Abstract
Proper cell division requires the formation of the microtubule-based mitotic spindle, which mediates the dynamic movement and alignment of chromosomes to the metaphase plate and their equal transmission to daughter cells. Kinesins are molecular motors that utilize ATP hydrolysis to perform their functions and are instrumental in spindle assembly and function. Of the over 45 kinesins encoded in the human genome, only two are specifically enriched at the centrioles, Kif24 at the mother centriole and STARD9/Kif16a at the daughter centriole. While Kif24 possesses centriolar microtubule-depolymerizing activity and has been implicated in regulating cilia formation, our recent study implicates STARD9 in maintaining pericentriolar material (PCM) cohesion during early mitosis. However, very little is known about how STARD9 performs its function, including the mechanisms that recruit or retain STARD9 at the centrioles and how it cooperates with centrosome components to regulate PCM stability. Additionally, the signals leading to apoptosis in the absence of STARD9 remain to be explored.
During interphase, mammalian centrosomes play critical roles in organizing the cells microtubule array that functions to define cell shape, polarity and motility.Citation1 Centrosomes are pivotal for the fidelity of karyokinesis (the division of the nuclear material during cell division), including the organization of the mitotic microtubule spindle, which carries out chromosome congression during prometaphase and chromosome segregation during anaphase of mitosis.Citation1 At the core of centrosomes are two microtubule-based barrel like structures known as centrioles (). Centrioles are composed of nine triplet microtubules that form hollow cylinders.Citation2 The two centrioles (mother and daughter) can be distinguished from each other, as the mother centriole has added protein-based appendages at its distal end.Citation2 Centrioles are surrounded by an amorphous protein rich matrix, the pericentriolar material (PCM), which is primarily composed of Pericentrin and CG-NAP ().Citation2 During interphase centrosomes contain a small PCM that expands and matures (recruits additional proteins) at mitotic entry, a process that is necessary to resist the centrosome directed forces that are generated by microtubule associated molecular motors, like dynein and kinesins.Citation2,Citation3 These forces are not only critical for nuclear envelope breakdown but also for pushing and pulling chromosomes to the metaphase plate. Thus, the PCM has important roles in organizing spindle assembly through microtubule nucleation, elongation, and for resisting forces applied to the centrosome by molecular motors.
Figure 1. The role of STARD9/Kif16a during cell division. (A) STARD9 is a large ~517 kDa modular protein with an N-terminal kinesin motor domain, an FHA phosphoprotein binding domain, and a C-terminal START lipid/sterol binding domain. (B) Localization of STARD9 to the centrosome. Note that STARD9 is enriched at the daughter centriole. (C) The localization of STARD9 to the daughter centriole occurs during centrosome maturation (after centrosome duplication but before centrosome separation) and is required for PCM cohesion and bipolar spindle assembly.
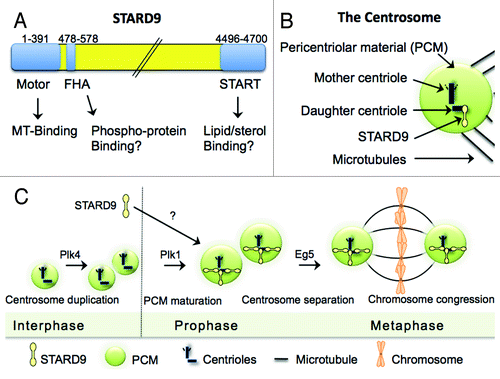
Proteomic, genetic, and bioinformatic approaches have been instrumental for defining the parts list of putative centrosome components and regulators of centrosome homeostasis.Citation4-Citation6 However, little is known about how these components localize to the centrosome, interact with each other, are regulated in a spatial-temporal manner, and coordinate the formation of the mature centrosomes. Recently, we set out to identify novel mitotic cancer targets by characterizing the proteome associated with mitotic microtubule asters, using biochemical purifications and mass spectrometry approaches, and by testing their importance for cell division, through genetic small interfering RNA (siRNA) screens for mitotic arrest and induction of apoptosis.Citation7 Interestingly, we identified STARD9 [steroidgenic acute regulatory