Abstract
Foods contaminated with Escherichia coli O157:H7 cause more than 63,000 foodborne illnesses in the United States every year, resulting in a significant economic impact on medical costs and product liabilities. Efforts to reduce contamination with E. coli O157:H7 have largely focused on washing, application of various antibacterial chemicals, and gamma-irradiation, each of which has practical and environmental drawbacks. A relatively recent, environmentally-friendly approach proposed for eliminating or significantly reducing E. coli O157:H7 contamination of foods is the use of lytic bacteriophages as biocontrol agents. We found that EcoShield™, a commercially available preparation composed of three lytic bacteriophages specific for E. coli O157:H7, significantly (p < 0.05) reduced the levels of the bacterium in experimentally contaminated beef by ≥ 94% and in lettuce by 87% after a five minute contact time. The reduced levels of bacteria were maintained for at least one week at refrigerated temperatures. However, the one-time application of EcoShield™ did not protect the foods from recontamination with E. coli O157:H7. Our results demonstrate that EcoShield™ is effective in significantly reducing contamination of beef and lettuce with E. coli O157:H7, but does not protect against potential later contamination due to, for example, unsanitary handling of the foods post processing.
Introduction
Escherichia coli O157:H7 is a Shiga toxin producing member of enterohemorrhagic E. coli (STEC). It is a major foodborne bacterial pathogen, which was first associated with human illness during an outbreak of hemorrhagic colitis in 1982, and has been listed by the Centers for Disease Control and Prevention as a national notifiable disease since 1994.Citation1,Citation2 This bacterium has been estimated to cause > 63,000 foodborne illnesses and approximately 61 deaths annually in the United States.Citation2,Citation3 E. coli O157:H7 infections are of particular concern in young children and elderly persons because it is associated with hemolytic uremic syndrome which may permanently damage the kidneys.Citation4,Citation5
In addition to being of significant importance to public health, the economic impact of E. coli O157:H7 contamination of foods is substantial. It has been estimated that hospitalizations and deaths due to E. coli O157:H7 infections in the United States may lead to $405 million in medical costs and lost productivity annually.Citation6 Furthermore, substantive costs to manufacturers and growers may be incurred in the form of product loss and brand-damaging publicity associated with recalling products contaminated with this bacterium. These costs significantly increase (due to additional legal fees and settlement agreements) if the consumption of those foods results in human illness or mortality, and may force the company out of business.Citation7-Citation9 For example, a single E. coli O157:H7 outbreak associated with contaminated spinach in 2006 cost the spinach industry between $37 and $74 million.Citation10 Thus, there are very strong public health and economic incentives to develop novel, environmentally-friendly, safe and effective approaches for managing E. coli O157:H7 contamination of a broad range of foods.
A variety of treatment strategies are currently employed to eliminate or significantly reduce E. coli O157:H7 contamination, ranging from simple washing of foods to chemical or physical decontamination of foods. These methods vary with regard to their efficacy, cost and impact on the flavor and aesthetic integrity of food. For example, gamma-irradiation is considered to be one of the most effective treatments, capable of reducing E. coli O157:H7, and various other bacteria, by 5 log10.Citation11 However, the process is very expensive and more effective (high) levels of gamma irradiation may adversely affect the organoleptic qualities of foods, including taste and appearance.Citation12 Other strategies involve the application of various antibacterial chemicals, such as calcium hypochlorite, which has been reported to reduce E. coli contamination by 1.5 – 2.5 logs,Citation13 but many of those chemicals have a negative environmental impact. In addition to targeting pathogenic bacteria, both gamma-irradiation and chemical antibacterials target beneficial bacteria, thus negatively impacting the availability of beneficial bacteria in foods.Citation14 A relatively new intervention strategy that has the potential to alleviate those problems involves using lytic bacteriophages to target specific foodborne bacterial pathogens in various foods.
Bacteriophages (or viruses that lyse bacteria) are the most ubiquitous life form on Earth and they are part of the normal microflora of all fresh foods.Citation15,Citation16 The concept of using lytic bacteriophages to improve food safety relies on application of an appropriate lytic phage preparation onto foods that may be contaminated with foodborne bacterial pathogens that are susceptible to those phages. If the foods happen to be contaminated with the targeted bacterial pathogen, the phages will eliminate or significantly reduce the contamination, thus making the foods safe to consume without deleterious effect on their normal, beneficial microflora. This strategy is referred to as bacteriophage-mediated biocontrol.
Interest in bacteriophage-mediated biocontrol has recently gained increased momentum, as a variety of laboratories are pursuing the development of applications that utilize bacteriophages for pathogen control.Citation17-Citation19 Several reports have detailed the successful use of bacteriophages in significantly reducing the levels of various foodborne pathogens in a range of foods.Citation20-Citation23 Also, a variety of phage-based preparations have been recently approved for direct food applications in the United States and Europe; e.g., ListShield™, Listex P-100™ and EcoShield™.Citation24 One of those FDA-cleared preparations, EcoShield™, is a bacteriophage cocktail composed of three E. coli O157:H7-specific lytic bacteriophages, which has been previously reported to significantly reduce the E. coli O157:H7 contamination on surfaces and various foods.Citation20 The studies presented in the current communication were performed to determine whether treatment with EcoShield™ can (1) safely and significantly reduce E. coli O157:H7 levels in lettuce and beef under conditions that mimic their usual storage conditions, and (2) protect the phage-treated foods from recontamination with E. coli O157:H7.
