Abstract
The PCI fold is based on a stack of α-helices topped with a winged-helix domain and is found in a range of proteins that form central parts of large complexes such as the proteasome lid, the COP9 signalosome, elongation factor eIF3, and the TREX-2 complex. Recent structural determinations have given intriguing insight into how these folds function both to facilitate the generation of larger proteinaceous assembles and also to interact functionally with nucleic acids.
Introduction
Although the PCI-fold motif was originally detected from homology between the sequences of the 26S proteasome lid, the COP9 signalosome (CSN), and the eukaryotic translation initiator eIF3 (ref. Citation1), more comprehensive analysis has indicated that this motif is found in a range of functional cellular complexes.Citation2 For example, PCI-folds have been detected by sequence analysis of Sac3 and Thp1 (ref. Citation2), components of the TREX-2 complex that functions to anchor a range of actively-transcribing genes to nuclear pore complexes (NPCs) in yeast, and also integrate mRNA export with earlier nuclear steps in the gene expression pathway.Citation3-Citation5 Structural information is now emerging that describes how PCI-domain proteins interact with one another and with nucleic acids, and how these interactions impact on key cellular functions.Citation6,Citation7
PCI-Domain Architecture
The six structures of PCI-proteins currently available [eIF3κ (1RZ4), CSN7 (3CHM), Rpn6 (3TXN), and the Sac3-Thp1-Sem1 (3T5V) and PCID2-DSS1 complexes (3T5X)] all display a bipartite fold () consisting of a structurally conserved C-terminal winged-helix domain (WH), and a more divergent N-terminal (HD) domain that is based on a stack of α-helices, albeit the number and arrangement of the α-helices varies considerably.Citation6-Citation10 The WH domain has a helix-turn-helix fold with an α-β-α-α-β-β topology, in which the β-strands form a short anti-parallel β-sheet on the protein surface. The loops formed between the second and third β-strands and between the third β-strand and the C-terminus of the molecule form the two “wings” of the WH domain.Citation11 In each structure, the first wing between the second and third β-strands is comparatively short (3–4 residues), whereas the longer second wing is arranged along the molecule and leads to a C-terminus that often forms short α-helices that interact on the surface of the HD domain. The structure of the WH domain is strongly conserved between these PCI-proteins, thus when considered in parallel with the relatively high sequence conservation observed across most members of this family, it is likely that this structural homology will be tightly preserved between all PCI-proteins.Citation9
Figure 1. (A) Structure of PCI proteins currently available. PCI-proteins contain a bipartite fold consisting of a N-terminal helical domain (blue) and a C-terminal winged-helix domain (green). The C-terminal winged-helix domain often leads to a C-terminal tail (orange) that interacts with the surface of the helical domain. (B) The Sac3-Thp1-Sem1 structure. The two PCI-proteins Thp1 (gray) and Sac3 (blue) share an extensive interaction interface marked by the insertion of the Thp1 WH “recognition helix” (red) into a cleft formed by the Sac3 WH and HD. The small Sem1 protein (yellow) wraps around the surface of Thp1 and makes little contact with Sac3. Blue spheres indicate the position of side-chains of conserved positive residues that when mutated to a negative charge, resulted in a loss of nucleic acid binding and mRNA export defects in yeast. (C) The role of Sem1 in modulating PCI complexes. Sem1 acts to stabilize the mini-PCI complex formed between Sac3-Thp1-Sem1 by binding to two sites on the Thp1 surface (Sites A and B). In doing so it may be acting as a capping protein shielding the surface of Thp1 from interacting with other PCI-proteins. Within the larger PCI-complex of the 26S proteasome lid Sem1 may act as a modulator of complex assembly, or subunit composition, by binding to and capping specific PCI-protein subunits. Within the schematic representations, the recognition helix (RH) of each WH domains is shown as a protruding tab.