protein-related lipid transfer (START) domain containing 9] as a novel mitotic kinesin that localizes to the daughter centriole and is necessary for PCM cohesion during bipolar spindle assembly.Citation7 STARD9 is a very large (~517 kDa) modular protein with an N-terminal kinesin motor domain, an FHA phosphoprotein binding domain, and a C-terminal START lipid/sterol-binding domain (). Based on sequence homology, STARD9 is a member of the kinesin-3 family, whose members predominantly have roles in the transport of vesicles and organelles.Citation8-Citation10 Of the over 45 human kinesins, STARD9 and Kif24 are the only two kinesins that are enriched at the mother and daughter centrioles respectively.Citation7,Citation11,Citation12 Interestingly, they are only conserved in vertebrates, indicating that they may perform specialized functions in higher organisms.Citation7,Citation11 Currently, we have a limited understanding of what recruits or retains STARD9 or Kif24 to the centrioles. Centrioles contain δ- and ε-tubulin isoforms not usually found in other microtubule based structures.Citation13-Citation16 In addition, unlike the bulk of polymerized microtubule arrays, centriolar microtubules are highly polyglutamylated and this modification is thought to regulate the association of centrosomal MAPs (microtubule-associated proteins) to the centrioles.Citation17-Citation21 However, these modifications apply to both centrioles and they are unlikely to be targeting signals for STARD9 or Kif24. Although the microtubule-binding and ATPase activities of the STARD9 motor domain are required for its localization to centrioles, other factors are likely to play a role.Citation7 Centrosomal components are highly phosphorylated by mitotic kinase families (like Cdk, Plk, Aurora and Nek) in early mitosis.Citation22 Therefore, phosphorylation of a daughter centriole specific protein and its recognition by the FHA phospho-protein binding domain of STARD9 could be a plausible targeting mechanism. However, only a handful of proteins have been identified enriched at daughter centrioles, most notably Centrobin.Citation6,Citation23 These proteins appear to be associated with the daughter centriole throughout the cell cycle.Citation6,Citation23 Conversely, STARD9 has a dynamic localization and only associates with centrioles from the onset of mitosis to late anaphase, indicating that its localization is regulated in a spatial-temporal manner ().Citation7 Thus, further experimentation will be necessary to define the mechanisms of STARD9 localization.
The expansion of the PCM during late G2-phase is thought to be critical for the absorption of microtubule and molecular motor-dependent forces during bipolar spindle assembly and the absence of an expanded PCM leads to centrosome fragmentation.Citation3 The bonds that keep the expanded PCM cohered are largely unknown. Recently, Kizuna was shown to be a “glue” that holds the expanded PCM together.Citation24 The depletion of Kizuna led to PCM fragmentation and multipolar spindle formation in human cells.Citation24 Similarly, depletion of STARD9 leads to PCM fragmentation, multipolar spindles, and mitotic cell death (apoptosis).Citation7 Like STARD9, Kizuna is only conserved in vertebrates, indicating that higher organisms have more sophisticated mechanisms that reinforce the PCM to deal with centrosome-directed forces.Citation24 Kizuna binds to Cep72 and this interaction is required for its localization to the PCM, where it associates with CG-NAP, pericentrin and γ-tubulin.Citation24,Citation25 The mechanism by which STARD9 functions to cohere the expanded PCM has not been determined. Unlike Kizuna that localizes to the PCM, STARD9 localizes to daughter centrioles.Citation7,Citation11 In addition, its been hypothesized that the mother centriole is predominantly responsible for the nucleation and tethering of microtubules.Citation26 Thus, how STARD9 contributes to PCM cohesion remains an enigma. It’s possible that the localization of STARD9 to centrioles per se is not important for PCM cohesion, but instead its association and/or stabilization of PCM components.
