilitating the fusion of TGN and late endosome-derived vesicles, is required for autophagosome formation during the process of ATG5- and ATG7-independent noncanonical autophagy.Citation40 This study showed that RAB9 localizes to LAMP2-positive autolysosomes. Knockdown of RAB9 leads to a decrease in the number of autophagic structures, but increases the number of phagophores, indicated by ultrastructural studies, suggesting that this GTPase is required for the early steps of autophagy. The exact molecular mechanism of the contribution of RAB9 to autophagosome formation remains unknown, although this study suggested that it directly participates in providing a TGN-derived membrane source for phagophore expansion. The upstream effectors of RAB9 are poorly characterized; its only known GEF is DENND2A,Citation99 but it has not yet been implicated in autophagy.
RAB33A and RAB33B are Golgi-resident RAB proteins with a well-defined role in the retrograde transport between the Golgi and ER.Citation100 Recently, it was described that expression of constitutively active RAB33B suppressed autophagy, whereas its depletion has no detectable effect on it.Citation51 This study suggests that RAB33B has a role in autophagosome formation, likely through its interaction with ATG16L1. Consistent with this, the RAB33 GAP TBC1D25/OATL1 was identified as an LC3-binding protein.Citation52 This study showed that OATL1 is recruited to the autophagosomes through its physical interaction with LC3. The GAP activity on RAB33B of this protein was found to be required for the proper fusion of autophagosomes with lysosomes. These results suggest a possible feedback loop in autophagy regulation by RAB33B, which is also involved in autophagosome maturation, since the overexpression of RAB33B or TBC1D25 inhibits the fusion of autophagosomes with lysosomes. However, the molecular mechanism by which RAB33B contributes to the regulation of autophagy is still unknown.
Further RABs involved in Autophagy: RAB8, RAB24, and RAB32
RAB8 has a well-defined role in vesicle transport toward the plasma membrane both in constitutive biosynthetic and RAB11-independent recycling pathways.Citation101-Citation103 Similarly to these functions, RAB8A was recently implicated in autophagy-based unconventional secretion the of proinflammatory cytokine, IL1B.Citation37 This study showed that autophagy-induction enhances the secretion of IL1B. This process depends on inflammasomes, ATG5, CTSD/cathepsin D, and RAB8A as well. Furthermore, through its effector, the innate immunity regulator TBK1 (TANK-binding kinase 1), RAB8B appears to have a role in the maturation of Mycobacterium-containing autophagosomes.Citation38 TBK1 is recruited to autophagic structures by RAB8B and it facilitates selective antibacterial autophagy through activating the autophagic adaptor protein SQSTM1/p62 by phosphorylation. How RAB8 is recruited to autophagosomes and whether it has other roles in the regulation of selective or nonselective autophagy requires further investigation.
RAB24 is detected on ER, cis-Golgi and their intermediate compartment and it colocalizes with the autophagy marker LC3.Citation104 The frequency of this colocalization increased upon starvation-induced autophagy and, moreover, overexpression of RAB24 results in an increased number of autophagic structures.Citation48 This observation suggests that RAB24 has a role in autophagy; however clarifying its exact function needs further research.
RAB32 has a key role in the biogenesis of lysosome-related organelles, such as melanosomesCitation105 and it was recently described as a regulator of ER-mitochondria interaction at mitochondria-associated membrane sitesCitation106 and also as a required factor for autophagosome formation.Citation49 This study reported that overexpression of RAB32 leads to an increase in the number of LC3-positive autophagic structures. They also found the ER localization of RAB32 to be required for autophagosome formation, suggesting that it has a role in the very early steps of autophagy by providing an ER-derived membrane source for the phagophore. In line with these, depletion of RAB32 results in the impaired autophagic clearance of SQSTM1-attached ubiquitinated proteins. These findings are in line with the recent observations of Hamasaki and colleagues, which suggested that autophagosome formation can occur at ER-mitochondria contact sites.Citation107 Taken together, these results raise the possibility that RAB32 may have a role in this mechanism. Supporting these findings, a further study showed that RAB32 and its GEF, Claret, are required for normal size of lipid droplets and also for autophagy in Drosophila.Citation50
Autophagic RABs in Pathological Conditions
Several RAB proteins described in the previous sections are implicated in various acquired and genetic diseases. Impairments in regulation or proper function of these proteins can result in autophagic failures and altered cell physiology.