Results and Discussion
Effect of phage contact time on E. coli O157:H7 levels
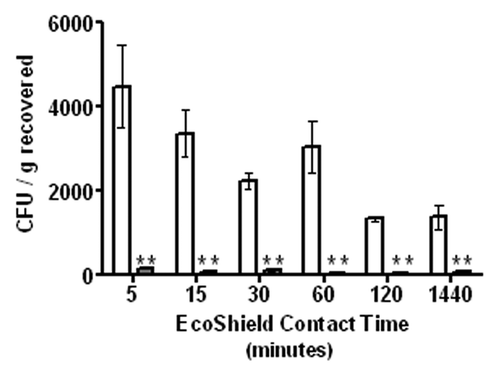
In an earlier study in which the efficacy of EcoShield™ on the E. coli O157:H7 levels on various foods and hard surfaces was evaluated, the standard incubation time between phage application and testing for the residual bacterial contamination levels (i.e., “contact time”) was 5 min.Citation20 To determine if longer contact times would increase the efficacy of EcoShield™, a time-course experiment was performed in which artificially contaminated beef steaks were treated with EcoShield™ (or PBS control) and held for 5, 15, 30, 60, 120 and 1440 min at 4°C. The levels of viable E. coli O157:H7 in the meat were measured immediately after sampling. EcoShield™ application significantly (p < 0.05, unpaired t-test) reduced the concentration of viable E. coli O157:H7 on beef samples at all contact times examined () compared with the PBS-treated control samples. The reductions were similar for all of the contact times examined, ranging from 94% to 98%. The 5 min contact time is considerably shorter than the time required for a full replication cycle of lytic bacteriophage (e.g., the replication cycle of a typical lytic phage such as T4 takes approximately 20–40 minCitation25), indicating our results support the idea that E. coli cells were infected within the first five minutes and there was no additional significant killing during the storage conditions we examined (). This observation may have some important practical implications for designing proper treatment strategies with bacteriophages in industrial food processing facilities.
Impact of food storage conditions on E. coli O157:H7 post phage treatment
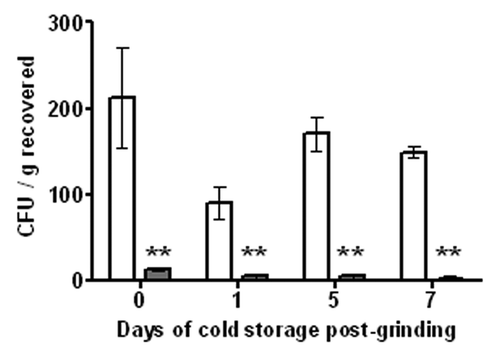
Most foods that are at high risk of contamination with E. coli O157:H7 (including ground beef) are typically stored and transported refrigerated or frozen. E. coli O157:H7 typically does not grow or grows very slowly at refrigerated (2–8°C) temperatures,Citation26 and growth may also occur during cooling from room to refrigeration temperatures. Thus, we were interested in determining if the decrease in bacterial load as the result of EcoShield™ treatment was consistent throughout a storage period of up to one week at refrigerated temperatures. This time period is consistent with the time it takes for beef processing, transport and sales. Artificially contaminated beef steaks were treated with EcoShield™ (or PBS control) then ground after 24 h at 4°C. The levels of viable E. coli O157:H7 and phage remaining in the ground meat were measured immediately (time = 0) and again at 1, 5 and 7 d of storage at 4°C. As in the previous experiment, EcoShield™ application significantly (p < 0.05, unpaired t-test) reduced the concentration of viable E. coli O157:H7 by 94% at time 0 (). After 1, 5 and 7 d at 4°C, the difference in levels of E. coli O157:H7 recovered from EcoShield™-treated and PBS-treated samples remained approximately the same as that observed at time 0 and were 94%, 97% and 98%, respectively. Thus, EcoShield™ application provided significant (p < 0.005) protection against the initial bacterial load and that reduction was maintained over a period of seven days of refrigeration, storage conditions likely to be encountered in real life settings. The concentration of phage recovered from the EcoShield™-treated samples at each time point ranged from 8 × 105–1 × 106 PFU/g (data not shown). The difference is within the 0.3 log titration error identified for the titration assay,Citation27 indicating a consistent phage concentration over 7 d.