Recently, microtubule associated enzymes with regulatory roles in spindle assembly, like kinases (Plk1, Aurora A and B) and kinesins (Kinesin-5, CENPE) have received considerable attention as potential cancer targets.Citation27,Citation28 Kinesins are microtubule-dependent molecular motors that possess ATPase activity. In mitosis, kinesins have critical roles in spindle assembly, chromosome congression, and cytokinesis.Citation29,Citation30 Not surprisingly, the deregulation of most kinesins can lead to the development and progression of many types of cancers.Citation31 In addition to being required for mitotic progression, kinases and kinesins require ATP hydrolysis to perform their functions. This feature makes them amenable to small molecule inhibition with ATP-competitive inhibitors or allosteric inhibitors. Indeed, clinical trials are underway to test inhibitors targeting these enzymes for their antiproliferation properties.Citation32,Citation33 However, inhibition of Plk1 or Kinesin-5 leads to a very stable mitotic arrest with a circular arrangement of chromosomes attached to a monopolar spindle.Citation34,Citation35 In the case of Plk1-inhibition, the PCM is not matured and the chromosomes lack proper microtubule-kinetochore attachments, whereas in Kinesin-5 inhibition the PCM is matured and syntelic kinetochore-microtubule attachments are observed.Citation34-Citation36 Nonetheless, these cells are able to remain arrested for prolonged periods of time, upwards of 18 h, before they activate an apoptotic response and die. Most importantly these arrests are reversible, if the drug is washed out of these cells they recover and continue to divide. These stable and reversible arrests are a critical problem in the treatment of cancer. With sufficient time, cells are able to slip out of mitosis or wait for drug clearance and can continue to divide. In contrast, STARD9-depleted cells display a fragmented PCM, form multipolar spindles, activate the spindle assembly checkpoint (SAC), arrest in mitosis, and undergo apoptosis.Citation7 Interestingly, STARD9-depleted cells undergo apoptosis with faster kinetics than Plk1 or Kinesin-5 inhibited cells, indicating that their mitotic arrests differ.Citation7 Although the triggers that activate the apoptotic pathway in response to antimitotics or STARD9-depletion are poorly defined, it is tempting to speculate that PCM fragmentation, defective microtubule dynamics, or DNA tearing observed in STARD9-depleted cells could be plausible apoptotic triggers. Centrosome expansion/maturation occurs in late G2 phase and prepares the centrosome for increased microtubule density required in mitotic spindle assembly.Citation37,Citation38 Stabilization of the expanded pericentriolar material only occurs during mitosis and agents selectively destabilizing this process would be expected to act only on cells undergoing cell division. Because STARD9 is essential, specifically for cohesion of the expanded PCM during mitosis, inhibition of STARD9 is likely to have fewer side effects than drugs that inhibit other essential microtubule-based processes outside of mitosis, like microtubule poisons that have unwarranted neurotoxicities.
Our studies with STARD9-depletions indicate that compromising the integrity of the PCM may be a trigger for apoptosis and that inhibition of STARD9’s function may be a viable approach to inhibiting cancer cell division. The PCM fragmentation and spindle assembly defects observed in STARD9-depleted cells are conserved among multiple types of cancer cell lines.Citation7 However, depletion of STARD9 in a panel of non-small cell lung carcinomas (NSCLC) indicated that only specific subtypes of NSCLC cell lines were sensitive to STARD9-depletion.Citation7 This raises the possibility that the genetic background of each type of cancer contributes to the cell death response. Genetic variation could modify the timing of mitotic progression, the presence or absence of mitotic checkpoints, or the activation of compensatory mechanisms that could render a cell resistant. The types and subtypes of cancers that are responsive to STARD9-depletion must be defined to fully assess if STARD9-inhibition will be an effective avenue to pursue in inhibiting cancer cell division and to identify markers that will predict anti-STARD9-inhibitor sensitivity. Nonetheless, several lines of evidence indicate that STARD9 should be pursued as a cancer target. STARD9-depleted cells apoptose during mitosis with faster kinetics than current antimitotic treatments and STARD9 possesses an ATP hydrolyzing activity that is essential for its function and amenable to small molecule inhibition.Citation7 In addition, depletion of STARD9 strongly synergizes with taxol treatment, making STARD9 an appealing candidate target for combined cancer therapies.Citation7
The study of STARD9 is in its infancy. Future studies aimed at understanding the mechanisms of STARD9s function and regulation and the factors that render cells sensitive to STARD9-depletion will be invaluable to understanding the mechanisms driving cancer cell division and how inhibition of these mechanisms can be used to treat cancer.