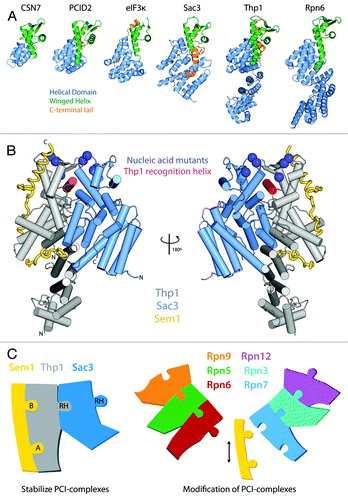
The N-terminal HD domain of PCI-proteins is more divergent, both at the structural and sequence level.Citation9 Although these domains have a common structural theme, based on a right-handed superhelical arrangement of α-helices that share characteristics of both tetratricopeptide (TPR) and HEAT repeats, there is considerable variation between them. Thus, for example, eIF3k (the first PCI-protein to be characterized structurally) contains 11 α-helices that form 3 HEAT analogous motifs in a right handed-superhelical arrangement,Citation8 whereas CSN7 contains a similar but smaller arrangement of only 6 α-helices forming a pair of HEAT repeats, one of which resembles an ARM-like repeat made up of 3 helices.Citation10 Although the way in which the α-helices are arranged at the base of the WH domains in the recently characterized Rpn6 and Thp1 structures have analogies to both eIF3k and CSN7 (refs. Citation6 and Citation7), these proteins contain much larger N-terminal extensions that are arranged in a right-handed superhelical configuration that projects away from the WH domain, and gives the molecules a twisted extended rod-like shape. Both Rpn6 and Thp1 contain TPR-like repeats within this superhelical arrangement, confirming previous prediction of their presence within the N-terminal region of multiple PCI-proteins.Citation9 Rpn6 contains five pairs of TPR-like repeats that form into a twisted α-solenoid arrangement, whereas Thp1 forms a straighter arrangement that is capped by a helical bundle at the N-terminus formed by four α-helices.Citation6,Citation7 In each structure, the N-terminal HD domains act as a buttress for the WH domains, with the elongated first helix of the WH domain providing a link between the two. Although within the Sac3-Thp1-Sem1 structure Sac3 was truncated to enable crystallization, sequence-based structure prediction suggests that its N-terminal HD probably extends further than the nine α-helices present within the domain, and most likely resembles the extended twisted rod-like character of Rpn6 and Thp1 (ref. Citation7). Furthermore, based on both sequence and structural conservation it is expected that full-length PCID2 (the human homolog of Thp1), which was also truncated at its N-terminus to enable crystallization, would form an elongated N-terminal extension resembling that of Thp1.
Taken together, these structures highlight the modular nature of the N-terminus of PCI-proteins; the structures vary in both length and arrangement of helical repeats from small helical bundles of HEAT-like repeats to an α-solenoid array containing TPR-like helical repeats. These variations at the N-terminus may give PCI-complexes adaptability in coordinating multiple protein-protein interactions, as is the case with the association of Sem1 with the TPR-like repeats of Thp1 in the Sac3-Thp1-Sem1 ternary complex.Citation7 In contrast, the C-terminal WH domain and the helical bundle that supports the WH are structurally much more similar across the six structures. This structural conservation is consistent with recent structural and mutational studies that demonstrate that the WH domain is a major site of interactions between PCI-proteins, as well as a region crucial in directing functionally important interactions with nucleic acids.Citation6,Citation7,Citation12
Interactions Between PCI Domains in the Sac3-Thp1-Sem1 Complex
The crystal structure of the Sac3-Thp1-Sem1 PCI-complex, which forms part of the larger TRanscription and EXport complex 2 (TREX-2) that couples mRNA transcription and processing with mRNA nuclear export,Citation7 provides a molecular understanding of the basis of the interactions between PCI-proteins that suggests how PCI components interact within larger PCI-complexes such as the 26S proteasome “lid,” the eIF3 complex, and the CSN complex. The TREX-2 complex is integral to gene-gating in which it associates with both the nuclear face of nuclear pore complexes (NPCs) and with the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex.Citation13-Citation16 This dual association is thought to enable TREX-2 to act as a NPC anchor point for the transcription