Abstract
High-resolution imaging of autophagy has been used intensively in cell culture studies, but so far it has been difficult to visualize this process in detail in whole animal models. In this study we present a versatile method for high-resolution imaging of microbial infection in zebrafish larvae by injecting pathogens into the tail fin. This allows visualization of autophagic compartments by light and electron microscopy, which makes it possible to correlate images acquired by the 2 techniques. Using this method we have studied the autophagy response against Mycobacterium marinum infection. We show that mycobacteria during the progress of infection are frequently associated with GFP-Lc3-positive vesicles, and that 2 types of GFP-Lc3-positive vesicles were observed. The majority of these vesicles were approximately 1 μm in size and in close vicinity of bacteria, and a smaller number of GFP-Lc3-positive vesicles was larger in size and were observed to contain bacteria. Quantitative data showed that these larger vesicles occurred significantly more in leukocytes than in other cell types, and that approximately 70% of these vesicles were positive for a lysosomal marker. Using electron microscopy, it was found that approximately 5% of intracellular bacteria were present in autophagic vacuoles and that the remaining intracellular bacteria were present in phagosomes, lysosomes, free inside the cytoplasm or occurred as large aggregates. Based on correlation of light and electron microscopy images, it was shown that GFP-Lc3-positive vesicles displayed autophagic morphology. This study provides a new approach for injection of pathogens into the tail fin, which allows combined light and electron microscopy imaging in vivo and opens new research directions for studying autophagy process related to infectious diseases.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a well-conserved cellular process that is aimed at targeting cytosolic components for lysosomal degradation. This process plays an important role in the maintenance of cellular homeostasis, inducing degradation of protein aggregates and damaged organelles.Citation1 Upon initiation of autophagy, a phagophore is expanded around cytosolic material to form double-membrane vesicles that are called autophagosomes, or initial autophagic vacuoles. After fusion with lysosomes, autolysosomes, also called degradative autophagic vacuoles, are formed and their content is degraded, which can be as large as mitochondria.Citation2-Citation5 Substrates can be targeted selectively for autophagic degradation by a molecular tag, like polyubiquitin. Subsequently, these tags are recognized by SQSTM1/p62-like receptors (SLRs), which link the ubiquitinated targets to autophagosome-associated proteins.Citation6 One of these proteins, microtubule-associated protein 1 light chain 3 (MAP1LC3, abbreviated as LC3), is involved in cargo recruitment and biogenesis of autophagosomes and has successfully been used as a marker for autophagic structures.Citation7-Citation9
In addition to its role in cytosolic homeostasis, autophagy is involved in the defense against intracellular microbes.Citation10,Citation11 Certain types of pathogenic bacterial species are able to escape from phagosomal compartments or inhibit the maturation of phagosomes by manipulating the cell’s molecular machinery. It has been shown that the autophagic mechanism can act as a secondary defense line against microbes that evade phagocytotic destruction.Citation12 Besides its role in microbial degradation, autophagy plays a role in the antibacterial defense mechanism by contributing to cytokine secretion and regulation of the immune response, and it may be involved in the clearance of cellular components that have been damaged as a result of the bacterial infection.Citation13,Citation14
Perhaps the most notorious of intracellular pathogens that can manipulate the host phagocytic process is Mycobacterium tuberculosis (Mtb), which is estimated to have infected a third of the world population and currently causes nearly 1.5 million deaths per year. In macrophages, Mtb prevents phagosome-lysosome fusion resulting in immature phagosomes, in which Mtb can survive and replicate.Citation15,Citation16 In some studies Mtb has been shown to escape from phagosomes into the cytoplasm.Citation17 This translocation requires the early secretory antigenic target 6 system 1 ESX-1, a type VII secretion system involved in general virulence of pathogenic mycobacterial species.Citation18 ESX-1 permeabilizes phagosomes triggering LC3 recruitment to Mtb-containing phagosomes and this secretion system is required for recognition of Mtb DNA by the host’s DNA sensing pathway initiating autophagy, which leads to degradation of Mtb in autolysosomes.Citation19 Induction of autophagy by starvation, inhibition of MTOR, or interferon-gamma treatment can stimulate phagosomal maturation and restrict mycobacterial replication.Citation11,Citation20,Citation21 In additional mechanistic studies, autophagy has been shown to produce autolysosomes containing antimicrobial peptides from specific ribosomal and ubiquitinated proteins capable of killing Mtb.Citation22,Citation23
Microscopy data on autophagy has mainly been generated using cell culture studies, and fewer studies have been performed on whole animal model systems. Besides the obvious increased validity of vertebrate animal models, these systems enable studying the autophagic response in different cell types and the interactions between these cell types. Ultimately, this may facilitate the development of novel therapies against autophagy-related disorders.Citation24-Citation26 In the present study, we have used the zebrafish animal model for high-resolution imaging of autophagy during bacterial infection. Zebrafish embryos and larvae are extensively used because of their visual transparency, allowing fluorescent imaging of a wide variety of disease processes.Citation27 The zebrafish is currently being used as a model system to study autophagy in different fields, such as development,Citation28,Citation29 neurodegeneration,Citation25,Citation30 and infection.Citation31 An important tool is the GFP-Lc3 transgenic zebrafish line, Tg(CMV:EGFP-map1lc3b), which enables the visualization of autophagosomal structures.Citation32 This line has previously been used to show the autophagy response against bacterial infection.Citation31,Citation33
The exact role of LC3 and its homologs in intracellular processes has not entirely been elucidated yet. It is clear that LC3 is not exclusively involved in biogenesis of autophagosomes, but is also involved in membrane expansion of other organelles.Citation34,Citation35 In innate immunity, it has been reported that the toll-like receptor signaling pathway triggers the recruitment of LC3 to phagosomal membranes.Citation36 In addition, LC3 is recruited to phagosome-enclosed apoptotic cells.Citation36,Citation37 In order to study autophagy in zebrafish, it is important to visualize this process by electron microscopy and to be able to correlate the obtained images to the images of GFP-Lc3 structures observed with light microscopy. This is especially important due to the current lack of antibodies that can be used as specific autophagy markers in the zebrafish model.
Zebrafish are a natural host for infection by M. marinum with similar phenotypes as Mtb infection in human tissues.Citation38,Citation39 Similar to Mtb, M. marinum can propagate in macrophages by preventing phagosome-lysosome fusion. The infected leukocytes subsequently attract other immune cells, leading to the formation of organized cellular aggregates called granulomas.Citation40 M. marinum can escape from the phagosome into the cytosol and develop actin-based motility.Citation41 Different procedures have been established to induce systemic M. marinum infection in zebrafish embryos and larvae, of which injection of bacteria into the caudal vein is most widely used.Citation42 However, high-resolution imaging of cellular processes during these systemic infections, such as autophagy, is complicated. Using light microscopy, the applicability of high numerical aperture (NA) lenses is limited when infected tissues are located deeper inside the organism, and out of focus interference greatly limits the resolution and contrast. Using electron microscopy, which is required to visualize cellular ultrastructures such as autophagic vacuoles, it is extremely labor-intensive to localize infected cells that are scattered throughout the body of the zebrafish larvae.