Autophagy is important in the clearance of toxic protein aggregates and damaged organelles, so under- or overactivity of this process often underlies the pathology of various neurodegenerative diseases. A well-characterized example is the involvement of autophagy in removing of SNCA in Parkinson disease (PD).Citation108 A previous study showed that overexpression of RAB1 is able to rescue the neurodegenerative phenotype in PD models associated with SNCA accumulation.Citation109 According to a recent observation, both SNCA overexpression and RAB1A-depletion result in increased accumulation of autophagic substrates and mislocalization of ATG9, suggesting the failure of autophagosome formation.Citation91 In line with these findings, SNCA aggregation leads to Golgi fragmentation, a known symptom of PD, due to disruption of RAB1 homeostasis.Citation110 Similarly, a protective role was revealed for Rab8A in the case of SNCA toxicity.Citation111
Huntington disease (HD) is another well-known neurodegenerative disorder characterized by the accumulation of HTT-containing protein aggregates. HTT was previously described as a GEF for RAB11.Citation112 Several studies showed that RAB11 dysfunction or loss of its activity underlies the cellular pathomechanism of HDCitation113-Citation115 and in various HD models, RAB11 is able to rescue the neuronal loss.Citation46,Citation116 Besides RAB11, RAB8 is also involved in HD and its mislocalization results in perturbation of post-Golgi trafficking to lysosomesCitation117 that may affect autophagic degradation. Dysregulation of the autophagic process is found in HD as well.Citation118 However, the issue of whether inefficient autophagy is a cause or an implication of neurodegenerative diseases still remains open. Further investigation of the role of autophagic RABs in these disorders may provide a deeper insight into this question.
Autophagy impairment has also been involved in neurodegeneration observed in lysosomal storage disorders, such as Niemann–Pick Type C disease, which is associated with cholesterol and sphingolipid accumulation in late endosomes and lysosomes.Citation119,Citation120 Interestingly, both RAB8Citation121 and RAB9Citation122,Citation123 are able to rescue the defects in lipid trafficking in various Niemann–Pick Type C disease models.
Concluding Remarks
The amount of data concerning the possible involvement of RAB proteins in the regulation of autophagy has grown almost exponentially during the past several years. These basic observations opened a new, promising and fascinating field of autophagy research. The results summarized above clearly show that a specific set of RAB small GTPases have a deep impact in the molecular mechanisms of the autophagic process (). RAB proteins implicated in autophagy can contribute to this process directly and indirectly as well. RAB5 and RAB7 participate in several steps of the autophagic process: both of them take part in the upstream regulation of autophagy induction and in the formation of autophagosomes. RAB7 has further roles in the maturation and trafficking process of autophagosomes and amphisomes. Although RAB5, as a part of the BECN1-PIK3C3 complex, regulates directly autophagosome formation, the contribution of RAB7 to the maturation process of autophagosomes seems to be more indirect and dependent on its endosomal functions.
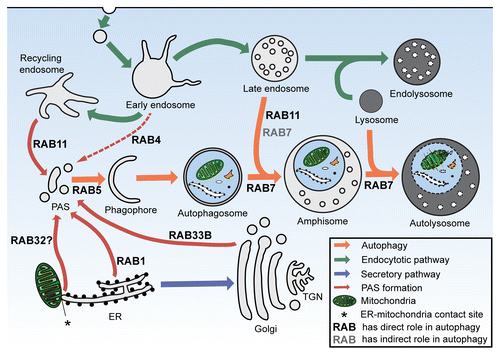
Figure 5. The functions of RAB proteins in autophagy. The schematic picture shows the interactions of autophagy with the endocytic and secretory pathways. A set of RAB small GTPases play a role in the earliest steps of autophagosome formation by providing various membrane sources for the PAS. RAB5 also participates in this autophagic stage, through its interactions with the PIK3C3-BECN1 complex. RAB7 has both direct roles—in the transport of autophagosomes and amphisomes—and indirect roles in the fusion process of autophagosomes with late endosomes. Besides its roles in PAS formation, RAB11 regulates amphisome formation and coordinates the autophagic and endosomal pathways. ER, endoplasmic reticulum; TGN, trans-Golgi network; PAS, phagophore assembly site.
RAB1 and RAB11 are undoubtedly among the key participants of the earliest steps of autophagosome formation through providing membrane from various sources for the expansion of the phagophore. On top of these, RAB1 has a well-characterized role in the recruitment of several ATG proteins and in the regulation of membrane transport toward the PAS. Furthermore, RAB11, through the regulation of the subcellular localization of Hook protein, may coordinate the maturation process of autophagic and endosomal pathways.
