Abstract
Antibacterial autophagy is understood to be a key cellular immune response to invading microbes. However, the mechanism(s) by which bacteria are selected as targets of autophagy remain unclear. We recently identified diacylglycerol as a novel signaling molecule that targets bacteria to the autophagy pathway, and show that it acts via protein kinase C activation. We also found that Pkc1 is required for autophagy in yeast, indicating that this kinase plays a conserved role in autophagy regulation.
The mechanism by which bacteria and other subcellular targets are identified and degraded by the autophagy pathway is an area of intense research. Ubiquitin has been recently found to act as an essential signal required for the autophagy of bacteria and proteins. We have previously observed ubiquitin on autophagy-targeted Salmonella enterica serovar Typhimurium (S. Typhimurium) but were surprised to see that only 50% of these bacteria were positive for ubiquitin. This indicated the possibility that an alternate signal was required for efficient autophagic targeting of the nonubiquitinated population of these bacteria.
We initially performed a screen quantifying the colocalization of different lipid second messengers (diacylglycerol (DAG), PtdIns(3)P, PtdIns(4,5)P2, PtdIns(3,4) P2, and PtdIns(3,4,5)P3) with autophagytargeted (i.e., LC3+) S. Typhimurium. We observed that DAG preferentially localizes with LC3+ bacteria. A kinetic analysis revealed that maximal DAG colocalization with bacteria (45 min post-infection) precedes maximal autophagy of the bacteria (60 min post-infection). Using pharmacological agents, siRNA and dominant negative constructs we were able to determine that DAG localization to the bacteria requires the action of phospholipase D (PLD; phosphatidylcholine to phosphatidic acid conversion) and phosphatidic acid phosphatase (PAP; phosphatidic acid to DAG conversion). We observed that inhibition of these pathways significantly reduces DAG localization to bacteria as well as concomitant autophagy of the bacteria, indicating a role for this lipid second messenger in the regulation of this process.
Having determined that DAG is necessary for autophagy of bacteria we subsequently wanted to identify the effector through which it was signaling. Conventional and novel isoforms of the protein kinase C (PKC) family contain DAG-binding C1 domains. Accordingly, we targeted PKC isoforms using pharmacological agents, siRNA and knockout cell lines and were able to determine that DAG is signaling through the δ isoform of PKC. Inhibition of this serine/threonine kinase results in significant inhibition of antibacterial autophagy. Furthermore, bacterial replication in PKCδ knockout mouse embryonic fibroblasts is significantly higher compared to control fibroblasts, consistent with previous observations demonstrating that autophagy impairs intracellular replication of S. Typhimurium (Birmingham et al. 2006).
We addressed the possibility that DAG and ubiquitin are functioning in a cooperative manner to target Salmonella for degradation by autophagy. We simultaneously inhibited both pathways using siRNA or pharmacological agents and observed additive inhibitory effects on autophagy of the bacteria. While this is indicative of two independent pathways, we cannot discount the possibility that there is still cooperation between the two pathways, especially as we did observe a small population of bacteria that were positive for both DAG and ubiquitin (). There are also a number of technical limitations in the methods we used, such as detection levels of the probes and antibodies that warrant caution in concluding that the two pathways are completely independent. Nonetheless, our studies clearly demonstrate a role for both DAG (Shahnazari et al. 2010) and ubiquitin (Zheng et al. 2009) in autophagy of S. Typhimurium. Future studies are required to further examine how these signals contribute to regulation of antibacterial autophagy.
Having characterized this pathway in antibacterial autophagy we were interested in determining whether these components were required for general autophagy. We therefore tested whether DAG localizes with rapamycin-induced autophagosomes. We observed DAG on these compartments and also found a requirement for PAP and PKCδ in this process. Other PKC isoforms are involved in alternate types of autophagy including ER stress-induced autophagy (Sakaki et al. 2008) as well as hypoxia-induced autophagy (Chen et al. 2009). As a result, we were interested in determining whether PKC function in autophagy was evolutionarily conserved. We therefore tested a role for the yeast ortholog, Pkc1, in this process and observed that it is required for starvation-induced autophagy in Saccharomyces cerevisiae.
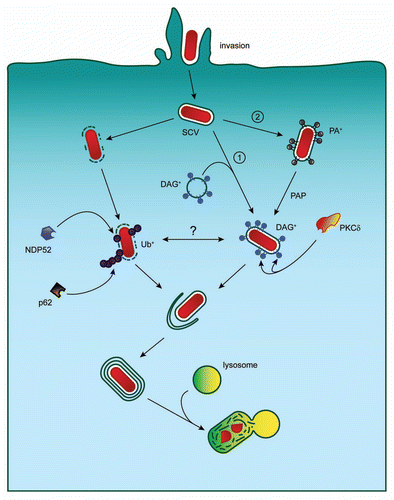
Having identified and characterized a novel signal and effector for antibacterial autophagy, further work still remains to be done in order to obtain a complete picture of this process. This includes additional study of the mechanism by which DAG is generated and the subcellular localization of PLD and PAP during this process. It is possible that DAG+ endocytic vesicles fuse with the Salmonella-containing vacuole (SCV) coating this compartment with DAG (pathway 1, see ). It is also possible that both PLD and PAP function directly on the SCV, converting phosphatidylcholine to DAG via the phosphatidic acid intermediate (pathway 2, ).
More work also needs to be done to dissect DAG and ubiquitin signaling contributions to this pathway. Questions to be answered include the identification of the ubiquitinated protein(s) on the SCV, which may be host or bacterial proteins. Additionally, while we know that DAG is present on the SCV we do not yet know the signal that induces its generation. One intriguing possibility is that DAG generation occurs in response to bacterial-induced damage to the SCV during invasion. To date, PKC has been implicated in at least three different types of autophagy, and the possibility exists that other PKC isoforms (DAG responsive or not) are also involved in this process.
Figures and Tables
Figure 1 Autophagic targeting of Salmonella Typhimurium. Invading S. Typhimurium can be targeted to the autophagy pathway by two independent signaling mechanisms. The first requires ubiquitin and the autophagy adaptors p62 and NDP52. The second requires DAG generation and PKCδ function. DAG generation on the SCV may occur through interaction of the SCV with DAG-positive endocytic vesicles (pathway 1) or through direct DAG production on the SCV (pathway 2). SCV, Salmonella-containing vacuole; PA, phosphatidic acid; DAG, diacylglycerol; PAP, phosphatidic acid phosphatase; PKCδ, protein kinase C delta; Ub, ubiquitin.
Punctum to: Shahnazari S, Yen WL, Birmingham CL, Shiu J, Namolovan A, Zheng YT, Nakayama K, Klionsky DJ, Brumell JH. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 2010; 8:137 - 146; PMID: 20674539; http://dx.doi.org/10.1016/j.chom.2010.07.002