In this study we present a novel infection model for pathogens in zebrafish larvae. Microinjection of M. marinum directly into the tail fin of zebrafish larvae results in a highly localized infection. This enables studying intracellular processes during the entire course of the infection process, from the infection of a few cells until formation of a granuloma. In particular, the role of autophagy during this process can be investigated using visualization of the autophagosomal structures by high-resolution light and electron microscopy. Using this approach we show that autophagy is induced during the course of infection by M. marinum. Using light microscopy, a large number of small GFP-Lc3-positive structures (~1 μm) and a small number (± 6 per granuloma) of larger (~3 μm) GFP-Lc3-positive structures (containing bacteria) were observed in the infected tissue. These larger GFP-Lc3-positive structures containing bacteria were significantly more common in leukocytes than in the remaining cell types and mostly positive for the acidicity marker LysoTracker Red (LyTR). Using electron microscopy it was shown that about 5% of intracellular bacteria were present in autophagic vacuoles, and that the majority of these compartments had degradative autophagic vacuole morphology. Correlation of light and electron microscopy images showed that the small GFP-Lc3-positive vesicles in the vicinity of bacteria, as well as the larger GFP-Lc3 structure containing sequestered bacteria have autophagic vacuole morphology. Thus, our results show that by using the presented tail fin infection method, the autophagy process during the course of infection can be studied by direct observations in vivo.
Results
The tail fin infection model
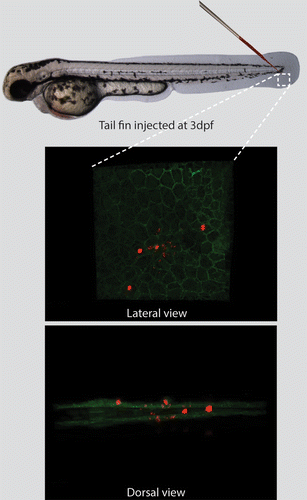
In the present study a model was set up which enabled studying autophagy during bacterial infection in zebrafish by visualizing intracellular structures using both light and electron microscopy. For this purpose a local infection was established using microinjection of bacteria into the tail fin of zebrafish larvae at 3 d post fertilization (dpf). At this stage the tail fin is approximately 20- to 50-μm thick. It consists of a thin layer containing mesenchymal cells, extracellular matrix, collagenous fibers (called actinotrichia), and an epidermis that consists of 2 cell layers covering the tail fin on both sides.Citation43 Several hours after injection of fluorescently labeled M. marinum in the tail fin (~500 colony-forming units), by confocal laser scanning microscopy (CLSM) a small number of bacteria can be visualized in the tail fins, which reside in epithelial cell layers and in the extracellular matrix. An overview of this model system is presented in , showing tail-fin infection of E2-Crimson labeled M. marinum in transgenic zebrafish larvae expressing membrane-bound GFP.Citation44 In order to visualize the ultrastructure of the infected tail fin, the infected larvae can subsequently be processed for transmission electron microscopy.
Figure 1. The tail-fin injection model, enabling the induction of a localized infection in zebrafish larvae. The needle indicates the location for injection in the tail fin of 3 dpf zebrafish larvae. The inset represents the region imaged by CLSM. The transgenic larva expressing membrane-bound GFP was injected with fluorescently labeled M. marinum (shown in red). The larva is imaged and presented from a lateral and dorsal perspective, showing the epithelial cell layers and the bacteria residing in these layers and in the extracellular space between these layers.
Injection of M. marinum in zebrafish larvae results in an innate immune response represented by recruitment of leukocytes to the site of infection and subsequent formation of granulomas.Citation40,Citation45 In order to study the course of infection and leukocyte recruitment in the tail fin infection model, we injected E2-Crimson labeled M. marinum into the tail fin of 3 dpf zebrafish larvae and visualized neutrophils and macrophages. This visualization of neutrophils was performed using a transgenic zebrafish line Tg(mpx:GFP), in which GFP is expressed in neutrophils.Citation46 Lcp1/L-plastin immunostaining was performed for visualization of all leukocytes, and Lcp1-positive cells without GFP expression were considered to be macrophages.Citation47 The course of infection was imaged using CLSM at 4 h postinfection (hpi) and 1, 3, and 5 d postinfection (dpi) (). At 4 hpi, small numbers of fluorescently labeled bacteria (< 50) were detected at and around the site of injection. At this time point a few neutrophils and macrophages (< 5) were already recruited to the infection site. Some of the bacteria were taken up by leukocytes (). The bacteria that did not show colocalization with leukocytes could be extracellular or reside in other cell types. After this time point the bacteria proliferate and most of them appeared to grow in aggregates, which are at least 5 μm in size. In addition, increasing numbers of both neutrophils and macrophages were attracted. At 4 to 5 dpi the infection in the tail fin resulted in formation of an initial stage granuloma, which we observed as a large local accumulation of macrophages (20 to 30) and neutrophils (20 to 30) at the site of the infection, close to the site of injection. At this stage in the center of the granuloma a pore in the tail fin had been formed representing the necrotic (caseous) center of a granuloma, which has been reported previously in the granulomas of human lungs and adult zebrafish.Citation16,Citation48 Due to the thin tissue of the tail fin this center was extruded, resulting in a pore at late stages of infection (see also ). The development of these granulomas appeared to be dependent on an ESX-1 secretion system, since the M. marinum Delta RD1 mutant strain,Citation49 which is deficient in this system, was cleared within 4 dpi (data not shown).
Figure 2.M. marinum infection attracts leukocytes forming an initial stage granuloma. The course of M. marinum (red) infection is shown in Tg(mpx:GFP) larvae. Larvae have been stained using Lcp1 immunohistochemistry for leukocytes. Recruitment of neutrophils (green) and leukocytes (blue) was observed in the tail fin. In the top panels (A–D) an overview image of the entire tail fin at the indicated time point is presented. In the lower panels (A’–D’) higher magnification images of the indicated regions are shown. The scale bars represent 100 μm for the images on the top panels and 20 μm for the bottom panels. For each time point approximately 20 larvae were imaged and representative images for each time point are shown.
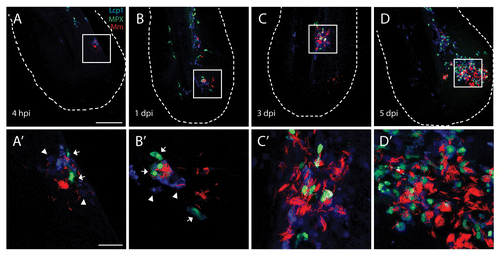
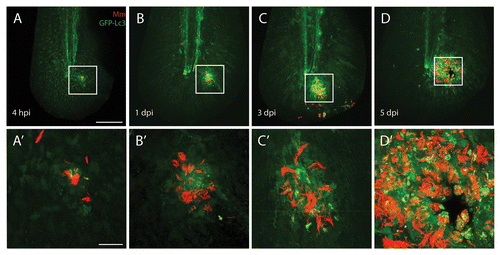
Figure 3. Autophagy is induced during M. marinum infection. M. marinum infected Tg(CMV:EGFP-map1lc3b) larvae were imaged at different time points after infection. GFP-Lc3-positive (green) vesicles were observed at the site of infection from 4 hpi to 5 dpi in the vicinity of the pathogens (red) by CLSM. In the top panels (A–D) an overview of the entire tail fin imaged at low magnification is shown. In the bottom panels (A’–D’) the indicated region imaged at higher magnification is presented. A necrotic center is formed at the center of the initial stage granuloma at 5 dpi. The scale bars represent 100 μm for the images in the top panels and 20 μm for images in the bottom panels. For each time point approximately 20 larvae were imaged and representative images for each time point are shown.