machinery, allowing transcription to occur in close proximity to NPCs, thereby increasing the efficiency of mRNA export. Furthermore, the TREX-2 complex associates transiently with the major mRNA nuclear export factor, the Mex67-Mtr2 complex, and this interaction appears critical for efficient nuclear transport.Citation14,Citation18 Deletion of the N-terminal region of Sac3 critical for both Mex67-Mtr2 and Thp1 interactions leads to mRNA export defects in yeast.Citation18 The large scaffolding protein Sac3 forms the core of the TREX-2 complex, to which the smaller proteins of Sus1, Cdc31, Sem1 and Thp1 bind.Citation14,Citation17-Citation19 Two Sus1 chains and one Cdc31 chain wrap around the extended α-helical CID domain of Sac3 (residues 723–805) and coordinate tethering of the complex to the nuclear face of the NPC.Citation18,Citation19 The N-terminal region of the Sac3 protein contains a PCI domain (~residues 140–563) that binds to the PCI-protein Thp1 and the small highly negatively charged Sem1 protein ().Citation7 The predominant interaction interface within the Sac3-Thp1-Sem1 complex is formed between the WH domains and the supporting helical bundle of the HD domain that interacts with the elongated first α-helix of the WH domain. This interface is formed by the insertion of the recognition helix of the Thp1 WH domain into a cleft formed by the Sac3 WH domain and the C-terminal helical bundle of the HD domain.Citation7 Interestingly, it is the recognition helix of the WH domains that is presented to the major groove of DNA in many WH-DNA interactions.Citation11 This helical insertion results in the juxtaposition of the two WH domains in the Sac3-Thp1 heterodimer to form a six stranded β-sheet at the surface of the complex.Citation7 Both the structure and sequence of WH domains and the C-terminus of the HD domain are highly conserved between different members of the PCI-protein family, indicating that these regions probably contribute to the majority of the interface between interacting PCI-proteins.Citation9 Mutations within Thp1 and Sac3 that impair their interaction result in significant mRNA export defects, illustrating the functional importance of this PCI-protein complex within the mRNA export pathway.Citation7 The structure of the PCID2-DSS1 complex, the human homologs of Thp1 and Sem1 respectively, is in a very similar conformation to the Thp1 and Sem1 chains suggesting that in addition to a structural conservation of this complex from yeast to humans, the Thp1 protein structure changes little upon Sac3 binding.Citation7
Nucleic Acid Binding by PCI-Proteins
WH motifs are well-documented mediators of DNA and RNA binding, with multiple identified modes of nucleic acid interaction, and so it has been hypothesized that PCI-proteins or PCI-complexes could modulate functionally important interactions with nucleic acids.Citation2,Citation8,Citation10 Support for this hypothesis is provided by the observation that the Sac3-Thp1-Sem1 complex binds to a range of nucleic acids in vitro, including RNA and both single and double stranded DNA.Citation7 Efficient recognition of nucleic acids required PCI-protein complex formation and neither the Sac3 protein nor the Thp1-Sem1 complex alone efficiently associated with nucleic acids. In the Sac3-Thp1-Sem1 structure, juxtaposition of the Sac3 and Thp1 WH domains generates a positively charged stripe on the surface of the complex, and mutations of conserved residues within this motif () impaired nucleic acid binding. Importantly, these mutations also displayed mRNA export defects in yeast as well as synthetic lethality with the mRNA adaptor Yra1 and the mRNA transport factor Mex67 (ref. Citation7). These data, combined with previous studies, suggest that the TREX-2 complex functions as a NPC-tethered docking station that assists in the loading of the transport factors Mex67 and Mtr2 onto mRNA at the nuclear face of the NPC.Citation7 Although further work will be needed to build upon this model and gain a structural understanding of the precise mechanism by which TREX-2 functions, these results represent a clear identification of a functional role for nucleic acid binding by PCI-proteins.