Recontamination of foods
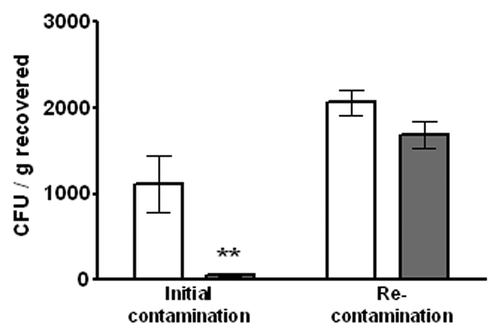
The reduction in E. coli O157:H7 levels we observed are in agreement with those previously reported for EcoShield™Citation20 and further support the idea that post-harvest application of EcoShield™ can significantly reduce the levels of E. coli O157:H7 in various foods. Our data also indicate that the foods treated with phages retain residual phages in them for at least one week of refrigerated storage. Thus, we sought to determine whether the residual phage would provide continued protection of the foods (or “continued technical effect,” Code of Federal regulations, 21 CFR § 101.100) from possible recontamination with E. coli O157:H7. To address this question, artificially contaminated beef steaks were treated with EcoShield™, stored at 10°C for 24 h, then ground. After grinding, the levels of viable E. coli O157:H7 in the ground meat were measured in half of the samples, while the other half were recontaminated with E. coli O157:H7, but not treated with EcoShield™. After an additional 24 h of storage at 10°C, the viable levels of E. coli O157:H7 in the recontaminated ground meat were measured. EcoShield™ significantly (p < 0.005, unpaired t-test) reduced the levels of the initial E. coli O157:H7 contamination in ground beef samples by 95% (). However, following recontamination, there was no statistically significant (p > 0.05) decrease in bacteria when compared with the PBS control (). Thus, EcoShield™ application provided significant protection against the initial bacterial contamination but no significant protection after recontamination with E. coli O157:H7. The results suggest that there is no continued protection by residual phage in ground beef.
Figure 3. Effect of EcoShield™ on recontamination of ground beef. White bars indicate PBS controls lacking EcoShield™, gray bars indicate EcoShield™ treated test groups. Error bars represent the SEM (n = 3). ** indicates statistical significance (p < 0.05 value).
To test if the lack of continued protection after recontamination was due to a decline in the bacteriophage concentrations during storage at 10°C, we assayed for the presence of EcoShield™ bacteriophages and found no change in their populations; the first set of challenged samples yielded a mean phage concentration of 1.5x105 PFU/g; the recontaminated set yielded a mean phage concentration of 8.5x104 PFU/g. As was seen at 4°C, the difference is within the 0.3 log titration error identified for the titration assay and, therefore, the levels of phage recovered can be considered approximately identicalCitation27 and the lack of continued protection is not due to a decrease in phage. Also, the surviving E. coli O157:H7 cells continued to be susceptible to EcoShield™ in vitro when randomly selected colonies were tested for susceptibility (data not shown), suggesting that the bacterial resistance to phage was not a factor in our observed inability of EcoShield™ to protect ground beef from recontamination. One possible explanation is that grinding substantially changes the surface area to volume of the matrix, thus dispersing both phage and bacteria and limiting phage access to the bacteria. The data suggest that while EcoShield™ can be very effective in reducing E. coli O157:H7 levels in red meat prior to grinding, it does not alleviate the need for subsequent proper and safe handling of foods because it does not protect against potential later contamination due to, for example, unsanitary handling of the foods by the end-customer.
EcoShield™ efficacy is concentration dependent
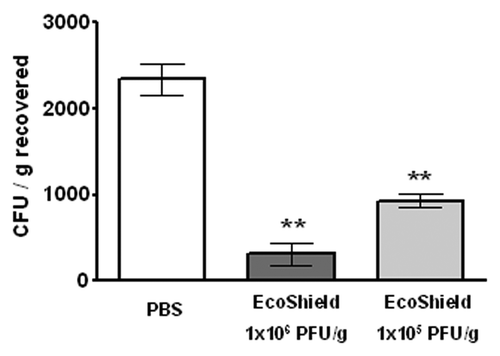
Several previous studies suggested that the efficacy of lytic phage preparations is dependent on the concentration of their constituent phages.Citation20,Citation21,Citation28,Citation29 Thus, we examined if EcoShield™ would still be effective if fewer phages were available to target E. coli O157:H7 cells in foods – a scenario which could occur, for example, if EcoShield™ was used to treat foods with high moisture content or residual water on its surface that might dilute the phages contained in EcoShield™. To test this hypothesis, artificially contaminated lettuce leaves were treated with two concentrations of EcoShield™ (1x106 PFU/g and 1x105 PFU/g) for five minutes and the levels of viable E. coli O157:H7 were determined. Both concentrations significantly (p < 0.05, one-way ANOVA) reduced the bacterial load recovered from the surface of the lettuce (87% and 60%, respectively (). The typical concentration (i.e., ca. 1x106 PFU/g) was statistically more effective than the lower concentrations (p < 0.05). The data suggest that further dilution of EcoShield™ by moisture on foods may slightly reduce the efficacy of the treatment, but that dilution will still result in a significant decrease in bacterial load. Foods to be treated with EcoShield™ are unlikely to have moisture content sufficient to dilute the preparation 10-fold, as was tested in our studies, but even in this hypothetical scenario, EcoShield™ should provide a significant reduction in E. coli O157:H7 levels.