Two Golgi-localized RABs, RAB9 and RAB33, are also implicated in autophagy. RAB9 has a well-defined role in autophagosome formation, although, the molecular mechanism is as yet poorly understood. However, as a previous study suggested, it may have a direct effect on autophagy by providing a TGN-derived membrane source for phagophore expansion. RAB33B and its GAP, TBC1D25, are involved both in the early and late stages of autophagy, but their exact roles in these processes are still unknown. Some observations suggest that other RABs (RAB8, RAB24, and RAB32) might also be involved in the regulation of autophagy, but their contribution requires further investigation. In spite of numerous studies focused on the role of small GTPases in autophagy, the role of their GAPs and GEFs in autophagy remained unclear. Recently, several observations showed that previously unknown RAB regulators are able to interact with core components of the autophagic machinery and have an important role in orchestrating this degradative route.Citation42,Citation124 However, the work to date represents only the first steps in understanding the molecular basis of the dynamic interactions between the RAB cascades and core autophagic machinery. As several RABs are implicated in various diseases, identifying unknown regulators and mechanisms may establish the promising development of more specific drugs and therapeutic approaches.
| Abbreviations: | ||
| ATG | = | autophagy-related |
| COG | = | conserved oligomeric Golgi |
| EE | = | early endosome |
| ER | = | endoplasmic reticulum |
| GAP | = | GTPase activating protein |
| GAS | = | Group A Streptococcus |
| GDP | = | guanosine diphosphate |
| GEF | = | guanosine nucleotide exchange factor |
| GTP | = | guanosine triphosphate |
| HD | = | Huntington disease |
| HOPS | = | homotypic fusion and vacuolar protein sorting complex |
| (M)TOR | = | (mechanistic) target of rapamycin |
| (M)TORC1 | = | (mechanistic) TOR complex 1 |
| PAS | = | phagophore assembly site |
| PD | = | Parkinson disease |
| PtdIns3P | = | phosphatidylinositol 3-phosphate |
| PIK3C3 | = | class III phosphatidylinositol 3 kinase |
| RE | = | recycling endosome |
| SNARE | = | SNAP (soluble NSF attachment protein) receptor |
| TRAPP | = | transport protein particle |
| ULK | = | unc-51 like autophagy activating kinase |
| UVRAG | = | UV radiation resistance associated |
| VPS | = | vacuolar protein sorting |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27 - 42; http://dx.doi.org/10.1016/j.cell.2007.12.018; PMID: 18191218
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107 - 32; http://dx.doi.org/10.1146/annurev-cellbio-092910-154005; PMID: 21801009
- Tong J, Yan X, Yu L. The late stage of autophagy: cellular events and molecular regulation. Protein Cell 2010; 1:907 - 15; http://dx.doi.org/10.1007/s13238-010-0121-z; PMID: 21204017
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009; 10:513 - 25; http://dx.doi.org/10.1038/nrm2728; PMID: 19603039
- Zoppino FCM, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 2010; 11:1246 - 61; http://dx.doi.org/10.1111/j.1600-0854.2010.01086.x; PMID: 20545908
- Wang J, Menon S, Yamasaki A, Chou H-T, Walz T, Jiang Y, Ferro-Novick S. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci U S A 2013; 110:9800 - 5; http://dx.doi.org/10.1073/pnas.1302337110; PMID: 23716696
- Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy 2005; 1:37 - 45; http://dx.doi.org/10.4161/auto.1.1.1512; PMID: 16874038
- Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen E-L, Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem 2005; 280:33669 - 78; http://dx.doi.org/10.1074/jbc.M501701200; PMID: 16079147
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A 2010; 107:7811 - 6; http://dx.doi.org/10.1073/pnas.1000063107; PMID: 20375281
- Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci U S A 2012; 109:6981 - 6; http://dx.doi.org/10.1073/pnas.1121299109; PMID: 22509044
- Yen W-L, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol 2010; 188:101 - 14; http://dx.doi.org/10.1083/jcb.200904075; PMID: 20065092
- Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan K-L. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem 2010; 285:19705 - 9; http://dx.doi.org/10.1074/jbc.C110.102483; PMID: 20457610
- Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 2008; 121:1649 - 60; http://dx.doi.org/10.1242/jcs.025726; PMID: 18430781
- Su W-C, Chao T-C, Huang Y-L, Weng S-C, Jeng K-S, Lai MMC. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol 2011; 85:10561 - 71; http://dx.doi.org/10.1128/JVI.00173-11; PMID: 21835792
- Dou Z, Chattopadhyay M, Pan J-A, Guerriero JL, Jiang Y-P, Ballou LM, Yue Z, Lin RZ, Zong W-X. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol 2010; 191:827 - 43; http://dx.doi.org/10.1083/jcb.201006056; PMID: 21059846
- Dou Z, Pan J-A, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, Chen J-S, Liang Z, Li G, Backer JM, et al. Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell 2013; 50:29 - 42; http://dx.doi.org/10.1016/j.molcel.2013.01.022; PMID: 23434372
- Bridges D, Fisher K, Zolov SN, Xiong T, Inoki K, Weisman LS, Saltiel AR. Rab5 proteins regulate activation and localization of target of rapamycin complex 1. J Biol Chem 2012; 287:20913 - 21; http://dx.doi.org/10.1074/jbc.M111.334060; PMID: 22547071
- Talaber G, Miklossy G, Oaks Z, Liu Y, Tooze SA, Chudakov DM, Banki K, Perl A. HRES-1/Rab4 promotes the formation of LC3(+) autophagosomes and the accumulation of mitochondria during autophagy. PLoS One 2014; 9:e84392; http://dx.doi.org/10.1371/journal.pone.0084392; PMID: 24404161
- Bains M, Zaegel V, Mize-Berge J, Heidenreich KA. IGF-I stimulates Rab7-RILP interaction during neuronal autophagy. Neurosci Lett 2011; 488:112 - 7; http://dx.doi.org/10.1016/j.neulet.2010.09.018; PMID: 20849920
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjørkøy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 2010; 188:253 - 69; http://dx.doi.org/10.1083/jcb.200907015; PMID: 20100911
- Maday S, Wallace KE, Holzbaur ELF. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 2012; 196:407 - 17; http://dx.doi.org/10.1083/jcb.201106120; PMID: 22331844
- Tabata K, Matsunaga K, Sakane A, Sasaki T, Noda T, Yoshimori T. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol Biol Cell 2010; 21:4162 - 72; http://dx.doi.org/10.1091/mbc.E10-06-0495; PMID: 20943950
- Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A 2010; 107:19338 - 43; http://dx.doi.org/10.1073/pnas.1010554107; PMID: 20974968
- Hyttinen JMT, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 2013; 1833:503 - 10; http://dx.doi.org/10.1016/j.bbamcr.2012.11.018; PMID: 23220125
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147:435 - 46; http://dx.doi.org/10.1083/jcb.147.2.435; PMID: 10525546
- Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 2004; 117:2687 - 97; http://dx.doi.org/10.1242/jcs.01114; PMID: 15138286
- Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen E-L. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 2004; 117:4837 - 48; http://dx.doi.org/10.1242/jcs.01370; PMID: 15340014
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3:452 - 60; PMID: 17534139
- Su H, Li F, Ranek MJ, Wei N, Wang X. COP9 signalosome regulates autophagosome maturation. Circulation 2011; 124:2117 - 28; http://dx.doi.org/10.1161/CIRCULATIONAHA.111.048934; PMID: 21986281
- Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell 2010; 21:1001 - 10; http://dx.doi.org/10.1091/mbc.E09-08-0693; PMID: 20089838
- Ganley IG, Wong P-M, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 2011; 42:731 - 43; http://dx.doi.org/10.1016/j.molcel.2011.04.024; PMID: 21700220
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010; 465:942 - 6; http://dx.doi.org/10.1038/nature09076; PMID: 20526321
- Lin W-J, Yang CY, Li LL, Yi YH, Chen KW, Lin Y-C, Liu CC, Lin CH. Lysosomal targeting of phafin1 mediated by Rab7 induces autophagosome formation. Biochem Biophys Res Commun 2012; 417:35 - 42; http://dx.doi.org/10.1016/j.bbrc.2011.11.043; PMID: 22115783