Evidence of the importance of nucleic acid PCI-complex interactions has also come from the yeast eIF3 translation initiation complex that contains two proteins with PCI folds. Truncation and alanine scanning mutations of the eIF3 subunit c/NIP1, demonstrated that mutants that encroached on the structure of the WH domain resulted in a loss of eIF3 binding to the 40S ribosomal subunit, with a corresponding decrease in translation initiation.Citation20 Interestingly, the c/NIP1 PCI domain binds RNA non-specifically in vitro, and this interaction was impaired following mutations within the WH domain.Citation20 Furthermore, in silico docking studies based on the RFX-1-DNA and the SelB-RNA WH complexes indicate that the CSN7 complex would be compatible with DNA and RNA binding.Citation10 Indeed, the CSN complex has been previously implicated in transcriptional regulation and can associate with chromatin.Citation21 Similarly, recent evidence suggests that two additional PCI-proteins CSN12 and Thp3, that also form a complex with Sem1, associate with chromatin and have important roles in the regulation of transcription.Citation17,Citation22 This function of CSN12 and Thp3 appears to be independent from the “core complex” of the CSN.Citation22 Numerous studies have also connected the 26S proteasome with roles in transcription and DNA repair, and the Rpn6 subunit structure appears compatible with DNA binding based on superposition with the transcription factor E2F-DP.Citation6,Citation23,Citation24
Currently there is scant structural understanding of the molecular basis of the interaction between PCI-proteins and nucleic acids. Classically, the canonical recognition of nucleic acids by WH domains is mediated through interactions involving the third helix of the domain, termed the “recognition helix,” and the major groove of DNA.Citation11 In the crystal structure of the transcription factor hepatocyte nuclear factor-3 (HNF-3γ) bound to DNA, the interaction causes a bending of the DNA by 13 degrees resulting in a narrowing of the major groove of the DNA.Citation11 However, within the Sac3-Thp1-Sem1 complex, the Thp1 recognition helix is largely buried and makes important conserved interactions within the PCI-dimer interface and so it is unlikely that the recognition helix of Thp1 plays a major role in DNA or RNA binding. Although the Sac3 recognition helix is exposed to the solvent on the surface of the TREX-2 complex, the reliance of PCI-proteins upon dimerization and the position of positively charged conserved residues within the “wings” of the winged helix, suggest that the canonical mechanism of recognition helix-nucleic acid interaction may not apply in this case. Indeed, WH domains show remarkable adaptability in their modes of nucleic acid interaction. This is highlighted by the Werner Helicase WH domain that mediates DNA interactions through the loop formed between helices α2 and α3 with little involvement of the recognition helix.Citation25,Citation26 Furthermore, the WH domain of the elongation factor SelB interacts with the backbone of a RNA hairpin and external flipped RNA bases, yet fails to interact with either the major or minor grooves of the RNA hairpin.Citation27 Determining the crystal structure of the Sac3-Thp1-Sem1 complex bound to nucleic acids would be invaluable in defining the molecular details of this interaction and provide much needed mechanistic data pertaining to the role of Sac3-Thp1-Sem1 in the mRNA export pathway. Future work is also needed to ascertain how widespread the interaction of PCI-proteins and PCI-complexes with nucleic acids is. Nevertheless, this recent Sac3-Thp1-Sem1 structure in conjunction with the mutagenesis studies extend the functional repertoire of PCI-proteins to encompass important protein-nucleic acid interactions in addition to their scaffolding role mediated by protein-protein interactions in large complexes.
Formation of Large PCI Domain Complexes
PCI-proteins have long been thought to have architectural roles within the 26S proteasome lid, the CSN complex and eIF3 (ref. Citation1). However, despite extensive work detailing subunit composition, interaction profiles, and assembly pathways, little is known about the molecular basis of interactions between PCI-proteins. Canonically, the three large PCI-complexes contain six subunits that have identifiable PCI-sequence motifs and two subunits that contain MPN domains.Citation2 The structure of the Sac3-Thp1-Sem1 complex provides the first high-resolution view of the molecular nature of the interactions formed between PCI-proteins and immediately suggests a mechanism of PCI-protein assembly within the larger PCI-complexes.
