Abstract
In a manifestation of the immunological autophagy termed xenophagy, autophagic adapter proteins such as p62 and NDP52 directly capture microbes for delivery to autophagosomal organelles where they are eliminated. In a mirror image phenomenon, which is also an immunological variant of the process termed decryption, p62 and autophagy contribute to the elimination of Mycobacterium tuberculosis. During decryption, p62 sequesters cytosolic proteins into autophagosomes where they are proteolytically converted into peptides termed cryptides. A subset of cryptides possesses antimicrobial peptide properties exhibited upon their delivery to parasitophorous vacuoles where they kill intracellular microbes.
Autophagy is an evolutionarily conserved cytoplasm-homeostatic process with a multitude of functions supporting, for the most part, cellular viability. During autophagy, cytoplasmic targets ranging from protein aggregates to whole organelles such as mitochondria and intracellular microbes are sequestered into a double-membrane bound organelle called the autophagosome. Autophagosomes mature into autolysosomes through fusion with lysosomes or their transport intermediates, bringing about acidification and acquisition of hydrolases leading to the digestion of the captured substrates. It is generally assumed that autophagy produces terminal degradative products such as free amino acids that are then used by the cell or the body as nutrients at times of starvation. Recently, we have discovered that autophagy generates, by proteolysis of captured cytosolic proteins, a mixture of peptides conferring potential cryptic biological functions, termed “cryptides.” Some of the cryptides with thus far assigned biological functions are the neo-antimicrobial peptides liberated from innocuous cytoplasmic proteins such as the ribosomal protein precursor FAU and ubiquitin.
Our study was motivated by the search for factors or ingredients that make autophagic organelles particularly mycobactericidal, as Mycobacterium tuberculosis can survive the environment of the conventional phagolysosome. This was shown in the 1970s by the classical work of Armstrong and D'Arcy Hart at the same time when these authors established the more broadly appreciated and well-ingrained reputation of the tubercle bacillus as inhibiting the conventional phagosome-lysosome fusion. The approach to identifying such hypothetical ingredients was to first examine the steps of the autophagic pathway that are necessary for the mycobactericidal nature of macrophages induced for autophagy by, for example, starvation. We have found that not only are all stages of autophagy (initiation, elongation/closure and maturation) required for full mycobactericidal potency, but that p62, the first autophagic adapter characterized by the Johansen group, and also known as sequestosome 1, is absolutely required for autophagic elimination of M. tuberculosis. Sequestosome 1/p62 recognizes ubiquitinated protein aggregates and possibly ubiquitinated depolarized mitochondria and other targets, and delivers them to nascent autophagosomes; p62 also binds to the mammalian Atg8 paralog LC3 via its LC3-interaction region (LIR), thus conveniently bridging the targets with forming phagophores.
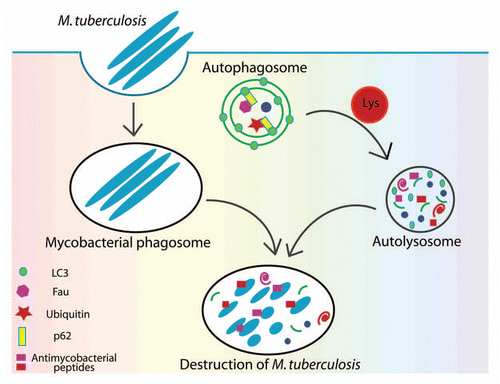
At first blush, it may seem that mycobacteria follow the same fate demonstrated for several other bacteria, whereby p62 or another autophagic adapter, NDP52, capture cytosolic microbes and deliver them to autophagosomes. For example, the fraction of Salmonellae that are no longer retained within phagosomes and are free in the cytosol, or Shigella and Listeria that actively escape into the cytosol, are associated with ubiquitinated material or become otherwise recognized by p62 or NDP52, and end up being sequestered into autophagosomes. However, we found no evidence for p62 acting directly to transfer intraphagosomal mycobacteria into autophagic vacuoles. Instead, we observed p62-positive organelles as periodically fusing with mycobacterial phagosomes. At the same time, we found by imaging and biochemical means that proteins recruited by p62 from the cytosol into conventional autophagic organelles are subsequently transferred to model (latex bead phagosomes formed upon feeding 1 µm beads to macrophages) or mycobacterial phagosomes, as they gradually acquire autolysosomal characteristics. Next, we established that p62-captured cytosolic proteins (ribosomal protein rpS30 precursor FAU and ubiquitin) are proteolytically degraded into smaller peptides, and that specific peptides from these complex mixtures show antimycobacterial activity. Thus, the emerging model posits that autophagy captures cytosolic proteins and converts them into neo-antimycobacterial peptides that can then kill M. tuberculosis upon delivery to mycobacteria-containing phagosomes, which in turn gradually acquire autolysosomal properties ().
In contrast to the direct mechanism of capturing bacteria employed in some instances described above, in the case of M. tuberculosis, an organism that resides within the phagosomes, the adapter molecule p62 exerts its anti-microbial action through an indirect, but rather sophisticated mechanism. By sequestering into autophagosomes the initially harmless cytosolic components and by proteolytically processing them within maturing autophagosomes, p62 and autophagy liberate antimicrobial peptides from the otherwise innocuous substrates. This amounts to a resourceful utilization by the cell of otherwise spent or to-be-discarded cytoplasmic proteins and gives them an after-function upon completion of their “day jobs” that they performed as whole proteins.
Our studies have uncovered a previously unappreciated function for autophagy in generating neo-antimicrobial peptides, and perhaps also opened the prospect for other biological functions potentially engendered by the products of autophagic proteolysis. Given that autophagy has the capacity to capture en masse and subject to digestion large sections of the cytoplasm, most cellular proteins are undergoing, or can undergo, processing into peptides or peptide intermediates within autophagic organelles. We postulate that the antimicrobial peptide production revealed in our studies thus far is only one manifestation of a spectrum of potential biological functions of cryptides generated by autophagy.
Figures and Tables
Figure 1 Elimination of M. tuberculosis by autophagy and p62. Mycobacteria are phagocytosed by macrophages and at least for some time reside within phagosomes. Upon induction of autophagy, p62, as a bifunctional agent interacting with autophagic substrates and with LC3, recruits into autophagosomes pre-antimicrobicidal cytosolic substrates. Autophagosome maturation including acquisition of lysosomal hydrolases leads to the proteolytic cleavage of p62 substrates and their conversion into peptides (cryptides) that can act as antimicrobial peptides.
Punctum to: Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by p62 endows autophagosomes with unique anti-microbial properties. Immunity 2010; 32:329 - 341; PMID: 20206555; http://dx.doi.org/10.1016/j.immuni.2010.02.009