Autophagy during M. marinum infection: confocal laser scanning microscopy
In order to study whether the Tg(CMV:EGFP-map1lc3b) fish lineCitation32 can be used in our infection model to study autophagy, we injected 3 dpf larvae from this line with fluorescently labeled M. marinum in the tail fin. At 4 hpi and 1, 3, and 5 dpi the GFP-Lc3 signal and the fluorescent bacteria were imaged using CLSM ().
At 4 hpi the response to M. marinum infection was observed as small (< 1 μm) GFP-Lc3-positive vesicles, not containing bacteria, in cells close to the site of injection. These vesicles were not specific to the antibacterial response since in a control experiment a similar GFP-Lc3 response was observed after injection of polystyrene beads (Fig. S1). In this control experiment the GFP-Lc3 response was no longer observed from 1 dpi onward, indicating that any GFP-Lc3 response after this time point is specific for the bacterial infection (). One day after bacterial injection, more bacteria were present and the increased infection results in more small GFP-Lc3-positive vesicles in infected cells. The vast majority of these vesicles was ~1 μm in diameter and did not contain bacteria (). This pattern was also observed at 3 dpi (). At 5 dpi in most larvae an initial stage granuloma had been formed, which could be observed as a large local accumulation of bacterial aggregates and GFP-Lc3-positive vesicles.
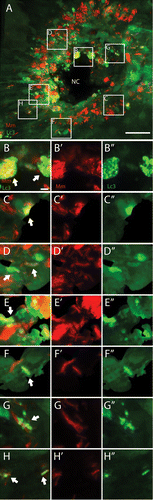
Different types of interaction between GFP-Lc3 structures and bacteria were observed at 5 dpi, and this is shown in more detail in . The vast majority of bacterial aggregates did not colocalize with any GFP-Lc3 signal, however in some cells accumulation of GFP-Lc3 vesicles occurred associated with these aggregates (i.e., the fluorescent signals from the bacteria and the vesicles are (partially) overlapping; ). Few cells contained larger GFP-Lc3-positive vesicles (~3 μm, ~6 per granuloma), which contained sequestered bacteria (). A large number (a few thousand) of small (~1 μm) GFP-Lc3-positive vesicles were present, which did not contain bacteria, although they could be observed in the vicinity of bacteria (). These small vesicles were able to fuse with the other compartments that contain bacteria. This process was monitored in a separate experiment, in which infected Tg(CMV:EGFP-map1lc3b) larvae were imaged alive (Vid. S1). In addition, GFP-Lc3-positive vesicles were observed in the vicinity of M. marinum without fusion with compartments containing bacteria (Vid. S2).
Figure 4. The GFP-Lc3 response observed during M. marinum infection. Tg(CMV:EGFP-map1lc3b) larvae infected with M. marinum at 5 dpi were imaged with CLSM. (A) Representative image of a granuloma in the infected tail fin. The GFP-Lc3 signal (green) and fluorescently labeled bacteria (red) are shown. (B–H) Magnified images of regions indicated in (A). (B) In highly infected cells near the necrotic center (NC) accumulation of GFP-Lc3-positive vesicles was observed (indicated by arrow). (C–F) In cells shown in these images larger GFP-Lc3-positive vesicles were observed, which entirely surround the bacteria (indicated by arrows). (G and H) In lowly infected cells GFP-Lc3-positive vesicles were observed in the vicinity of bacteria (indicated by arrows). Red and green signals from (B–H) are presented separately in (B’–H’) and (B”–H”), respectively. Scale bars: (A) 30 μm and (B–H) 3 μm.
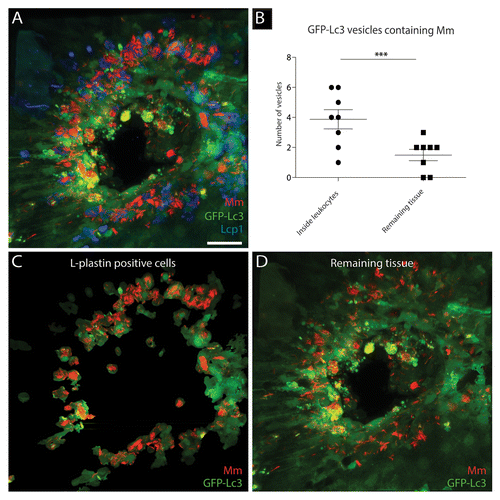
In order to differentiate between the GFP-Lc3 signal in leukocytes and the remaining cell types, infected Tg(CMV:EGFP-map1lc3b) larvae were immuno-stained using an antibody against Lcp1, a pan-leukocytic marker (). By segmentation, based on the Lcp1 signal, of the 3D rendered images 2 images were generated. One image showed the bacteria and GFP-Lc3-positive structures inside Lcp1-positive cells () and the other image showed this in the remaining cell types (). Quantitative analysis of 13 granulomas (1 per larva) showed that the majority of larger GFP-Lc3-positive vesicles containing bacteria were located inside leukocytes (). Each granuloma contained on average 5.4 (± 1.0) of these large vesicles containing bacteria. Of these vesicles, 3.9 (± 0.6) were located inside leukocytes and 1.5 (± 0.4) were located outside Lcp1-positive cells. No statistically significant difference in the number of small (~1 μm) GFP-Lc3-positive vesicles was observed between leukocytes and the remaining cell types (data not shown).
Figure 5.M. marinum containing GFP-Lc3-positive vesicles differ between leukocytes and other cell types. Tg(CMV:EGFP-map1lc3b) larvae infected with M. marinum at 5 dpi were immunostained for Lcp1, and their tail fins were imaged using CLSM. (A) Representative image of a granuloma in the infected tail fin. The GFP-Lc3 signal (green), Lcp1 immunostaining (blue) and fluorescently labeled bacteria (red) are shown. (B) Quantification of GFP-Lc3-positive vesicles for 13 granulomas (1 per tail fin) having sequestered M. marinum inside and outside Lcp1-positive leukocytes. The data (mean ± SEM) were analyzed using a paired 2-tailed Student t test (n = 13). *** indicates P < 0.001. (C) The GFP-Lc3 signal (green) and bacteria (red) in Lcp1-positive cells. (D) The GFP-Lc3 signal (green) and bacteria (red) outside Lcp1-positive cells. Scale bar: 30 μm.
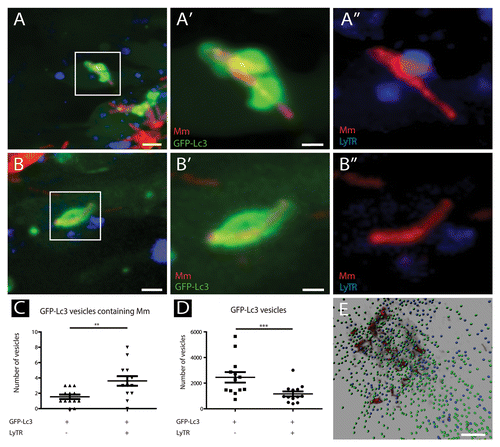
The lysosomal marker LyTR was used to study which fraction of the GFP-Lc3-positive vesicles had undergone fusion with lysosomal compartments (). The number of GFP-Lc3 vesicles positive for this marker was quantified, and approximately 30% of the small GFP-Lc3 vesicles were shown to be LyTR positive (). Approximately 70% of the larger GFP-Lc3 vesicles containing bacteria were positive for this lysosomal marker, indicating that the majority of these vesicles had undergone fusion with a lysosomal compartment ().