- Sakurai A, Maruyama F, Funao J, Nozawa T, Aikawa C, Okahashi N, Shintani S, Hamada S, Ooshima T, Nakagawa I. Specific behavior of intracellular Streptococcus pyogenes that has undergone autophagic degradation is associated with bacterial streptolysin O and host small G proteins Rab5 and Rab7. J Biol Chem 2010; 285:22666 - 75; http://dx.doi.org/10.1074/jbc.M109.100131; PMID: 20472552
- Yamaguchi H, Nakagawa I, Yamamoto A, Amano A, Noda T, Yoshimori T. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog 2009; 5:e1000670; http://dx.doi.org/10.1371/journal.ppat.1000670; PMID: 19956673
- Flinn RJ, Yan Y, Goswami S, Parker PJ, Backer JM. The late endosome is essential for mTORC1 signaling. Mol Biol Cell 2010; 21:833 - 41; http://dx.doi.org/10.1091/mbc.E09-09-0756; PMID: 20053679
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J 2011; 30:4701 - 11; http://dx.doi.org/10.1038/emboj.2011.398; PMID: 22068051
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012; 37:223 - 34; http://dx.doi.org/10.1016/j.immuni.2012.04.015; PMID: 22921120
- Nozawa T, Aikawa C, Goda A, Maruyama F, Hamada S, Nakagawa I. The small GTPases Rab9A and Rab23 function at distinct steps in autophagy during Group A Streptococcus infection. Cell Microbiol 2012; 14:1149 - 65; http://dx.doi.org/10.1111/j.1462-5822.2012.01792.x; PMID: 22452336
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009; 461:654 - 8; http://dx.doi.org/10.1038/nature08455; PMID: 19794493
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 2012; 197:659 - 75; http://dx.doi.org/10.1083/jcb.201111079; PMID: 22613832
- Knævelsrud H, Søreng K, Raiborg C, Håberg K, Rasmuson F, Brech A, Liestøl K, Rusten TE, Stenmark H, Neufeld TP, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol 2013; 202:331 - 49; http://dx.doi.org/10.1083/jcb.201205129; PMID: 23878278
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013; 154:1285 - 99; http://dx.doi.org/10.1016/j.cell.2013.08.044; PMID: 24034251
- Fader CM, Sánchez D, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 2008; 9:230 - 50; http://dx.doi.org/10.1111/j.1600-0854.2007.00677.x; PMID: 17999726
- Fader CM, Sánchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta 2009; 1793:1901 - 16; http://dx.doi.org/10.1016/j.bbamcr.2009.09.011; PMID: 19781582
- Richards P, Didszun C, Campesan S, Simpson A, Horley B, Young KW, Glynn P, Cain K, Kyriacou CP, Giorgini F, et al. Dendritic spine loss and neurodegeneration is rescued by Rab11 in models of Huntington’s disease. Cell Death Differ 2011; 18:191 - 200; http://dx.doi.org/10.1038/cdd.2010.127; PMID: 21217767
- Szatmári Z, Kis V, Lippai M, Hegedus K, Faragó T, Lorincz P, Tanaka T, Juhász G, Sass M. Rab11 facilitates cross-talk between autophagy and endosomal pathway through regulation of Hook localization. Mol Biol Cell 2014; 25:522 - 31; http://dx.doi.org/10.1091/mbc.E13-10-0574; PMID: 24356450
- Munafó DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic 2002; 3:472 - 82; http://dx.doi.org/10.1034/j.1600-0854.2002.30704.x; PMID: 12047555
- Hirota Y, Tanaka Y. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell Mol Life Sci 2009; 66:2913 - 32; http://dx.doi.org/10.1007/s00018-009-0080-9; PMID: 19593531
- Wang C, Liu Z, Huang X.. Rab32 Is Important for Autophagy and Lipid Storage in Drosophila. 2012; 7:1 - 9
- Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell 2008; 19:2916 - 25; http://dx.doi.org/10.1091/mbc.E07-12-1231; PMID: 18448665
- Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol 2011; 192:839 - 53; http://dx.doi.org/10.1083/jcb.201008107; PMID: 21383079
- Numrich J, Ungermann C. Endocytic Rabs in membrane trafficking and signaling. Biol Chem 2014; 395:327 - 33; http://dx.doi.org/10.1515/hsz-2013-0258; PMID: 24158421
- Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic 2002; 3:416 - 27; http://dx.doi.org/10.1034/j.1600-0854.2002.30605.x; PMID: 12010460
- van der Sluijs P, Hull M, Webster P, Mâle P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 1992; 70:729 - 40; http://dx.doi.org/10.1016/0092-8674(92)90307-X; PMID: 1516131
- Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol 2000; 149:901 - 14; http://dx.doi.org/10.1083/jcb.149.4.901; PMID: 10811830
- Huotari J, Helenius A. Endosome maturation. EMBO J 2011; 30:3481 - 500; http://dx.doi.org/10.1038/emboj.2011.286; PMID: 21878991
