Abstract
Main Escherichia coli cytosolic chaperones such as DnaK are key components
of the control quality network designed to minimize the prevalence of
polypeptides with aberrant conformations. This is achieved by both favouring
refolding activities but also stimulating proteolytic degradation of folding
reluctant species. This last activity is responsible for the decrease of the
proteolytic stability of recombinant proteins when co-produced along with DnaK,
where an increase in solubility might be associated to a decrease in protein
yield. However, when DnaK and its co-chaperone DnaJ are co-produced in
cultured insect cells or whole insect larvae (and expectedly, in other
heterologous hosts), only positive, folding-related effects of these chaperones
are observed, in absence of proteolysis-mediated reduction of recombinant
protein yield.
In living cells, the protein quality control system surveys the folding of nascent polypeptides and full-length proteins in a kinetically driven process. The folding machinery acts in a set of ATP-dependent, coordinated activities intended to avoid the accumulation and aggregation of misfolded protein species, as protein deposits might be unusable reservoirs of amino acidsCitation1 and the amyloid-like stretches contained in them might be cytotoxic. Chaperones, included in the set of heat shock proteins, have high affinity for hydrophobic surfaces displayed in partially folded polypeptides leading them to either a proteolytic pathway, or a refolding pathway by modifying misfolded structures to convert them into a closer to native-like conformation. In fact, conformational deficiencies are associated to degenerative conditions and aging,Citation2,Citation3 being chaperones proposed as therapeutic agents against accumulation of these protein aggregates.Citation4,Citation5
In biotechnology, the production of soluble, biologically active recombinant proteins in prokaryotic gene expression systems such as Escherichia coli is limited by several factors, including saturation of the folding machinery, differential post-translational modifications or the absolute incapability to perform some of them.Citation6 In many cases, overexpression of recombinant genes leads to the accumulation of misfolded, aggregated protein forms known as inclusion bodies,Citation7 a fact that drastically reduces the yield of soluble protein. However, in prokaryotic systems, protein solubility can be improved by modulating cell growth conditions (i.e., lowering growth temperatureCitation8), fusing highly soluble protein partners (i.e., GST and MBPCitation9), creating a more oxidative environment (i.e., Origami strains,Citation10 fusion to DsbC,Citation11 periplasmic secretionCitation12), optimizing codon usage,Citation13 introducing eukaryotic glycosylated moieties in produced recombinant proteinsCitation14 or co-expressing protein factors involved in the protein folding control system (i.e., DnaK/J, GroEL/S and others).Citation15–Citation17
The genetic supplementation of folding modulators in recombinant protein production driven by E. coli can alleviate the saturation of the folding system caused by the input of newly synthesized polypeptides in the overproducing stressed cell. However, in some cases increasing the amount of folding modulators in over expressing cells to improve solubility results in decreased or constant protein yields or an increase only in aggregated soluble conformers.Citation15,Citation18,Citation19 This dual and contradictory effect reflects the complex equilibrium that drives misfolded polypeptides to either folding attempts or to proteolytic degradation. In that sense, it has been demonstrated that defined sets of several folding modulators might increase the success rate of recombinant protein production and global solubility,Citation17 although, as discussed later, gaining solubility is not always accompanied by a real increase in the soluble protein yield and quality.
In eukaryotic cells, several families of chaperones have been identified.Citation20 Among them, the eukaryotic Hsp70 chaperone family and its co-factors Hsp40 and HsdJ (DnaJ family) are the orthologs of the prokaryotic DnaK and its co-chaperone DnaJ. In the use of eukaryotic cells (namely insect cells, mammalian cells and yeast) as protein factories, co-expression of eukaryotic chaperones also increases solubility.Citation21–Citation23 Unfortunately, an assessment of the effect of such strategy on the conformational quality of the target proteins has not been performed side by side, making it difficult to completely evaluate the convenience of using such folding modulators as a general approach. In E. coli, DnaK (the homologous counterpart of the eukaryotic HSP70) is the main cytosolic chaperone exhibiting