Figure 6. The majority of GFP-Lc3-positive vesicles having sequestered M. marinum are LyTR-positive. Tg(CMV:EGFP-map1lc3b) larvae infected with M. marinum at 5 dpi were stained with LyTR and their tail fins were imaged using CLSM. (A and B) Representative images of M. marinum in GFP-Lc3-positive vesicles that were positive (A) or negative (B) for LyTR. Magnified images of the regions indicated in (A and B) are presented separately for M. marinum (red) and GFP-Lc3 (green) in (A’ and B’), and for M. marinum (red) and LyTR (blue) in (A” and B”). (C) Quantification of M. marinum containing GFP-Lc3-positive vesicles, positive or negative for LyTR. The data (mean ± SEM) were analyzed using a paired 2-tailed Student t test (n = 13). *** indicates P < 0.001 and ** P < 0.01. (D) Quantification of small GFP-Lc3 vesicles positive and negative for LyTR. The data (mean ± SEM) were analyzed using a paired 2-tailed Student t test (n = 13). (E) Representative image of a 3D-rendered representation of small (~1μm) GFP-Lc3 vesicles, negative (green) or positive (blue) for LyTR. Scale bars: (A and B) 5 μm, (A’ and B’) 1 μm, and (E) 25 μm.
Autophagy during M. marinum infection: transmission electron microscopy
Transmission electron microscopy (TEM) was performed at 5 dpi of M. marinum infection in the tail fin of wild type larvae, in order to study the autophagy response and ultrastructure in more detail (). The tail fin infection model is very suitable for TEM analyses because the localization of infected cells is facilitated by the limited amount of tissue that needs to be analyzed in this local infection model. As described above, at 5 dpi a granuloma has been formed and a representative TEM image of such a granuloma is shown in . At the left side of the image a part of the necrotic center of the granuloma is visible. This center is surrounded by a large number of cells infected with bacteria, which represent a variety of cell-bacteria interactions.
Figure 7. TEM images of tail fin of zebrafish larva infected with M. marinum. (A) Overview image of a granuloma with necrotic center (NC) in the tail fin at 5 dpi. The tail fin is oriented anterior to the top and ventral to the left and the position of the notochord (NTO) is indicated. (B) Higher magnification of the region indicated in (A), showing part of a macrophage with bacteria of which one in a double-membrane autophagic vacuole (black asterisk) and 2 were cytoplasmic (white asterisk). (B’) Higher magnification of bacteria in an initial autophagic vacuole in (B), with arrows indicating the typical double membrane with electron-lucent cleft and arrowheads indicating ribosomes inside (white) and outside (black) of the vacuole. The ruffled appearance of the double membrane could be a fixation or sectioning artifact. (C) Higher magnification of region indicated in (A), showing a single bacterium (asterisk) in a degradative autophagic vacuole. (C’) Higher magnification of the degradative autophagic vacuole containing bacteria in (C), showing a lysosome fused with this vacuole (arrow), which contains partially degraded cytoplasmic material (arrowhead). (D) Higher magnification of indicated region, showing a macrophage with several phagosomal compartments containing bacteria. (E) Higher magnification of indicated region, showing infected epithelial cells. Scale bars: (A) 5 μm, (B and C) 1 μm, (B’) 250 nm, (C’) 500 nm, and (D and E) 10 μm.
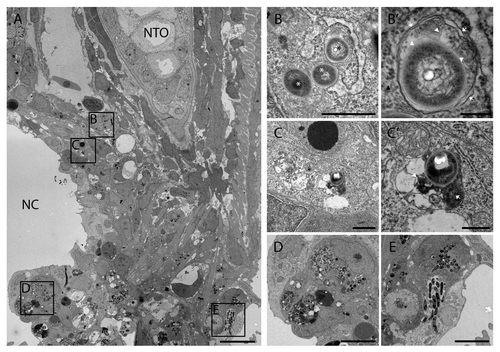
Four regions of this image are presented at higher magnification illustrating different cell-bacteria interactions (). The images show that some cells contain bacteria that are encapsulated by a double-membrane vesicle. Such an initial autophagic vacuole inside a macrophage is presented in ’. Other bacteria residing in the cytoplasm of the same macrophage were not enclosed by any membrane (). Some bacteria were encapsulated by single membrane structures with degradative autophagic vacuole morphology. An example is presented in , which shows a cell, most probably a macrophage, with an autophagic vacuole containing a bacterium and the remains of partially degraded cytoplasmic material. Many other macrophages contained larger aggregates of bacteria enclosed by a single membrane. Examples of these bacteria containing phagosomes in macrophages are shown in . Moreover, infected epithelial cells with bacteria were observed, and an example is shown in .
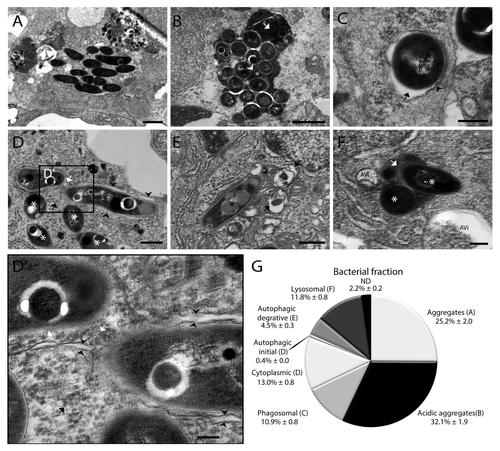
In order to determine the frequency of different cell-bacteria interactions we quantified the occurrence of 7 different types of interaction. This quantitative analysis and representative images of each type of interaction are presented in . The majority (~57%) of intracellular bacteria was found in aggregates, and approximately half of these aggregates had lysosomal morphology with uniform electron dense content in the compartment (~32%) (). Individual bacteria occurred in phagosomal compartments characterized by a single membrane with an electron-transparent zone or tightly surrounding the bacteria without any cytoplasmic material (~11%, ). In addition bacteria were found in the cytoplasm not enclosed by any membrane (~13%, ). Approximately 5% of bacteria were found in autophagic compartments, either in a typical double-membrane compartment also enclosing cytoplasmic content such as ribosomes (~0.4%, ) or in a compartment with late autophagic morphology (~4.5%). These compartments were characterized by partially degraded cytoplasmic content and organelles (). Finally, ~12% of bacteria were located inside lysosomal compartment with regular electron dense content ().