- Wang T, Ming Z, Xiaochun W, Hong W. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal 2011; 23:516 - 21; http://dx.doi.org/10.1016/j.cellsig.2010.09.012; PMID: 20851765
- Johansson M, Lehto M, Tanhuanpää K, Cover TL, Olkkonen VM. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell 2005; 16:5480 - 92; http://dx.doi.org/10.1091/mbc.E05-03-0189; PMID: 16176980
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol 2007; 176:459 - 71; http://dx.doi.org/10.1083/jcb.200606077; PMID: 17283181
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 2001; 11:1680 - 5; http://dx.doi.org/10.1016/S0960-9822(01)00531-0; PMID: 11696325
- Kim BY, Krämer H, Yamamoto A, Kominami E, Kohsaka S, Akazawa C. Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J Biol Chem 2001; 276:29393 - 402; http://dx.doi.org/10.1074/jbc.M101778200; PMID: 11382755
- Starai VJ, Hickey CM, Wickner W. HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. Mol Biol Cell 2008; 19:2500 - 8; http://dx.doi.org/10.1091/mbc.E08-01-0077; PMID: 18385512
- Mizuno K, Kitamura A, Sasaki T. Rabring7, a novel Rab7 target protein with a RING finger motif. Mol Biol Cell 2003; 14:3741 - 52; http://dx.doi.org/10.1091/mbc.E02-08-0495; PMID: 12972561
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008; 10:776 - 87; http://dx.doi.org/10.1038/ncb1740; PMID: 18552835
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11:385 - 96; http://dx.doi.org/10.1038/ncb1846; PMID: 19270696
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009; 11:468 - 76; http://dx.doi.org/10.1038/ncb1854; PMID: 19270693
- Lin MG, Zhong Q. Interaction between small GTPase Rab7 and PI3KC3 links autophagy and endocytosis: A new Rab7 effector protein sheds light on membrane trafficking pathways. Small GTPases 2011; 2:85 - 8; http://dx.doi.org/10.4161/sgtp.2.2.15256; PMID: 21776407
- Razi M, Chan EYW, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol 2009; 185:305 - 21; http://dx.doi.org/10.1083/jcb.200810098; PMID: 19364919
- Köchl R, Hu XW, Chan EYW, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006; 7:129 - 45; http://dx.doi.org/10.1111/j.1600-0854.2005.00368.x; PMID: 16420522
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EMC, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 2007; 179:485 - 500; http://dx.doi.org/10.1083/jcb.200702115; PMID: 17984323
- Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun 1988; 151:40 - 7; http://dx.doi.org/10.1016/0006-291X(88)90556-6; PMID: 3126737
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012; 151:1256 - 69; http://dx.doi.org/10.1016/j.cell.2012.11.001; PMID: 23217709
- Takáts S, Nagy P, Varga Á, Pircs K, Kárpáti M, Varga K, Kovács AL, Hegedűs K, Juhász G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol 2013; 201:531 - 9; http://dx.doi.org/10.1083/jcb.201211160; PMID: 23671310
- Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 2008; 4:851 - 65; PMID: 18670193
- Jewell JL, Russell RC, Guan K-L. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 2013; 14:133 - 9; http://dx.doi.org/10.1038/nrm3522; PMID: 23361334
- Dwivedi M, Sung H, Shen H, Park B-J, Lee S. Disruption of endocytic pathway regulatory genes activates autophagy in C. elegans. Mol Cells 2011; 31:477 - 81; http://dx.doi.org/10.1007/s10059-011-1035-1; PMID: 21618079
- Otomo A, Kunita R, Suzuki-Utsunomiya K, Ikeda J-E, Hadano S. Defective relocalization of ALS2/alsin missense mutants to Rac1-induced macropinosomes accounts for loss of their cellular function and leads to disturbed amphisome formation. FEBS Lett 2011; 585:730 - 6; http://dx.doi.org/10.1016/j.febslet.2011.01.045; PMID: 21300063
- Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis--Golgi tethering. Traffic 2001; 2:268 - 76; http://dx.doi.org/10.1034/j.1600-0854.2001.1o007.x; PMID: 11285137
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685 - 701; http://dx.doi.org/10.1083/jcb.200803137; PMID: 18725538
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009; 11:1433 - 7; http://dx.doi.org/10.1038/ncb1991; PMID: 19898463
- Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009; 5:1180 - 5; http://dx.doi.org/10.4161/auto.5.8.10274; PMID: 19855179