foldase activity in cooperation with its DnaJ and GrpE co-chaperones. However, in IB-forming cells, DnaK species are found associated to the IB surfacesCitation24 and involved in the dissagregation of proteins from IBs, in cooperation with DnaJ, GrpE, ClpB, IbpA and IbpB.Citation25–Citation27 On the other hand, DnaK is a negative modulator of the heat-shock response through the proteolytic inactivation of the stress-activated RNA polymerase subunit σ32. In a similar way, it has been shown that DnaK promotes ClpP- and probably also Lon-mediated proteolysis of recombinant proteins.Citation28 When producing recombinant enzymes and fluorescent proteins in DnaK-deficient E. coli cells, there is an increase in protein aggregation and yield compared to a wild type genetic background, in concordance with the loss of both chaperone and proteolytic enhancer activities.Citation18,Citation29 Surprisingly, an increase in specific activity is observed in the produced recombinant protein mainly in the insoluble cell fraction.Citation8 This observation seems to be also related to the absence of DnaK-induced proteolytic activity since ClpP− cells with a wild type DnaK also produce protein conformers with higher specific activity associated to protein deposited in inclusion bodies.Citation8 Therefore, in DnaK− cells, protein yield and quality increase at expenses of protein solubility. In agreement, in cells overexpressing DnaK and its immediate co-chaperone DnaJ along with the target recombinant protein, solubility increases with a concomitant loss of conformational quality and protein yield, being these parameters mutually exclusive.Citation18
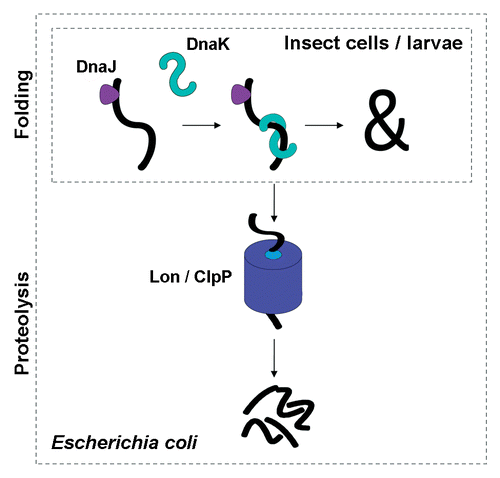
In the context of the antagonistic activities of the DnaK and DnaJ pair observed in bacteria, the introduction of these prokaryotic folding modulators in heterologous expression systems such as cultured insect cells,Citation30 insect larvaeCitation31 or others can benefit from their conserved foldase activities while avoiding the undesirable adverse proteolytic effect mediated by ClpP and Lon proteases, which is not expected to be conserved in eukaryotes. This principle () has been proved by the production of a modified aggregation-prone GFP (mGFP) along with DnaK and DnaJ in insect cells, where the chaperones compensate for the conformational stress during recombinant protein production in absence of proteolysis.Citation30 In fact, the recombinant GFP is proteolytically stabilized in the presence of DnaK and DnaJ,Citation30 contrarily to what had been observed in E. coli, where the half life of this protein is dramatically reduced under co-expression conditions.Citation18 Moreover, when analyzing protein fractioning in cultured cells, protein yield is significantly favoured in the soluble fraction and also in the insoluble fraction. When extending the chaperone rehosting concept to the production of other recombinant proteins in insect cell culture, these are better produced under co-production conditions than in absence of bacterial DnaK and DnaJ,Citation30 showing an increase in the yield of extractable protein and a reduction of oligomer formation depending on the model protein.Citation30 When rehosting the chaperones to a whole insect larvae system, total protein levels were not significantly affected by chaperone co-production, but soluble protein amounts represented almost the total of the recombinant protein and solubility values higher than 90% were obtained.Citation31 However, in both cultured insect cells and larvae, the recombinant GFP exhibits slightly lower functional quality (measured through specific fluorescence) in presence of bacterial chaperones,Citation30,Citation31 which can probably be accounted for by the increased yield. This is in agreement with previous observations in bacteria indicating that increasing protein yield is accompanied by a reduction of the conformational quality,Citation8 probably associated to the formation of soluble aggregates in the soluble cell fraction.Citation19
All these observations add further evidences to the fact that, taken independently, solubility is a poor indicator of protein quality in protein production processes,Citation32 and show that a more accurate case by case analysis is an inexcusable need for a proper control of conformational and functional protein quality, as recently claimed.Citation33,Citation34
Figures and Tables
Acknowledgements
The authors appreciate the financial support through grants BIO2007-61194 and EUI2008-03610 (MICINN). We also acknowledge the support of the CIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Spain. M.M.A. is recipient of a predoctoral fellowship from MEC, Spain. A.V. has been distinguished with an ICREA ACADEMIA award (Catalonia, Spain).