Figure 8. Quantification of intracellular M. marinum shows the distribution of bacteria in different compartments. (A–F) Representative TEM images of M. marinum in different compartments. (A) Aggregates were observed as a compact cluster of bacteria (< 5) without any electron dense areas. (B) Acidic aggregates were observed as a compact number of bacteria in a compartment having a uniform electron density between the bacteria (arrowhead) and/or electron dense regions (arrow). (C) Phagosomal compartment containing bacteria surrounded by a single membrane (arrowhead) with an electron-transparent zone (arrow), without any cytoplasmic material in the compartment. (D) Initial autophagic vacuoles containing bacteria (black asterisk), with the typical double membrane with electron-lucent cleft (arrowheads) and arrows indicating ribosomes inside (white) and outside (black) of the vacuole. Also in this image cytoplasmic bacteria are shown not enclosed by any membrane, indicated by a white asterisk. (D’) Higher magnification of the region indicated in (D), showing part of the bacteria and the vacuole in more detail. (E) Degradative autophagic vacuoles with partially degraded content (arrowhead) and other fused vacuoles (arrows). (F) Lysosomal compartment containing bacteria (asterisk) with uniform electron dense content (arrow). Near this compartment 2 autophagic vacuoles were observed indicated by AVi. (G) The fractions of intracellular M. marinum found in different compartments and free cytoplasmic M. marinum are presented in a pie chart. The bacteria for which their compartment could not be determined are indicated as ND. For this experiment 3 larvae with initial stage granulomas were used, with 3 regions for each larva at ~10 μm distance in order to prevent double counting of bacteria. In total 9 sections were analyzed containing over 2400 intracellular bacteria. Data shown are mean ± SEM. Scale bars: (A and B) 500 nm, (C and D) 250 nm, and (E and F) 1 μm.
Correlating light and electron microscopy images
Since we could obtain both CLSM and TEM images of the same specimen, the tail fin infection model provided the opportunity to correlate images generated by these 2 types of microscopy. For this purpose, infected larvae were first imaged alive using CLSM, and directly after imaging chemically fixed and prepared for electron microscopy (). This approach enabled the visualization of the ultrastructure of the GFP-Lc3-positive and negative structures containing bacteria that first had been identified by CLSM.
Figure 9. Correlative light and electron microscopy shows the ultrastructure of GFP-Lc3-positive structures. (A) CLSM image of infected Tg(CMV:EGFP-map1lc3b) zebrafish tail fin at 3 dpi. (B) Higher magnification of region indicated in (A), showing the projection view and the surface of the bacteria in red and of the GFP-Lc3 signal in green. (C) TEM image of the same area shown in (B). (D) Segmentation of adjacent TEM images showing the surface area of bacteria in blue. (E) Alignment of bacterial surfaces. The fluorescent signal (imaged by CLSM) is shown in red and the segmented surface (imaged by TEM) in blue. (F) 3D representation of CLSM and TEM images based on alignment shown in (E). (G) Magnified image of GFP-Lc3-positive structure enclosed 2 bacteria. (H) TEM image of the GFP-Lc3-positive compartment with bacteria shown in (G). (I) Magnified image with GFP-Lc3 signal in vicinity of bacteria. (J) TEM image of the same region, showing an initial autophagic vacuole, (indicated by arrow), at the tip of bacteria at same position as GFP-Lc3 signal in (H). The magnified inset shows the double membrane (arrowheads) and the ribosomes (asterisk) inside this vacuole. Scale bars: (A) 20 μm, (B–F) 5 μm, and (G and H) 1 μm.
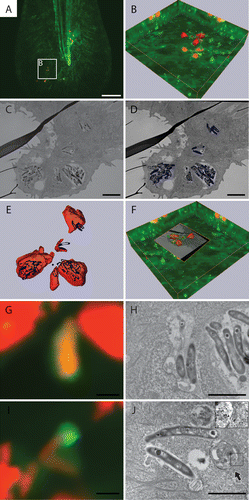
The tail fin of a Tg(CMV:EGFP-map1lc3b) larva infected with E2-crimson labeled bacteria (3 dpi) was first imaged using low magnification CLSM (), followed by higher magnification imaging of the region of interest and 3D rendering (). In this image (like in the images shown in and ), the majority of bacteria was not sequestered by GFP-Lc3-positive membranes. However, some bacteria were observed in close proximity to GFP-Lc3-positive vesicles and a small number of bacteria were inside a GFP-Lc3-positive structure. Subsequently, samples were sectioned and 2 adjacent sections were imaged by TEM (). The TEM images showed that in this region all bacteria were located inside epithelial cells. The 2 TEM images were aligned and the surface area covered by the bacteria was segmented (). This surface area was fitted into the 3D rendered image of the bacteria created from the CLSM images ().
Using this approach, different cell-bacteria interactions were observed. First, bacteria inside a GFP-Lc3-positive structure were observed using CLSM (). In the correlated TEM image these bacteria were present in a degradative autophagic vacuole that also contained partially degraded cytoplasmic material (). Second, in the same CLSM image other bacteria were observed which were not inside a GFP-Lc3-positive structure. However, they were observed to be associated with small GFP-Lc3-positive vesicles (). In the correlated TEM image this vesicle correlates with an autophagic vacuole containing cytoplasmic material and having a double membrane ().
Discussion
In this paper we provide a novel infection method that enables visualization of autophagic structures by both light and electron microscopy in vivo using a vertebrate infectious disease model. We show that this opens up the possibility to correlate light and electron microscopic images. In our method the pathogens are injected into the tail fin of zebrafish larvae, which results in a localized infection in this thin part of the body. We utilized this model for the study of infection by M. marinum showing that injection of this pathogen in the tail fin results in a local infection, which leads to the formation of single granuloma-like structure. The tail fin is very suitable for high-resolution light microscopy imaging, because the tissue consists of only a few cell layers and the infected cells are therefore located at a relatively short distance from the objective. This provides the opportunity to use high NA lenses, which are generally designed with a short free working distance. The relative absence of out of focus light enhances the contrast and the resolution of images of this tissue. For transmission electron microscopy this localized infection model has the major advantage that the site of infection can easily be found and subsequently imaged, due to the small tissue volume that needs to be investigated. When this tissue is imaged in live larvae using light microscopy before TEM imaging, the images obtained by these 2 different techniques can be correlated. This was performed based on the localization of the bacteria, which can be easily recognized in TEM images and were fluorescently labeled for detection in light microscopy. This model will be very valuable for high resolution in vivo imaging of autophagy, which until now has mainly been performed in cell cultures.
The applicability of studying autophagy in the tail fin infection model was demonstrated using M. marinum infection. Using the Tg(CMV:EGFP-map1lc3b) line, we showed highly active mobilization of GFP-Lc3-positive vesicles upon infection of M. marinum in the tail fin. Two types of GFP-Lc3-positive structures were distinguished. First, numerous relatively small vesicles (~1 μm) were observed that did not contain bacteria. These small vesicles are highly dynamic and can fuse with other compartments containing bacteria. Correlation of light and electron microscopy images showed that the presented small GFP-Lc3-positive vesicle in the vicinity of bacteria indeed has the appearance of an initial autophagic vacuole (). Second, larger structures (~3 μm) were present that often contained sequestered bacteria. The GFP-Lc3 signal of these larger structures could either originate from an autophagosome, which has taken up cytosolic bacteria, or from an autophagosome/autolysosome after fusion with a phagosomal compartment containing bacteria as has been shown in cell culture studies.Citation11,Citation21,Citation50 It has also been shown in cell cultures that GFP-LC3 can be associated with early phagosomes and phagosomes that have taken up bacteria of apoptotic bodies.Citation37,Citation51 In our system the majority of larger bacteria containing GFP-Lc3 vesicles were stained positive with LyTR indicating that they had undergone fusion with a lysosomal compartment. These vesicles are most probably GFP-Lc3-containing autolysosomes that have emerged through the autophagic pathway, although it could not be excluded that they are fused with lysosomes as part of LC3-associated phagocytosis. The autophagosomal nature of these vesicles was supported by correlative light and electron microscopy data showing that the larger GFP-Lc3 vesicle containing bacteria had the morphology of a degradative autophagic vacuole.