Safety considerations: Genomic and Chemical Composition
Each of the component bacteriophages present in EcoShield™ was sequenced and genome maps were constructed (Fig. S1).Citation30 Each genome was examined for the presence of “undesirable genes,” including bacterial toxin genes listed in 40 CFR § 725.421, antibiotic resistance encoding genes, and bacterial 16 s rRNA genes. At a cutoff E-value ≤ 0.001 (coverage > 60%),Citation31 no genes corresponding to known antibiotic resistance determinants or toxins were found in any of the three phages included in EcoShield™. There was also no evidence of bacterial 16s rRNA genes in any of the component phages examined, indicating that none of the phages are capable of high-frequency transduction of host genes during the propagation in their host E. coli strains during phage production.
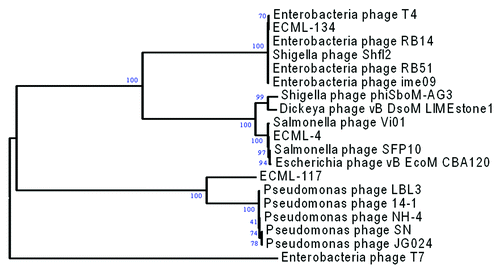
The neighbor-joining distance tree in shows the evolutionary relationship between EcoShield™ phages and several previously characterized phages. Whole genome comparisons suggest that these phages are similar to other bacteriophages also in the Myoviridae family. ECML-4 is most closely related to Salmonella phage Vi01, being 92.1% identical, based on EMBOSS stretcher analysis.Citation32 Other investigators have found bacteriophages lytic for E. coli O157:H7 to be also similar to Salmonella phage Vi01.Citation33 ECML-134 is most similar to the T4 bacteriophage (92.1%, MegaBLAST). ECML-117 is weakly similar to Pseudomonas phage LBL3 (FM201281). The global comparison of phage genomes to reference sequences from genome databases is shown in Figure S2.
Figure 5. Phylogenetic relationship between the predicted protein sequences of DNA polymerase genes from EcoShield™ phages and their homologs. The percentage of replicate trees in which > 50% of the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.
Chemical analysis of the EcoShield™ preparation indicates very low levels of non-phage ingredients, such as metals, salts and endotoxin (Table S1). Compared with the amount of these chemicals naturally present in foods, the contribution to an individual’s diet due to the application of EcoShield™ would be negligible. For example, normal saliva contains approximately 1 mg endotoxin per ml.Citation34 The levels of endotoxin normally present in EcoShield™ represent less than 2% of the endotoxin levels commonly found in the saliva of healthy adult humans. Therefore treating food with EcoShield™ is expected to be safe.
In summary, application of EcoShield™ at a typical concentration of 1 × 106 to 5 × 106 PFU per gram of foods significantly reduced E. coli O157:H7 contamination, by ca. 94% in beef and ca. 87% in lettuce, in as little as 5 min contact time. That reduction was maintained for at least 7 d of refrigeration, which mimics typical food storage conditions. However, the one-time application of EcoShield™ did not protect the foods from recontamination with E. coli O157:H7, which highlights the importance of continued safe handling of foods post-treatment. While the reductions of E. coli O157:H7 contamination provided by EcoShield™ are significant, they are lower than those obtained when using some other intervention strategies, such as those that utilize harsh chemicals. However, despite the common use of harsh chemicals, bacterial contamination of foods is still a major food safety concern and additional treatment modalities that can be safely added directly to foods to further reduce their contamination with pathogenic bacteria can be invaluable. EcoShield™, and similar phage-based preparations, may offer one such safe and environmentally-friendly approach.
Materials and Methods
Bacteriophage preparation
The bacteriophage product used in these studies is EcoShield™ (formerly ECP-100™), a cocktail of lytic phages developed and produced by Intralytix, Inc. The formulation includes three lytic phages: ECML-4 (ATCC #PTA-7948), ECML-117 (ATCC #PTA-7950) and ECML-134 (ATCC #PTA-7949). The phages contained in EcoShield (“component phages”) are subject of US Patents 7635584 and 7625741. EcoShield™ was cleared by the FDA for direct applications on red meat prior to grinding in 2011 (FCN #1018). EcoShield™ lots 0706K270409 (ca. 2 × 1010 PFU/mL), 0708C120181 (ca. 2 × 1010 PFU/mL), 0708J130181 (ca. 1 × 1010 PFU/mL) and 0709K170114 (ca. 2 × 1010 PFU/mL) were used during our studies. The lots were diluted with sterile saline as indicated before the experiments. A third party fee-for-service laboratory, under Good Laboratory Practice (GLP) conditions, performed chemical analyses of the preparations.