Addendum to:
References
- Kubota H. Quality control against misfolded proteins in the cytosol: a network for cell survival. J Biochem 2009; 146:609 - 616
- Sharma SK, Christen P, Goloubinoff P. Disaggregating chaperones: An unfolding story. Curr Protein Pept Sci 2009; 10:432 - 446
- Chaudhuri TK, Paul S. Protein-misfolding diseases and chaperone-based therapeutic approaches. Febs J 2006; 273:1331 - 1349
- Leandro P, Gomes CM. Protein misfolding in conformational disorders: Rescue of folding defects and chemical chaperoning. Mini Rev Med Chem 2008; 8:901 - 911
- Di Napoli M. New Molecular avenues in Parkinson's disease therapy. Curr Top Med Chem 2009; 9:913 - 948
- Ferrer-Miralles N, Domingo-Espin J, Corchero JL, Vazquez E, Villaverde A. Microbial factories for recombinant pharmaceuticals. Microb Cell Fact 2009; 8:17
- Villaverde A, Carrio MM. Protein aggregation in recombinant bacteria: biological role of inclusion bodies. Biotechnol Lett 2003; 25:1385 - 1395
- Martinez-Alonso M, Garcia-Fruitos E, Villaverde A. Yield, solubility and conformational quality of soluble proteins are not simultaneously favored in recombinant Escherichia coli. Biotechnol Bioeng 2008; 101:1353 - 1358
- Tolia NH, Joshua-Tor L. Strategies for protein coexpression in Escherichia coli. Nat Methods 2006; 3:55 - 64
- Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 2006; 72:211 - 222
- Zeng JM, Wang S. Enhanced gene delivery to PC12 cells by a cationic polypeptide. Biomaterials 2005; 26:679 - 686
- Ni Y, Chen R. Extracellular recombinant protein production from Escherichia coli. Biotechnol Lett 2009; 31:1661 - 1670
- Rosano GL, Ceccarelli EA. Rare codon content affects the solubility of recombinant proteins in a codon bias-adjusted Escherichia coli strain. Microb Cell Fact 2009; 8:41
- Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 2002; 298:1790 - 1793
- Haacke A, Fendrich G, Ramage P, Geiser M. Chaperone overexpression in Escherichia coli: Apparent increased yields of soluble recombinant protein kinases are due mainly to soluble aggregates. Protein Expr Purif 2009; 64:185 - 193
- Gupta P, Ghosalkar A, Mishra S, Chaudhuri TK. Enhancement of overexpression and chaperone assisted yield of folded recombinant aconitase in Escherichia coli in bioreactor cultures. J Biosci Bioeng 2009; 107:102 - 107
- de Marco A. Protocol for preparing proteins with improved solubility by co-expressing with molecular chaperones in Escherichia coli. Nat Protoc 2007; 2:2632 - 2639
- Garcia-Fruitos E, Martinez-Alonso M, Gonzalez-Montalban N, Valli M, Mattanovich D, Villaverde A. Divergent genetic control of protein solubility and conformational quality in Escherichia coli. J Mol Biol 2007; 374:195 - 205
- Martinez-Alonso M, Gonzalez-Montalban N, Garcia-Fruitos E, Villaverde A. The functional quality of soluble recombinant polypeptides produced in Escherichia coli is defined by a wide conformational spectrum. Appl Environ Microbiol 2008; 74:7431 - 7433
- Large AT, Goldberg MD, Lund PA. Chaperones and protein folding in the archaea. Biochem Soc Trans 2009; 37:46 - 51
- Yokoyama N, Hirata M, Ohtsuka K, Nishiyama Y, Fujii K, Fujita M, et al. Co-expression of human chaperone Hsp70 and Hsdj or Hsp40 co-factor increases solubility of overexpressed target proteins in insect cells. Biochim Biophys Acta 2000; 1493:119 - 124
- Carlage T, Hincapie M, Zang L, Lyubarskaya Y, Madden H, Mhatre R, et al. Proteomic profiling of a high-producing Chinese hamster ovary cell culture. Anal Chem 2009; 81:7357 - 7362
- Payne T, Finnis C, Evans LR, Mead DJ, Avery SV, Archer DB, et al. Modulation of chaperone gene expression in mutagenized Saccharomyces cerevisiae strains developed for recombinant human albumin production results in increased production of multiple heterologous proteins. Appl Environ Microbiol 2008; 4:7759 - 7766
- Carrio MM, Villaverde A. Localization of chaperones DnaK and GroEL in bacterial inclusion bodies. J Bacteriol 2005; 187:3599 - 3601
- Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol 2003; 50:585 - 595
- Schlieker C, Tews I, Bukau B, Mogk A. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. Febs Lett 2004; 578:351 - 356
- Thomas JG, Baneyx F. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol Microbiol 2000; 36:1360 - 1370
- Jubete Y, Maurizi MR, Gottesman S. Role of the heat shock protein DnaJ in the Lon-dependent degradation of naturally unstable proteins. J Biol Chem 1996; 271:30798 - 30803
- Martinez-Alonso M, Gonzalez-Montalban N, Garcia-Fruitos E, Villaverde A. Learning about protein solubility from bacterial inclusion bodies. Microb Cell Fact 2009; 8:4
- Martinez-Alonso M, Toledo-Rubio V, Noad R, Unzueta U, Ferrer-Miralles N, Roy P, et al. Rehosting of bacterial chaperones for high-quality protein production. Appl Environ Microbiol 2009; 75:7850 - 7854
- Martinez-Alonso M, Gomez-Sebastian S, Escribano JM, Saiz JC, Ferrer-Miralles N, Villaverde A. DnaK/DnaJ-assisted recombinant protein production in Trichoplusia ni larvae. Appl Microbiol Biotechnol 2009; 86:633 - 639
- Gonzalez-Montalban N, Garcia-Fruitos E, Villaverden A. Recombinant protein solubility—does more mean better?. Nat Biotechnol 2007; 25:718 - 720
- de Marco A, Sevastsyanovich YR, Cole JA. Minimal information for protein functional evaluation (MIPFE) workshop. N Biotechnol 2009; 25:170
- de Marco A. Minimal information: an urgent need to assess the functional reliability of recombinant proteins used in biological experiments. Microb Cell Fact 2008; 7:20