Abstract
Vps34 is the ancestral phosphatidylinositol 3-kinase (PtdIns3K) isoform and is essential for endosomal trafficking of proteins to the vacuole/lysosome, autophagy and phagocytosis. Vps34-containing complexes associate with specific cellular compartments to produce PtdIns(3)P. Understanding the roles of Vps34 has been hampered by the lack of potent, specific inhibitors. To boost development of Vps34 inhibitors, we determined the crystal structures of Vps34 alone and in complexes with multitargeted PtdIns3K inhibitors. These structures provided a first glimpse into the uniquely constricted ATP-binding site of Vps34 and enabled us to model Vps34 regulation. We showed that the substrate-binding "activation" loop and the flexibly attached amphipathic C-terminal helix are crucial for catalysis on membranes. The C-terminal helix also suppresses ATP hydrolysis in the absence of membranes. We propose that membrane binding shifts the C-terminal helix to orient the enzyme for catalysis, and the Vps15 regulatory subunit, which binds to this and the preceding helix, may facilitate this process. This C-terminal region may also represent a target for specific, non-ATP-competitive PtdIns3K inhibitors.
PtdIns3Ks phosphorylate their lipid substrates at the 3-hydroxyl position of the inositol headgroup. Vps34 is the primordial PtdIns3K present in all eukaryotes and the only PtdIns3K in fungi and plants. This Cinderella of the PtdIns3Ks is responsible for much of a cell's cleaning and self-feeding: It is essential for multivesicular body formation, autophagy and phagocytosis. It associates with endosomes, omegasomes and phagosomes producing PtdIns(3)P, the most abundant 3-phosphoinositide in resting mammalian cells, which is essential for recruiting a range of complexes to intracellular membranes, including the autophagy machinery, ESCRTs, the retromer, motor proteins and components necessary for abscission in cytokinesis. In cells, Vps34 is at the core of larger complexes that also contain two regulatory proteins, Vps15 and Beclin 1, which bind directly to Vps34. The N-terminally myristoylated putative Ser/Thr protein kinase p150/Vps15 increases the lipid kinase activity of Vps34 and facilitates its translocation to endosomal membranes and the phagophore assembly site (PAS) or phagophore ().
We have determined the structure of the catalytic core of Vps34 (PDB ID 2X6H) (), which consists of a helical solenoid domain forming an extensive interface with a bilobal catalytic domain. The catalytic domain reveals key features that are important for the catalytic mechanism of all PtdIns3Ks: A phosphate-binding loop (P-loop) that interacts with the phosphates of ATP, a substrate-binding loop or “activation” loop that recognizes the PtdIns substrate, and a catalytic loop that is required for the transfer of the ATP γ-phosphate to the 3-hydroxyl of PtdIns. For the first time in any PtdIns3K structure, all three of these elements are completely ordered. The C-terminal helix (kα12) was previously shown to be required for Vps34 catalytic activity. However, the molecular basis for its function was unknown. The Vps34 structure suggests that the C-terminal helix closely associates with the substrate-binding loop and catalytic loop in the closed conformation. Site-specific mutagenesis guided by the crystal structure provides key insights into mechanisms of enzymatic regulation of Vps34 by this C-terminal helix. Deletion of the last 10 residues or point mutations within this helix, dramatically impairs lipid kinase activity in the presence of substrate lipids, but increases basal ATPase activity in the absence of substrate. These results suggest that in the closed form of the enzyme, the amphipathic C-terminal helix acts as a lid on the catalytic site to suppress activity in the absence of substrate lipid. Hydrophobic residues in this helix are also important for membrane interaction. Enzymatic activity and membrane binding measurements are consistent with a model whereby the C-terminal helix shifts to facilitate membrane interaction and orientation of the enzyme on the membrane interface for optimal catalysis (). The amphipathic character of the C-terminal region is conserved in all of the PtdIns3Ks, and it probably represents a common regulatory element in the entire family of enzymes. This may also extend to the PtdIns3Krelated enzymes such as TOR where the equivalent region has been denoted as the “FATC” domain, which also associates with membranes.