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010; 6:764 - 76; http://dx.doi.org/10.4161/auto.6.6.12709; PMID: 20639694
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell 2012; 151:1501 - 12; http://dx.doi.org/10.1016/j.cell.2012.11.028; PMID: 23219485
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 2010; 190:1005 - 22; http://dx.doi.org/10.1083/jcb.200912089; PMID: 20855505
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 2012; 198:219 - 33; http://dx.doi.org/10.1083/jcb.201202061; PMID: 22826123
- Yorimitsu T, Klionsky DJ. Atg11 Links Cargo to the Vesicle-forming Machinery in the Cytoplasm to Vacuole Targeting Pathway. 2005; 16:1593 - 605
- He C, Song H, Yorimitsu T, Monastyrska I, Yen W-L, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol 2006; 175:925 - 35; http://dx.doi.org/10.1083/jcb.200606084; PMID: 17178909
- Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol 2002; 157:631 - 43; http://dx.doi.org/10.1083/jcb.200111081; PMID: 12011112
- Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, Campellone KG, Heo WD, Gruenheid S, Meyer T, et al. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 2011; 7:17 - 26; http://dx.doi.org/10.4161/auto.7.1.13840; PMID: 20980813
- Winslow AR, Chen C, Corrochano S, Acevedo-arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, et al. Alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. 2010; 190:1023 - 37
- Lipatova Z, Tokarev AA, Jin Y, Mulholland J, Weisman LS, Segev N. Direct Interaction between a Myosin V Motor and the Rab GTPases Ypt31 / 32 Is Required for Polarized Secretion. 2008; 19:4177 - 87
- Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol 2010; 22:528 - 34; http://dx.doi.org/10.1016/j.ceb.2010.05.008; PMID: 20541925
- Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell 2000; 11:4403 - 11; http://dx.doi.org/10.1091/mbc.11.12.4403; PMID: 11102533
- Morozova N, Liang Y, Tokarev AA, Chen SH, Cox R, Andrejic J, Lipatova Z, Sciorra VA, Emr SD, Segev N. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat Cell Biol 2006; 8:1263 - 9; http://dx.doi.org/10.1038/ncb1489; PMID: 17041589
- Wang W, Ferro-Novick S. A. Ypt32p Exchange Factor Is a Putative Effector of Ypt1p. 2002; 13:3336 - 43
- Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc Natl Acad Sci U S A 2009; 106:14408 - 13; http://dx.doi.org/10.1073/pnas.0906536106; PMID: 19666511
- Barbero P, Bittova L, Pfeffer SR. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J Cell Biol 2002; 156:511 - 8; http://dx.doi.org/10.1083/jcb.200109030; PMID: 11827983
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol 2010; 191:367 - 81; http://dx.doi.org/10.1083/jcb.201008051; PMID: 20937701
- Valsdottir R, Hashimoto H, Ashman K, Koda T, Storrie B, Nilsson T. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett 2001; 508:201 - 9; http://dx.doi.org/10.1016/S0014-5793(01)02993-3; PMID: 11718716
- Bravo-Cordero JJ, Marrero-Diaz R, Megías D, Genís L, García-Grande A, García MA, Arroyo AG, Montoya MC. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J 2007; 26:1499 - 510; http://dx.doi.org/10.1038/sj.emboj.7601606; PMID: 17332756
- Henry L, Sheff DR. Rab8 regulates basolateral secretory, but not recycling, traffic at the recycling endosome. Mol Biol Cell 2008; 19:2059 - 68; http://dx.doi.org/10.1091/mbc.E07-09-0902; PMID: 18287531
- Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell 2007; 18:2828 - 37; http://dx.doi.org/10.1091/mbc.E07-02-0169; PMID: 17507647
- Olkkonen VM, Dupree P, Killisch I, Lütcke A, Zerial M, Simons K. Molecular cloning and subcellular localization of three GTP-binding proteins of the rab subfamily. J Cell Sci 1993; 106:1249 - 61; PMID: 8126105
- Bultema JJ, Di Pietro SM. Cell type-specific Rab32 and Rab38 cooperate with the ubiquitous lysosome biogenesis machinery to synthesize specialized lysosome-related organelles. Small GTPases 2013; 4:16 - 21; http://dx.doi.org/10.4161/sgtp.22349; PMID: 23247405
- Bui M, Gilady SY, Fitzsimmons REB, Benson MD, Lynes EM, Gesson K, Alto NM, Strack S, Scott JD, Simmen T. Rab32 modulates apoptosis onset and mitochondria-associated membrane (MAM) properties. J Biol Chem 2010; 285:31590 - 602; http://dx.doi.org/10.1074/jbc.M110.101584; PMID: 20670942