| Abbreviations: | ||
| ATP | = | adenosine tri-phosphate |
| STARD9 | = | steroidgenic acute regulatory protein-related lipid transfer (START) domain containing 9 |
| MT | = | microtubule |
| MAP | = | microtubule-associated protein |
| PCM | = | pericentriolar material |
| siRNA | = | small interfering RNA |
Acknowledgments
Research conducted in the laboratory of J.Z.T. is supported by a Jonsson Cancer Center Foundation Seed Grant (J.Z.T.), The V Foundation for Cancer Research V Scholar Award (J.Z.T.), a UC-CRCC (University of California Cancer Research Coordinating Committee) Grant (J.Z.T.) and a March of Dimes Basil O’Connor Starter Scholar Research Award (J.Z.T.).
References
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 2011; 13:1154 - 60; http://dx.doi.org/10.1038/ncb2345; PMID: 21968988
- Azimzadeh J, Marshall WF. Building the centriole. Curr Biol 2010; 20:R816 - 25; http://dx.doi.org/10.1016/j.cub.2010.08.010; PMID: 20869612
- Abal M, Keryer G, Bornens M. Centrioles resist forces applied on centrosomes during G2/M transition. Biol Cell 2005; 97:425 - 34; http://dx.doi.org/10.1042/BC20040112; PMID: 15898952
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol 2007; 8:451 - 63; http://dx.doi.org/10.1038/nrm2180; PMID: 17505520
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003; 426:570 - 4; http://dx.doi.org/10.1038/nature02166; PMID: 14654843
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J 2011; 30:1520 - 35; http://dx.doi.org/10.1038/emboj.2011.63; PMID: 21399614
- Torres JZ, Summers MK, Peterson D, Brauer MJ, Lee J, Senese S, et al. The STARD9/Kif16a kinesin associates with mitotic microtubules and regulates spindle pole assembly. Cell 2011; 147:1309 - 23; http://dx.doi.org/10.1016/j.cell.2011.11.020; PMID: 22153075
- Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, et al. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell 2005; 121:437 - 50; http://dx.doi.org/10.1016/j.cell.2005.02.017; PMID: 15882625
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol 2005; 15:467 - 76; http://dx.doi.org/10.1016/j.tcb.2005.07.006; PMID: 16084724
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, et al. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell 1994; 79:1209 - 20; http://dx.doi.org/10.1016/0092-8674(94)90012-4; PMID: 7528108
- Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell 2011; 145:914 - 25; http://dx.doi.org/10.1016/j.cell.2011.04.028; PMID: 21620453
- Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A 2001; 98:7004 - 11; http://dx.doi.org/10.1073/pnas.111145398; PMID: 11416179
- Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. Epsilon-tubulin is an essential component of the centriole. Mol Biol Cell 2002; 13:3859 - 69; http://dx.doi.org/10.1091/mbc.E02-04-0205; PMID: 12429830
- Dutcher SK, Trabuco EC. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol Biol Cell 1998; 9:1293 - 308; PMID: 9614175
- Garreau de Loubresse N, Ruiz F, Beisson J, Klotz C. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol 2001; 2:4; http://dx.doi.org/10.1186/1471-2121-2-4; PMID: 11255590
- Dupuis-Williams P, Fleury-Aubusson A, de Loubresse NG, Geoffroy H, Vayssié L, Galvani A, et al. Functional role of epsilon-tubulin in the assembly of the centriolar microtubule scaffold. J Cell Biol 2002; 158:1183 - 93; http://dx.doi.org/10.1083/jcb.200205028; PMID: 12356863
- Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol 2003; 4:938 - 47; http://dx.doi.org/10.1038/nrm1260; PMID: 14685172
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Eddé B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol 1998; 143:1575 - 89; http://dx.doi.org/10.1083/jcb.143.6.1575; PMID: 9852152