The presented observations of larger and small GFP-Lc3-positive vesicles reflect the occurrence of different pathways of autophagy induced during infection. The larger GFP-Lc3 vesicles containing bacteria may correspond to bacterial autophagy, whereas the smaller vesicles in the vicinity of bacteria may correspond to nonbacterial autophagy, reviewed in refs. Citation12 and Citation52. The latter process may be involved in the clearance of membranes that have been damaged during phagosomal escapes of M. marinum. Alternatively, these vesicles may fuse with other autophagic or heterophagic compartments containing bacteria.Citation22,Citation53 Furthermore, the presented data show that during M. marinum infection the larger bacteria-containing GFP-Lc3 vesicles occur more often in leukocytes than in other cell types (mainly epithelial cells in the tail fin). This suggest that different cell types show different autophagic responses, illustrating the advantages of studying the infection process in a whole animal model, in which multiple cell types and their interactions can be studied at the same time. In future studies these experiments can be performed in other transgenic fish lines, expressing GFP-Lc3 in specific cell types, or using the line ubiquitously expressing GFP-Lc3 in combination with other cell-specific fluorescent markers. In addition, in future research it will be important to determine which fraction of GFP-Lc3-positive vesicles without bacteria are autophagosomes.
The suitability of the tail fin infection model for electron microscopy was demonstrated by analysis of granuloma structures. It was confirmed that at this stage the bacteria resided in different cell types and that they could occur in the extracellular matrix. We quantified the number of intracellular bacteria residing individually in phagosomes, the cytoplasm, autophagic vacuoles, or lysosomes, or being present in aggregates or acidic aggregates. Only a very small fraction (~0.4%) of bacteria was found inside an initial autophagic vacuole with a double membrane, which is most likely due to the highly transient nature of these structures.Citation54,Citation55 These autophagosomes generally contain a single bacterium. The fraction of bacteria in degradative autophagic vacuoles is considerably larger (~4.5%). Another population of bacteria resides in phagosomal compartments and this fraction of bacteria (~11%) has been taken up most recently or has succeeded in blocking lysosomal fusion. For M. marinum it has been shown in cell cultures that they are able to escape the phagosomal compartment,Citation41 which was shown in our model to result in ~13% of bacteria residing freely in the cytoplasm. This fraction of bacteria is an obvious target for autophagy.Citation21,Citation50
In summary, our model offers new possibilities for future studies on the role of autophagy during infection in vivo. Recently, Mostowy et al., have studied the response of zebrafish larvae toward another pathogen, Shigella flexneri, showing that escape of this pathogen into the cytosol induces septin caging and targeting to autophagy.Citation31 It would be highly interesting to compare the infection process by different pathogens in the model we have developed. The zebrafish has many advantages for genetic studies that make it highly suitable to provide new insights into the relation between cellular structures and the molecular mechanisms of autophagy. With the advancement of medical translational studies in zebrafish disease models this will provide new opportunities to develop possible therapies against autophagy-related disorders.Citation30,Citation56
Materials and Methods
Zebrafish strains and maintenance
Zebrafish were handled in compliance with the local animal welfare regulations and maintained according to standard protocols (www.zfin.org). The ABTL wild-type zebrafish strain and the transgenic lines, Tg(Ola.Actb:Hsa.HRAS-EGFP),Citation44 Tg(mpx:GFP),Citation46 Tg(CMV:EGFP-map1lc3b),Citation32 strains were used for this study. All fish were raised and grown at 28.5 °C on a 14 h light: 10 h dark cycle. Embryos were obtained from natural spawning at the beginning of the light period and kept in egg water (60 µg/ml Instant Ocean sea salts). At 1 dpf 0.003% N-phenylthiourea (PTU; Sigma-Aldrich, P7629) was added in order to prevent pigmentation.
Zebrafish tail fin infection
The M. marinum M strain fluorescently labeled with E2-crimsonCitation57 was used and prepared at ~500 colony-forming units per 1 nl as previously described.Citation42 Borosilicate glass microcapillaries (Harvard Apparatus, 300038) were used with a micropipette puller device (Sutter Instruments Inc.) for preparing microinjection needles. Zebrafish larvae were injected in the tail fin at 3 dpf using the Eppendorf microinjection system with a fine (~5 to 10 micron) needle tip broken off with tweezers and mounted at a 30-degree angle. Larvae were anesthetized in egg water with 200 μg/mL 3-aminobenzoic acid (Tricaine; Sigma-Aldrich, E10521) and injected between the 2 epidermal layers at the ventral part of the tail fin (). Larvae were fixed at desired time points after infection with 4% paraformaldehyde in PBS-T (phosphate-buffered saline; NaCl 150 mM, K2HPO4 15 mM, KH2PO4 5 mM) with 0.05% Tween 20 (Merck Millipore, 8221840500) with gentle agitation for 18 h at 4 °C. The larvae were washed the next day with PBS-T and stored at 4 °C for further staining or until imaging.
Immunohistochemistry
Lcp1 immunostaining was performed for at least 20 larvae from each time point after fixation. The larvae were rinsed in PBS-DTx (phosphate-buffered saline with 0.5% DMSO and 0.3% Triton X-100) and treated with proteinase K (10 µg/ml in PBS-DTx; Roche, 03115879001) for 10 min at 37 °C. The larvae were blocked in 5% normal sheep serum (Sigma-Aldrich, S2263) in PBS-DTx for 2 h at room temperature, incubated with Lcp1/L-Plastin antibody (a gift from Anna Huttenlocher, University of Wisconsin, USA) in 1:1000 dilution at 4 °C overnight and subsequently incubated with Alexa-405 conjugated secondary antibody (1:200; Invitrogen, A-31556) for 2 h at room temperature. The larvae were washed with PBS-DTx and stored at 4 °C until imaging.
LysoTracker Red staining
LysoTracker Red (LyTR; Invitrogen, L-7528) was used for visualization of acidic compartments. Larvae were incubated for 1 h in 10 μM LyTR solution (in egg water) at 28 °C and rinsed several times with fresh egg water before imaging by CLSM.
Confocal laser scanning microscopy
Fixed larvae were mounted in 1% low melting agarose (Sigma-Aldrich, A9414) and imaged with a Leica TCS SPE (Wetzlar, Germany) or Zeiss Exciter (Oberkochen, Germany) confocal laser scanning microscope using the 405, the 488, and the 641 laser lines with 20× (NA 0.7) and 63× (NA 1.2) objectives. Larvae shown in and were imaged with the Nikon A1 confocal laser scanning microscope (Tokyo, Japan) using the 405, 488, and 561 laser lines with 20× (NA 0.75) and 60× objectives (NA 1.49). Images were analyzed using Fiji software,Citation58 and Imaris 5.5 software (Bitplane AG) was utilized in order to develop 3-dimensional models of the infected sites to quantify the number of GFP-Lc3-positive vesicles and their colocalization with LyTR signals. Amira 2.3 software (FEI, Visualization Sciences Group) was used for 3D rendering of CLSM images, segmentation of TEM images and aligning the 2 surfaces for correlative visualization.