Bacteriophage sequencing and computational analysis
The DNA sequence of each of the three phages included in EcoShield™ was reoriented to render the genomes comparable with related bacteriophages. They were then annotated with myRAST available from http://blog.theseed.org/servers/installation/distribution-of-the-seed-server-packages.html.Citation30 The GenBank flatfile generated by this program was manually proofread in Kodon (Applied Maths) with the functional annotations checked against PfamCitation35 at http://pfam.sanger.ac.uk/ and by BLASTP at http://greengene.uml.edu/programs/NCBI_Blast.html. Genomes were screened for the presence of 16s rRNA genes using the BLASTN algorithmCitation36 comparing genomes to 16S rRNA databases to check for high frequency transduction activities. The BLAST databases created from the genomic sequences of the phages were screened for the presence of toxins using the TBLASTN algorithmCitation36 with the amino acid sequences of the toxin proteins listed in 40 CFR § 725.421 as a query. Genes encoding tRNAs were identified using tRNAscan-SE.Citation37 Rho-independent terminators and stem-loop structures were identified in intergenic regions using ARNold.Citation38 Comparative genomic analyses were performed using Artemis Comparison tool, progressive Mauve and CoreGenes.Citation39-Citation42 Genome diagrams were prepared using CGview.Citation43 The GenBank accession numbers for the phage genome sequences described in this manuscript are JX128257 (ECML-4), JX128258 (ECML-117) and JX128259 (ECML-134).
The relationship between the predicted protein sequence of DNA polymerase genes from EcoShield™ phages and other previously characterized phages was inferred using the Neighbor-Joining method.Citation44 A neighbor-joining distance tree was constructed using the translated amino acid sequence of genes annotated as DNA polymerases to compare them to those of other known bacteriophages.Citation44-Citation47 A DNA polymerase gene is present in all phages included in EcoShield™ and it serves as a useful marker for comparison to other phages.Citation48 The T7 DNA polymerase was included as an out-group. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed.Citation45 Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. All positions containing gaps and missing data were eliminated. There were a total of 700 positions in the final data set. Evolutionary analyses were conducted in MEGA5.Citation49 The following are accession number of proteins used in the construction of the neighbor joining tree: YP_004895353, YP_004957888, YP_004327561, YP_003358682, CCD57739, CCD57743, YP_002154198, YP_002364364, YP_002418863, ADF29350, AFD10641, YP_002854004, AEK12310, YP_002854382, YP_004414949, NP_049662.
Media and reagents
Challenge cultures were grown in LB, Miller agar and broth (Neogen), supplemented with nalidixic acid (Fisher Scientific). Peptone water (Becton, Dickinson and Company) was used during recovery of both bacteria and phage. Each study used specific amounts of diluents or aliquots depending on dissolution of the test matrix and countable recoveries. These factors were each accounted for when calculating the results (CFU/g recovered) for each experiment. Homogenized contaminant/peptone water mixtures were plated onto CR-SMAC (cefixime and rhamnose supplemented sorbitol MacConkey agar, Remel, Inc.) plates with nalidixic acid and cefixime supplements (Fisher Scientific).
Bacterial strains and challenge culture
The three E. coli O157:H7 strains, Ec229, Ec230 and Ec231, used in our studies were nalidixic acid resistant mutants of the EHEC strains 2886-75, G5101 and 93-111, respectively. The original strains were human isolates received from the STEC Center at Michigan State University. The strains were grown in LB media supplemented with nalidixic acid (25 μg/mL) at 37 ± 2°C for up to 24 h to a target concentration of 1–8 × 108 CFU/mL and mixed to prepare the challenge cultures. Challenge cultures were diluted in LB for each experiment as necessary to achieve a specific contaminating dose specified in the sections below.
Food samples
Beef chuck roasts and Romaine lettuce hearts were purchased at a local supermarket in Baltimore, Maryland. Each food sample was weighed and measured whole and, in some cases, cut to a particular size for challenge and/or treatment. Ground meat samples were prepared by grinding beef slices using a meat grinder (Northern Industrial Tools).
Phage application
All foods were treated by applying either PBS (control) or EcoShield™ with a spray gun (Badger Air-Brush Co., Franklin Park, IL; Basic Spray Gun, model #250-2) pre-calibrated to deliver 100 ± 20 µl in 4 sec.
Effect of EcoShield™ treatment time on beef steaks
Weighed and measured beef samples were placed into two groups, A and B. Both groups were contaminated with ca. 2 × 103 CFU/g of the challenge culture, then incubated (covered) at room temperature for 60 min. EcoShield™ (1 × 109 PFU/mL) was applied to one contaminated sample (Group A) at a rate of 2.7 µL/g (ca. 3 × 106 PFU/g;) the same volume of sterile PBS was applied to Group B, and samples were then refrigerated (covered) at 4 ± 2°C. After 5 min of contact time, three 25 g sections were cut from each steak with a sterile knife and the remaining portions were returned to refrigerated storage. The triplicate sections were placed into sterile plastic bags containing peptone water (10 mL/bag) and stomached, at medium setting for ca. 30 sec, using Stomacher 400 (Seward). Aliquots (0.1 mL) of the resulting suspensions were plated onto CR-SMAC plates supplemented with 25 μg/mL nalidixic acid and incubated at 35 ± 2°C for 16–24 h, at which time the colonies were counted and CFU/g were calculated, taking the sample size and diluent volume into account. The microbial analysis was repeated for all samples after 15 min, 30 min, 60 min, 120 min and 24 h post-treatment.