- Bobinnec Y, Moudjou M, Fouquet JP, Desbruyères E, Eddé B, Bornens M. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton 1998; 39:223 - 32; http://dx.doi.org/10.1002/(SICI)1097-0169(1998)39:3<223::AID-CM5>3.0.CO;2-5; PMID: 9519903
- Boucher D, Larcher JC, Gros F, Denoulet P. Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry 1994; 33:12471 - 7; http://dx.doi.org/10.1021/bi00207a014; PMID: 7522559
- Larcher JC, Boucher D, Lazereg S, Gros F, Denoulet P. Interaction of kinesin motor domains with alpha- and beta-tubulin subunits at a tau-independent binding site. Regulation by polyglutamylation. J Biol Chem 1996; 271:22117 - 24; http://dx.doi.org/10.1074/jbc.271.36.22117; PMID: 8703022
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer 2007; 7:911 - 24; http://dx.doi.org/10.1038/nrc2249; PMID: 18004399
- Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, et al. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol 2005; 171:437 - 45; http://dx.doi.org/10.1083/jcb.200506185; PMID: 16275750
- Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol 2006; 8:1095 - 101; http://dx.doi.org/10.1038/ncb1474; PMID: 16980960
- Oshimori N, Li X, Ohsugi M, Yamamoto T. Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J 2009; 28:2066 - 76; http://dx.doi.org/10.1038/emboj.2009.161; PMID: 19536135
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol 2000; 149:317 - 30; http://dx.doi.org/10.1083/jcb.149.2.317; PMID: 10769025
- Lansing TJ, McConnell RT, Duckett DR, Spehar GM, Knick VB, Hassler DF, et al. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol Cancer Ther 2007; 6:450 - 9; http://dx.doi.org/10.1158/1535-7163.MCT-06-0543; PMID: 17267659
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 2000; 150:975 - 88; http://dx.doi.org/10.1083/jcb.150.5.975; PMID: 10973989
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell 2005; 16:3187 - 99; http://dx.doi.org/10.1091/mbc.E05-02-0167; PMID: 15843429
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol 2008; 265:111 - 58; http://dx.doi.org/10.1016/S0074-7696(07)65003-7; PMID: 18275887
- Yu Y, Feng YM. The role of kinesin family proteins in tumorigenesis and progression: potential biomarkers and molecular targets for cancer therapy. Cancer 2010; 116:5150 - 60; http://dx.doi.org/10.1002/cncr.25461; PMID: 20661912
- Pérez de Castro I, de Cárcer G, Montoya G, Malumbres M. Emerging cancer therapeutic opportunities by inhibiting mitotic kinases. Curr Opin Pharmacol 2008; 8:375 - 83; http://dx.doi.org/10.1016/j.coph.2008.06.013; PMID: 18644252
- Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev 2009; 28:197 - 208; http://dx.doi.org/10.1007/s10555-009-9185-8; PMID: 19156502
- Steegmaier M, Hoffmann M, Baum A, Léńrt P, Petronczki M, Krssák M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol 2007; 17:316 - 22; http://dx.doi.org/10.1016/j.cub.2006.12.037; PMID: 17291758
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 1999; 286:971 - 4; http://dx.doi.org/10.1126/science.286.5441.971; PMID: 10542155
- Léńrt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol 2007; 17:304 - 15; http://dx.doi.org/10.1016/j.cub.2006.12.046; PMID: 17291761
- Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, Wu X. Centrosome maturation. Curr Top Dev Biol 2000; 49:449 - 70; http://dx.doi.org/10.1016/S0070-2153(99)49021-0; PMID: 11005031
- Blagden SP, Glover DM. Polar expeditions--provisioning the centrosome for mitosis. Nat Cell Biol 2003; 5:505 - 11; http://dx.doi.org/10.1038/ncb0603-505; PMID: 12776127