Transmission electron microscopy
Before being used for electron microscopy the zebrafish larvae were anesthetized with 200 µg/ml tricaine, imaged alive by CLSM and afterwards immediately fixated in 2% glutaraldehyde and 2% paraformaldehyde in sodium cacodylate buffer (pH 7.2) for 3 h at room temperature followed by fixation for 16 h at 4 °C. Postfixation was performed in 1% osmium tetroxide in sodium cacodylate buffer for 1 h at room temperature. After dehydration through a graded series of ethanol all specimens were kept in epoxy resin (Agar Scientific, AGR1043) for 16 h before embedding. Ultrathin sections were collected on Formvar coated 200 mesh or one hole copper grids (Agar Scientific, AGS162) stained with 2% uranyl acetate in 50% ethanol and lead citrate for 10 min each. Electron microscopy images were obtained with a JEOL JEM-1010 transmission electron microscope (Tokyo, Japan) equipped with an Olympus Megaview camera (Tokyo, Japan).
Statistical analysis
All data (mean ± SEM) were analyzed (Prism 5.0) using paired, 2-tailed Student t tests for comparing the 2 conditions.
| Abbreviations: | ||
| ATG | = | autophagy related |
| CLSM | = | confocal laser scanning microscopy |
| dpf | = | days postfertilization |
| dpi | = | days postinfection |
| ESX-1 | = | the early secretory antigenic target 6 system 1 |
| hpf | = | hours postfertilization |
| hpi | = | hours postinfection |
| GFP | = | green fluorescent protein |
| MAP1LC3 (LC3) | = | microtubule-associated protein 1 light chain 3 |
| Map1lc3 (Lc3) | = | microtubule-associated protein 1 light chain 3b |
| Mm | = | Mycobacterium marinum |
| Mtb | = | Mycobacterium tuberculosis |
| LysoTracker Red | = | LyTR |
| Lcp1 | = | lymphocyte cytosolic protein 1 |
Additional material
Download Zip (6.5 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dan Klionsky (University of Michigan) for providing the GFP-Lc3 zebrafish line and Ulrike Nehrdich, Laura van Hulst, and Davy de Witt for fish care. We thank Serge Mostowy (Imperial College London) for critical reading of the manuscript, and Bram Koster and Erik Bos (Leiden University Medical Center) for helpful discussion. Infectious disease research in our laboratory is supported by the SmartMix Program of the Netherlands Ministry of Economic Affairs and the Ministry of Education, Culture and Science, the European Commission 7th framework project ZF-HEALTH (HEALTH-F4-2010-242048) and the Cyttron II Program (LSH framework: FES0908).
References
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069 - 75; http://dx.doi.org/10.1038/nature06639; PMID: 18305538
- Dunn WA Jr.. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol 1990; 110:1923 - 33; http://dx.doi.org/10.1083/jcb.110.6.1923; PMID: 2351689
- Dunn WA Jr.. Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol 1990; 110:1935 - 45; http://dx.doi.org/10.1083/jcb.110.6.1935; PMID: 2161853
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814 - 22; http://dx.doi.org/10.1038/ncb0910-814; PMID: 20811353
- Eskelinen E-L. New insights into the mechanisms of macroautophagy in mammalian cells. Int Rev Cell Mol Biol 2008; 266:207 - 47; http://dx.doi.org/10.1016/S1937-6448(07)66005-5; PMID: 18544495
- Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 2010; 12:836 - 41; http://dx.doi.org/10.1038/ncb0910-836; PMID: 20811356
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720 - 8; http://dx.doi.org/10.1093/emboj/19.21.5720; PMID: 11060023
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15:1101 - 11; http://dx.doi.org/10.1091/mbc.E03-09-0704; PMID: 14699058
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445 - 544; http://dx.doi.org/10.4161/auto.19496; PMID: 22966490
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Autophagy defends cells against invading group A Streptococcus. Science 2004; 306:1037 - 40; http://dx.doi.org/10.1126/science.1103966; PMID: 15528445
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753 - 66; http://dx.doi.org/10.1016/j.cell.2004.11.038; PMID: 15607973
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323 - 35; http://dx.doi.org/10.1038/nature09782; PMID: 21248839
- Deretic V. Multiple regulatory and effector roles of autophagy in immunity. Curr Opin Immunol 2009; 21:53 - 62; http://dx.doi.org/10.1016/j.coi.2009.02.002; PMID: 19269148
- Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol 2012; 30:611 - 46; http://dx.doi.org/10.1146/annurev-immunol-020711-074948; PMID: 22449030
- Armstrong JA, Hart PD. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med 1971; 134:713 - 40; http://dx.doi.org/10.1084/jem.134.3.713; PMID: 15776571
- Russell DG. Who puts the tubercle in tuberculosis?. Nat Rev Microbiol 2007; 5:39 - 47; http://dx.doi.org/10.1038/nrmicro1538; PMID: 17160001
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 2007; 129:1287 - 98; http://dx.doi.org/10.1016/j.cell.2007.05.059; PMID: 17604718
- Houben D, Demangel C, van Ingen J, Perez J, Baldeón L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, et al. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 2012; 14:1287 - 98; http://dx.doi.org/10.1111/j.1462-5822.2012.01799.x; PMID: 22524898
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012; 150:803 - 15; http://dx.doi.org/10.1016/j.cell.2012.06.040; PMID: 22901810
- Fabri M, Stenger S, Shin D-M, Yuk J-M, Liu PT, Realegeno S, Lee H-M, Krutzik SR, Schenk M, Sieling PA, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci Transl Med 2011; 3:ra102; http://dx.doi.org/10.1126/scitranslmed.3003045; PMID: 21998409
- Bradfute SB, Castillo EF, Arko-Mensah J, Chauhan S, Jiang S, Mandell M, Deretic V. Autophagy as an immune effector against tuberculosis. Curr Opin Microbiol 2013; 16:355 - 65; http://dx.doi.org/10.1016/j.mib.2013.05.003; PMID: 23790398
- Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HW 4th, Kyei GB, Johansen T, Vergne I, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 2010; 32:329 - 41; http://dx.doi.org/10.1016/j.immuni.2010.02.009; PMID: 20206555
- Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A 2007; 104:6031 - 6; http://dx.doi.org/10.1073/pnas.0700036104; PMID: 17389386
- Dowling JJ, Low SE, Busta AS, Feldman EL. Zebrafish MTMR14 is required for excitation-contraction coupling, developmental motor function and the regulation of autophagy. Hum Mol Genet 2010; 19:2668 - 81; http://dx.doi.org/10.1093/hmg/ddq153; PMID: 20400459