Storage time effect study in ground beef
Weighed and measured beef samples were placed into two groups, A and B. Both groups were contaminated with ca. 3 × 103 CFU/g of the challenge culture, then incubated (covered) at room temperature for 60 min. EcoShield™ (2 × 109 PFU/mL) was applied to one contaminated sample (Group A) at a rate of 2.3 μL /g (ca. 5 × 106 PFU/g;) the same volume of sterile PBS was applied to Group B, and samples were incubated (covered) at approximately 4 ± 2°C. After 24 h of refrigerated storage, both samples were ground, three 25 g aliquots of each were immediately removed for analysis, and the remaining portions were returned to refrigerated storage. Triplicate aliquots of the remaining portions from groups A and B were analyzed at 1, 5 and 7 d, using the bacterial load and phage methods indicated above. The 25 g aliquots were placed in bags containing 100mL sterile peptone water supplemented with 25 μg/mL nalidixic acid and stomached for 30 sec at medium setting. Bacterial loads were determined by plating 0.5 mL suspensions onto CR-SMAC plates supplemented with 25 μg/ml nalidixic acid and incubating at 35 ± 2°C for 16–24 h, after which the colonies were counted. Phage titer was determined by a standard plaque-counting technique. CFU/g and PFU/g recovered were calculated taking the sample size and diluent volume into account.
Recontamination study in ground beef
Six beef samples (ca. 350 g each) were contaminated with ca. 1 × 103 CFU/g of challenge culture, then incubated (covered) at room temperature for 60 min. EcoShield™ (1 × 109 PFU/mL) was applied to three samples at a rate of 3.0 µL/g (ca. 3 × 106 PFU/g;) the same volume of sterile PBS was applied to the other three samples, and all samples were incubated (covered) at approximately 10 ± 2°C. After 24 h of refrigerated storage, all samples were ground and divided into two approximately equal portions, A and B. The A and B portions were transferred to sterile plastic bags and weighed. Portion A was analyzed immediately for bacterial levels and phage titer. Portion B was recontaminated with ca. 5 × 103 CFU/g, hand mixed, incubated at 10 ± 2°C for an additional 24 h, and then analyzed. Analysis of all samples included adding 125 mL of sterile peptone water to each bag containing ground meat sample and homogenizing the samples. Bacterial loads were determined by plating 0.1 mL suspensions onto CR-SMAC plates supplemented with 25 μg/ml nalidixic acid and incubating at 35 ± 2°C for 16–24 h, after which the colonies were counted. Phage titer was determined by a standard plaque-counting technique. CFU/g and PFU/g recovered were calculated taking specific sample size and diluent volume into account.
Studies to examine the impact of phage concentration on efficacy
Cut, weighed and measured leaf lettuce samples were placed into three groups, A, B and C. All groups were contaminated with ca. 4 × 103 CFU/g of challenge culture, then incubated (covered) at room temperature for 60 min. EcoShield™ (1 × 108 PFU/mL) was applied to one contaminated sample (Group A) at a rate of 10.0 µL/g (ca. 1 × 106 PFU/g;) the same volume of EcoShield™ (1 × 107 PFU/mL) was applied to Group B (ca. 1 × 105 PFU/g;) and the same volume of sterile PBS was applied to Group C. Samples were covered and incubated at room temperature for 5 min. Following the 5 min contact time, triplicate 25 g samples were removed, added to bags of 225 mL sterile peptone water, hand mashed and homogenized with a stomacher for 30 sec at medium setting. Bacterial levels were determined by plating 0.5 mL suspensions onto CR-SMAC plates supplemented with 25 μg/ml nalidixic acid and incubating at 35 ± 2°C for 16–24 h. At the end of incubation, the colonies were counted and the CFU/g of sample was calculated, taking the sample size and diluent volume into account.
Statistical analyses
The efficacy of EcoShield™ treatment in reducing the number of viable E. coli O157:H7 in all experimental conditions was evaluated by comparing the data obtained with the PBS-treated control samples to the EcoShield™ treated samples. Specific statistical tests performed are noted in each experimental discussion. Statistical analyses were performed with the GraphPad InStat (version 3.05) and/or the GraphPad Prism (version 4.0) programs (GraphPad Software). A p value of < 0.05 indicated a statistically significant difference.
| Abbreviations: | ||
| SEM | = | Standard Error of the Mean |
| STEC | = | Shiga-like toxin producing E. coli |
| ca. | = | approximately |
Additional material
Download Zip (310.4 KB)Acknowledgments
Arnold Kreger is gratefully acknowledged for his editorial assistance. The E. coli strains, 2886-75, G5101 and 93-111, were obtained from the STEC Center at Michigan State University. The study was supported, in part, by SBIR award W911QY-07-C-0125 from the US Army (to A.S.).