Early reports showed that methylated adenosine derivatives can inhibit autophagy. It was later demonstrated that the autophagy inhibitor 3-methyladenine (3-MA) inhibits PtdIns3Ks and that other general PtdIns3K inhibitors, such as wortmannin also inhibit autophagy. Although 3-MA shows some limited Vps34 preference in vitro, with an IC50 of 25 µM for Vps34 as compared with 60 µM for PtdIns3Kγ it is typically employed in cellular studies at a concentration of 10 mM, which can inhibit all PtdIns3Ks. Specific, potent inhibitors of Vps34 are acutely needed. All current PtdIns3K inhibitors are ATP-competitive, i.e., they target the ATP-binding site that is conserved among various PtdIns3K isotypes. The Vps34 structure suggests that the lack of potent Vps34 inhibitors could be accounted for by the uniquely constricted conformation of the Vps34 ATP-binding site in comparison with other PtdIns3Ks. Our structures of Vps34 in complexes with 3-MA and multitargeted PtdIns3K inhibitors (PIK-90, PIK-93 and PI-103) have provided insight into how this enzyme might be specifically inhibited. The slight preference for Vps34 inhibition by 3-MA probably arises from a hydrophobic ring specific to Vps34, which encircles the 3-methyl group of 3-MA. The insights arising from these structures have enabled us to develop a first generation of inhibitors with improved potency and Vps34 selectivity, e.g., the compound PT210 that has an IC50 of 0.45 µM as compared with 4.5 µM for PtdIns3Kγ. Further development of inhibitors guided by structures could lead to a new generation of improved inhibitors with applications as chemical tools to investigate PtdIns(3) P-dependendent pathways and as therapeutic agents.
Figures and Tables
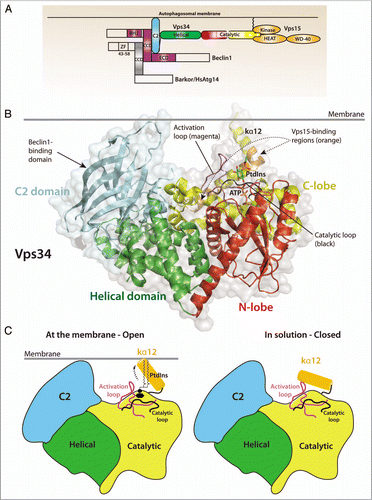
Figure 1 (A) Domain organization of Vps34, its regulatory subunit Vps15 and the adaptor proteins required for autophagy induction in mammalian cells, Beclin 1 and Atg14L/Barkor (Beclin1-associated autophagy-related key regulator). (B) Structure of Drosophila Vps34 helical (green) and catalytic (red/yellow) domains. A PtdIns substrate molecule has been modeled between the activation loop (magenta) and the catalytic loop (black) and ATP was modeled based on the p110γ/ATP structure (PDB ID 1E8X). The C2 domain (cyan) was also modeled from the p110γ/ATP structure. The enzyme is oriented so that the C2 domain and C-terminal helix interact with the membrane. Two regulatory proteins bind directly to Vps34: Vps15 binds to helices kα11 and kα12 (orange), and Beclin 1 binds to the C2 domain. Both Vps15 and Beclin 1 stimulate Vps34 activity. (C) A schematic representation of the Vps34 domains and the putative change in conformation of the kα12 helix. In solution (right), the helix is closed and interacts with residues in the substrate-binding and catalytic loops to exclude water. At the membrane (left), the kα12 helix undergoes a conformational change and interacts with the membrane, enabling productive substrate binding and catalysis.
Punctum to: Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, Williams RL. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science 2010; 327:1638 - 1642; PMID: 20339072; http://dx.doi.org/10.1126/science.1184429