- Fleming A, Rubinsztein DC. Zebrafish as a model to understand autophagy and its role in neurological disease. Biochim Biophys Acta 2011; 1812:520 - 6; http://dx.doi.org/10.1016/j.bbadis.2011.01.004; PMID: 21256213
- Boglev Y, Badrock AP, Trotter AJ, Du Q, Richardson EJ, Parslow AC, Markmiller SJ, Hall NE, de Jong-Curtain TA, Ng AY, et al. Autophagy induction is a Tor- and Tp53-independent cell survival response in a zebrafish model of disrupted ribosome biogenesis. PLoS Genet 2013; 9:e1003279; http://dx.doi.org/10.1371/journal.pgen.1003279; PMID: 23408911
- Meeker ND, Trede NS. Immunology and zebrafish: spawning new models of human disease. Dev Comp Immunol 2008; 32:745 - 57; http://dx.doi.org/10.1016/j.dci.2007.11.011; PMID: 18222541
- Hu Z, Zhang J, Zhang Q. Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis. Autophagy 2011; 7:1514 - 27; http://dx.doi.org/10.4161/auto.7.12.18040; PMID: 22082871
- Benato F, Skobo T, Gioacchini G, Moro I, Ciccosanti F, Piacentini M, Fimia GM, Carnevali O, Dalla Valle L. Ambra1 knockdown in zebrafish leads to incomplete development due to severe defects in organogenesis. Autophagy 2013; 9:476 - 95; http://dx.doi.org/10.4161/auto.23278; PMID: 23348054
- Wager K, Russell C. Mitophagy and neurodegeneration: the zebrafish model system. Autophagy 2013; 9:1693 - 709; http://dx.doi.org/10.4161/auto.25082; PMID: 23939015
- Mostowy S, Boucontet L, Mazon Moya MJ, Sirianni A, Boudinot P, Hollinshead M, Cossart P, Herbomel P, Levraud J-P, Colucci-Guyon E. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog 2013; 9:e1003588; http://dx.doi.org/10.1371/journal.ppat.1003588; PMID: 24039575
- He C, Bartholomew CR, Zhou W, Klionsky DJ. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy 2009; 5:520 - 6; http://dx.doi.org/10.4161/auto.5.4.7768; PMID: 19221467
- van der Vaart M, Spaink HP, Meijer AH. Pathogen recognition and activation of the innate immune response in zebrafish. Adv Hematol 2012; 2012:159807; http://dx.doi.org/10.1155/2012/159807; PMID: 22811714
- Hanson HH, Kang S, Fernández-Monreal M, Oung T, Yildirim M, Lee R, Suyama K, Hazan RB, Phillips GR. LC3-dependent intracellular membrane tubules induced by gamma-protocadherins A3 and B2: a role for intraluminal interactions. J Biol Chem 2010; 285:20982 - 92; http://dx.doi.org/10.1074/jbc.M109.092031; PMID: 20439459
- Lai SC, Devenish RJ. LC3-Associated Phagocytosis (LAP): Connections with Host Autophagy. Cells 2012; 1:396 - 408; http://dx.doi.org/10.3390/cells1030396; PMID: 24710482
- Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007; 450:1253 - 7; http://dx.doi.org/10.1038/nature06421; PMID: 18097414
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 2011; 13:1335 - 43; http://dx.doi.org/10.1038/ncb2363; PMID: 22002674
- Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol 2008; 10:1027 - 39; http://dx.doi.org/10.1111/j.1462-5822.2008.01133.x; PMID: 18298637
- Stamm LM, Brown EJ. Mycobacterium marinum: the generalization and specialization of a pathogenic mycobacterium. Microbes Infect 2004; 6:1418 - 28; http://dx.doi.org/10.1016/j.micinf.2004.10.003; PMID: 15596129
- Berg RD, Ramakrishnan L. Insights into tuberculosis from the zebrafish model. Trends Mol Med 2012; 18:689 - 90; http://dx.doi.org/10.1016/j.molmed.2012.10.002; PMID: 23084762
- Stamm LM, Morisaki JH, Gao L-Y, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med 2003; 198:1361 - 8; http://dx.doi.org/10.1084/jem.20031072; PMID: 14597736
- Benard EL, van der Sar AM, Ellett F, Lieschke GJ, Spaink HP, Meijer AH. Infection of zebrafish embryos with intracellular bacterial pathogens. J Vis Exp 2012; •••:3781; PMID: 22453760
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203:253 - 310; http://dx.doi.org/10.1002/aja.1002030302; PMID: 8589427
- Cooper MS, Szeto DP, Sommers-Herivel G, Topczewski J, Solnica-Krezel L, Kang H-C, Johnson I, Kimelman D. Visualizing morphogenesis in transgenic zebrafish embryos using BODIPY TR methyl ester dye as a vital counterstain for GFP. Dev Dyn 2005; 232:359 - 68; http://dx.doi.org/10.1002/dvdy.20252; PMID: 15614774
- Meijer AH, Spaink HP. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets 2011; 12:1000 - 17; http://dx.doi.org/10.2174/138945011795677809; PMID: 21366518
- Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, Whyte MKB. A transgenic zebrafish model of neutrophilic inflammation. Blood 2006; 108:3976 - 8; http://dx.doi.org/10.1182/blood-2006-05-024075; PMID: 16926288
- Mathias JR, Dodd ME, Walters KB, Yoo SK, Ranheim EA, Huttenlocher A. Characterization of zebrafish larval inflammatory macrophages. Dev Comp Immunol 2009; 33:1212 - 7; http://dx.doi.org/10.1016/j.dci.2009.07.003; PMID: 19619578
- Pozos TC, Ramakrishnan L, Ramakrishan L. New models for the study of Mycobacterium-host interactions. Curr Opin Immunol 2004; 16:499 - 505; http://dx.doi.org/10.1016/j.coi.2004.05.011; PMID: 15245746
- McLaughlin B, Chon JS, MacGurn JA, Carlsson F, Cheng TL, Cox JS, Brown EJ. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog 2007; 3:e105; http://dx.doi.org/10.1371/journal.ppat.0030105; PMID: 17676952
- Lerena MC, Colombo MI. Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system. Cell Microbiol 2011; 13:814 - 35; http://dx.doi.org/10.1111/j.1462-5822.2011.01581.x; PMID: 21447143
- Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunol Rev 2009; 227:203 - 20; http://dx.doi.org/10.1111/j.1600-065X.2008.00732.x; PMID: 19120486
- Mostowy S. Autophagy and bacterial clearance: a not so clear picture. Cell Microbiol 2013; 15:395 - 402; http://dx.doi.org/10.1111/cmi.12063; PMID: 23121192
- Ponpuak M, Deretic V. Autophagy and p62/sequestosome 1 generate neo-antimicrobial peptides (cryptides) from cytosolic proteins. Autophagy 2011; 7:336 - 7; http://dx.doi.org/10.4161/auto.7.3.14500; PMID: 21187720
- Pfeifer U. Inhibition by insulin of the formation of autophagic vacuoles in rat liver. A morphometric approach to the kinetics of intracellular degradation by autophagy. J Cell Biol 1978; 78:152 - 67; http://dx.doi.org/10.1083/jcb.78.1.152; PMID: 670291
- Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem 1981; 256:7652 - 8; PMID: 7019210
- Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 2012; 11:709 - 30; http://dx.doi.org/10.1038/nrd3802; PMID: 22935804
- Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry 2009; 48:8279 - 81; http://dx.doi.org/10.1021/bi900870u; PMID: 19658435
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9:676 - 82; http://dx.doi.org/10.1038/nmeth.2019; PMID: 22743772