Disclosure of Potential Conflicts of Interest
C.C., T.A., M.L., J.W. and A.S. hold an equity stake in Intralytix, Inc., a Maryland corporation involved with the development of phage preparations (including EcoShield™) for food safety applications.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/bacteriophage/article/22825
References
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, et al. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis 2011; 17:7 - 15; PMID: 21192848
- Bell BP, Goldoft M, Griffin PM, Davis MA, Gordon DC, Tarr PI, et al. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA 1994; 272:1349 - 53; http://dx.doi.org/10.1001/jama.1994.03520170059036; PMID: 7933395
- Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg Infect Dis 2005; 11:603 - 9; http://dx.doi.org/10.3201/eid1104.040739; PMID: 15829201
- Buzby JC. Children and microbial foodborne illness. FoodReview. 2001; 24:32
- Neill MA, Tarr PI, Clausen CR, Christie DL, Hickman RO. Escherichia coli O157:H7 as the predominant pathogen associated with the hemolytic uremic syndrome: a prospective study in the Pacific Northwest. Pediatrics 1987; 80:37 - 40; PMID: 3299236
- Frenzen PD, Drake A, Angulo FJ, Emerging Infections Program FoodNet Working Group. Economic cost of illness due to Escherichia coli O157 infections in the United States. J Food Prot 2005; 68:2623 - 30; PMID: 16355834
- Buzby JC, Roberts T. Economic costs and trade impacts of microbial foodborne illness. World Health Stat Q 1997; 50:57 - 66; PMID: 9282387
- Buzby JC, Roberts T. The economics of enteric infections: human foodborne disease costs. Gastroenterology 2009; 136:1851 - 62; http://dx.doi.org/10.1053/j.gastro.2009.01.074; PMID: 19457414
- Pennington H. Escherichia coli O157. Lancet 2010; 376:1428 - 35; http://dx.doi.org/10.1016/S0140-6736(10)60963-4; PMID: 20971366
- Shames, L. FDA’s food protection plan proposes positive first steps, but capacity to carry them out is critical. US Government Accountability Office 2008; GAO-08-435T.
- Aymerich T, Picouet PA, Monfort JM. Decontamination technologies for meat products. Meat Sci 2008; 78:114 - 29; http://dx.doi.org/10.1016/j.meatsci.2007.07.007; PMID: 22062101
- Wheeler TL, Shackelford SD, Koohmaraie M. Trained sensory panel and consumer evaluation of the effects of gamma irradiation on palatability of vacuum-packaged frozen ground beef patties. J Anim Sci 1999; 77:3219 - 24; PMID: 10641867
- Behrsing J, Winkler S, Franz P, Premier R. Efficacy of chlorine for inactivation of Escherichia coli on vegetables. Postharvest Biol Technol 2000; 19:187 - 92; http://dx.doi.org/10.1016/S0925-5214(00)00092-2
- Shi HN, Walker A. Bacterial colonization and the development of intestinal defences. Can J Gastroenterol 2004; 18:493 - 500; PMID: 15372112
- Sulakvelidze A, Barrow PA. Phage therapy in animals and agribusiness. In: Kutter E and Sulakvelidze A, ed(s). Bacteriophages: Biology and applications. Boca Raton, FL:CRC Press, 2005:335-380.
- Brüssow H, Hendrix RW. Phage genomics: small is beautiful. Cell 2002; 108:13 - 6; PMID: 11792317
- Hunter P. The return of the phage. EMBO Rep 2011; 13:20 - 3; http://dx.doi.org/10.1038/embor.2011.234; PMID: 22134545
- Goodridge LD, Bisha B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage 2011; 1:130 - 7; http://dx.doi.org/10.4161/bact.1.3.17629; PMID: 22164346
- Sulakvelidze A, Kutter EM. Bacteriophage therapy in humans. In: Kutter E and Sulakvelidze A, ed(s). Bacteriophages: Biology and applications. Boca Raton, FL:CRC Press, 2005:381-436.
- Abuladze T, Li M, Menetrez MY, Dean T, Senecal A, Sulakvelidze A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli, and ground beef by Escherichia coli O157:H7. Appl Environ Microbiol 2008; 74:6230 - 8; http://dx.doi.org/10.1128/AEM.01465-08; PMID: 18723643
- O’Flynn G, Ross RP, Fitzgerald GF, Coffey A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl Environ Microbiol 2004; 70:3417 - 24; http://dx.doi.org/10.1128/AEM.70.6.3417-3424.2004; PMID: 15184139
- Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul Toxicol Pharmacol 2005; 43:301 - 12; http://dx.doi.org/10.1016/j.yrtph.2005.08.005; PMID: 16188359
- Sharma M, Patel JR, Conway WS, Ferguson S, Sulakvelidze A. Effectiveness of bacteriophages in reducing Escherichia coli O157:H7 on fresh-cut cantaloupes and lettucet. J Food Prot 2009; 72:1481 - 5; PMID: 19681274
- Sulakvelidze A, Pasternack G. Industrial and regulatory issues in bacteriophage applications in food production and processing. In: Sabour PM and Griffiths M, ed(s). Bacteriophages in the control of food- and waterborne pathogens. Washington, DC:ASM Press, 2010:297-326.