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013; 495:389 - 93; http://dx.doi.org/10.1038/nature11910; PMID: 23455425
- Chua CEL, Gan BQ, Tang BL. Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci 2011; 68:3349 - 58; http://dx.doi.org/10.1007/s00018-011-0748-9; PMID: 21687989
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science 2006; 313:324 - 8; http://dx.doi.org/10.1126/science.1129462; PMID: 16794039
- Rendón WO, Martínez-Alonso E, Tomás M, Martínez-Martínez N, Martínez-Menárguez JA. Golgi fragmentation is Rab and SNARE dependent in cellular models of Parkinson’s disease. Histochem Cell Biol 2013; 139:671 - 84; http://dx.doi.org/10.1007/s00418-012-1059-4; PMID: 23212845
- Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, et al. The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci U S A 2008; 105:145 - 50; http://dx.doi.org/10.1073/pnas.0710685105; PMID: 18162536
- Li X, Sapp E, Valencia A, Kegel KB, Qin Z-H, Alexander J, Masso N, Reeves P, Ritch JJ, Zeitlin S, et al. A function of huntingtin in guanine nucleotide exchange on Rab11. Neuroreport 2008; 19:1643 - 7; http://dx.doi.org/10.1097/WNR.0b013e328315cd4c; PMID: 18845944
- Li X, Standley C, Sapp E, Valencia A, Qin Z-H, Kegel KB, Yoder J, Comer-Tierney LA, Esteves M, Chase K, et al. Mutant huntingtin impairs vesicle formation from recycling endosomes by interfering with Rab11 activity. Mol Cell Biol 2009; 29:6106 - 16; http://dx.doi.org/10.1128/MCB.00420-09; PMID: 19752198
- Li X, Valencia A, Sapp E, Masso N, Alexander J, Reeves P, Kegel KB, Aronin N, Difiglia M. Aberrant Rab11-dependent trafficking of the neuronal glutamate transporter EAAC1 causes oxidative stress and cell death in Huntington’s disease. J Neurosci 2010; 30:4552 - 61; http://dx.doi.org/10.1523/JNEUROSCI.5865-09.2010; PMID: 20357106
- Li X, Valencia A, McClory H, Sapp E, Kegel KB, Difiglia M. Deficient Rab11 activity underlies glucose hypometabolism in primary neurons of Huntington’s disease mice. Biochem Biophys Res Commun 2012; 421:727 - 30; http://dx.doi.org/10.1016/j.bbrc.2012.04.070; PMID: 22542623
- Steinert JR, Campesan S, Richards P, Kyriacou CP, Forsythe ID, Giorgini F. Rab11 rescues synaptic dysfunction and behavioural deficits in a Drosophila model of Huntington’s disease. Hum Mol Genet 2012; 21:2912 - 22; http://dx.doi.org/10.1093/hmg/dds117; PMID: 22466800
- Toro D, Alberch J, La F, Xifro X, Egea G, Canals JM. Mutant Huntingtin Impairs Post-Golgi Trafficking to Lysosomes by Delocalizing Optineurin / Rab8 Complex from the Golgi Apparatus. 2009; 20:1478 - 92
- Martinez-Vicente M, Tallóczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci 2010; 13:567 - 76; http://dx.doi.org/10.1038/nn.2528; PMID: 20383138
- Ko DC, Milenkovic L, Beier SM, Manuel H, Buchanan J, Scott MP. Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet 2005; 1:81 - 95; PMID: 16103921
- Sarkar S, Carroll B, Buganim Y, Maetzel D, Ng AHM, Cassady JP, Cohen MA, Chakraborty S, Wang H, Spooner E, et al. Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell Rep 2013; 5:1302 - 15; http://dx.doi.org/10.1016/j.celrep.2013.10.042; PMID: 24290752
- Linder MD, Uronen R, Ho M, Sluijs P, Van Der, Pera J, Ikonen E. Rab8-dependent Recycling Promotes Endosomal Cholesterol Removal in Normal and Sphingolipidosis Cells. 2007; 18:47 - 56
- Narita K, Choudhury A, Dobrenis K, Sharma DK, Holicky EL, Marks DL, Walkley SU, Pagano RE, Clinic M, Neuroscience M, et al. Protein transduction of Rab9 in Niemann-Pick C cells reduces cholesterol storage. 2005; 17:1 - 17
- Kaptzan T, West SA, Holicky EL, Wheatley CL, Marks DL, Wang T, Peake KB, Vance J, Walkley SU, Pagano RE. Development of a Rab9 transgenic mouse and its ability to increase the lifespan of a murine model of Niemann-Pick type C disease. Am J Pathol 2009; 174:14 - 20; http://dx.doi.org/10.2353/ajpath.2009.080660; PMID: 19056848
- Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol 2012; 32:1733 - 44; http://dx.doi.org/10.1128/MCB.06717-11; PMID: 22354992