- Mathews CK. An overview of the T4 developmental program. In: Karam JD, et al., ed(s). Molecular biology of bacteriophage T4. Washington, DC:ASM Press, 1994:1-10.
- Palumbo SA, Call JE, Schultz FJ, Williams AC. Minimum and maximum temperatures for growth and verotoxin production by hemorrhagic strains of Escherichia coli.. J Food Prot 1995; 58:352 - 6
- Anderson B, Rashid MH, Carter C, Pasternack G, Rajanna C, Revazishvili T, et al. Enumeration of bacteriophage particles: Comparative analysis of the traditional plaque assay and real-time QPCR- and nanosight-based assays. Bacteriophage 2011; 1:86 - 93; http://dx.doi.org/10.4161/bact.1.2.15456; PMID: 22334864
- Leverentz B, Conway WS, Camp MJ, Janisiewicz WJ, Abuladze T, Yang M, et al. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl Environ Microbiol 2003; 69:4519 - 26; http://dx.doi.org/10.1128/AEM.69.8.4519-4526.2003; PMID: 12902237
- Kudva IT, Jelacic S, Tarr PI, Youderian P, Hovde CJ. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl Environ Microbiol 1999; 65:3767 - 73; PMID: 10473373
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9:75; http://dx.doi.org/10.1186/1471-2164-9-75; PMID: 18261238
- Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, et al. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J Bacteriol 2003; 185:5220 - 33; http://dx.doi.org/10.1128/JB.185.17.5220-5233.2003; PMID: 12923095
- Lamprecht A-L, Naujokat S, Margaria T, Steffen B. Semantics-based composition of EMBOSS services. J Biomed Semantics 2011; 2:Suppl 1 S5 - 5; http://dx.doi.org/10.1186/2041-1480-2-S1-S5; PMID: 21388574
- Kutter EM, Skutt-Kakaria K, Blasdel B, El-Shibiny A, Castano A, Bryan D, et al. Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol J 2011; 8:430 - 430; http://dx.doi.org/10.1186/1743-422X-8-430; PMID: 21899740
- Leenstra TS, van Saene JJ, van Saene HK, Martin MV. Oral endotoxin in healthy adults. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1996; 82:637 - 43; http://dx.doi.org/10.1016/S1079-2104(96)80438-0; PMID: 8974136
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res 2010; 38:Database issue D211 - 22; http://dx.doi.org/10.1093/nar/gkp985; PMID: 19920124
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389 - 402; http://dx.doi.org/10.1093/nar/25.17.3389; PMID: 9254694
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955 - 64; PMID: 9023104
- Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol 2011; 8:11 - 3; http://dx.doi.org/10.4161/rna.8.1.13346; PMID: 21282983
- Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008; 24:2672 - 6; http://dx.doi.org/10.1093/bioinformatics/btn529; PMID: 18845581
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 2010; 5:e11147; http://dx.doi.org/10.1371/journal.pone.0011147; PMID: 20593022
- Kropinski AM, Borodovsky M, Carver TJ, Cerdeño-Tárraga AM, Darling A, Lomsadze A, et al. In silico identification of genes in bacteriophage DNA. Methods Mol Biol 2009; 502:57 - 89; http://dx.doi.org/10.1007/978-1-60327-565-1_6; PMID: 19082552
- Zafar N, Mazumder R, Seto D. CoreGenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinformatics 2002; 3:12; http://dx.doi.org/10.1186/1471-2105-3-12; PMID: 11972896
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics 2005; 21:537 - 9; http://dx.doi.org/10.1093/bioinformatics/bti054; PMID: 15479716
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406 - 25; PMID: 3447015
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985; 39:783 - 91; http://dx.doi.org/10.2307/2408678
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. Evolving Genes and Proteins. 1965; 97:97 - 166
- Sohpal VK, Dey A, Singh A. MEGA biocentric software for sequence and phylogenetic analysis: a review. Int J Bioinform Res Appl 2010; 6:230 - 40; http://dx.doi.org/10.1504/IJBRA.2010.034072; PMID: 20615832
- Rohwer F, Edwards R. The Phage Proteomic Tree: a genome-based taxonomy for phage. J Bacteriol 2002; 184:4529 - 35; http://dx.doi.org/10.1128/JB.184.16.4529-4535.2002; PMID: 12142423
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731 - 9; http://dx.doi.org/10.1093/molbev/msr121; PMID: 21546353