The Sac3-Thp1-Sem1 structure indicates that PCI-domain containing proteins may generally align in a parallel fashion by the intercalation of the WH recognition helix with the WH and HD cleft of a neighboring PCI-protein. The interaction interface observed between Sac3 and Thp1 would be repeated for the remaining four PCI-proteins in the proteasome lid generating an alignment of the WH domains at the interaction interface. In turn, the N-terminal α-solenoid HDs would point away from this interaction interface and would thus be free to instigate further protein-protein interactions analogous to those observed between other TPR-repeat containing proteins. Indeed, the Sac3-Thp1 interaction interface is compatible with recent progress on the electron microscopy structure of the 26S proteasome.Citation24,Citation28 The low-resolution structure of the “lid” sub-complex consists of multiple proteins including six that contain PCI-domains. Within the reported structures, the PCI-domain containing proteins could be consistently mapped to the side of the 26S proteasome and make contact to both the AAA-ATPase and CP regions.Citation24,Citation28 In the structural model generated from these micrographs, the PCI-proteins are arranged in a horseshoe shape whereby the C-terminal WH domains align laterally, and the N-terminal domains extend out from the interaction interface like fingers on an open hand.Citation24,Citation28
Previous mutational analysis of the interaction of two of the proteasome lid PCI-proteins, Rpn6 and Rpn7, based on the Rpn6 structure and sequence conservation, is also consistent with this model. These studies showed that the recognition helix of the Rpn6 WH domain is critical for modulating the interaction with Rpn7 (ref. Citation6), consistent with the electron microscopy reconstruction, which placed the Rpn6 recognition helix within the cleft formed by the WH and HD of Rpn7 (ref. Citation24). As such, it is highly likely that the Rpn6 and Rpn7 interaction resembles the Sac3-Thp1 interface, whereby the recognition helix of Rpn6 would insert into the cleft formed by the WH and HD of Rpn7. Mutations that target the HD of CSN7 that negatively affect its interaction with CSN8 are also compatible with this general interaction paradigm being applied to the larger PCI-complexes. Together these data are consistent with there being a conserved PCI interaction mechanism mediated by crucial protein-protein interactions between the WH recognition helix and a cleft formed between the WH and HD domains. Future mutational studies and high-resolution crystal structures of these larger PCI-complexes should confirm whether the Sac3-Thp1 interaction interface holds across the multiple PCI interactions within larger PCI-complexes.
Function of Sem1 in PCI Proteins
The Sac3-Thp1-Sem1 structure, together with the recent progress on the structure of the 26S proteasome lid complex, raises the question of the functional role of Sem1 in PCI-complexes. Sem1 is present independently in a host of different complexes. In addition to associating with the PCI-complexes of Thp1-Sac3, CSN12-Thp3 and Rpn3-Rpn7, Sem1 also interacts with the structurally unrelated Integrator and BRCA-2 complexes.Citation2 Sem1 is a small highly negatively charged protein that has been suggested to act as a multitasking organizer of PCI-protein complexes.Citation2 However there is currently very little understanding of its functional mechanism. Within the Sac3-Thp1-Sem1 structure, Sem1 appeared to function primarily to stabilize Thp1, and a broad stabilizing role is consistent with previous studies that suggest Sem1 stabilizes both the 26S proteasome lid and the BRCA2-RAD51 complexes.Citation2,Citation29 Pick et al. have suggested that Sem1 could act to stabilize small PCI complexes by acting as a shield to block the interaction surface that would normally be utilized to bind the PCI-proteins, such as Rpn3-Rpn-7, to the larger hexameric PCI-complex of the 26S proteasome lid.Citation2 The Sac3-Thp1-Sem1 structure is compatible with such a mechanism, as the Sem1 C-terminus forms a highly conserved C-terminal helix that binds to the cleft formed by the WH and HD of Thp1. As discussed above, this site is critical on the corresponding Sac3 PCI-structure for binding of the Thp1 WH recognition helix, and thus PCI-complex formation. As such, these data reinforce previous suggestions which support a mechanism whereby Sem1 could modulate the structure of the larger PCI-domain complexes by binding to and shielding one-side of the PCI-interaction interface, effectively acting as a “capping” protein. The logistics of this possible function would need to be experimentally investigated, however it is conceivable that Sem1 could stabilize small-PCI complexes prior to assembly, inhibit complete PCI-complex formation, and even rapidly modulate the subunit composition of a pre-formed complex ().
Questions Outstanding
Although there is now considerable structural information available on PCI folds in a number of different proteins, atomic resolution information on the precise manner in which they interact in larger complexes is only available for the Sac3-Thp1-Sem1 complex,Citation7 albeit the fitting of the Rpn6 crystal structure into 9 Å resolution EM reconstructions of the 26S proteasome indicates that it may interact with components such as Rpn7 in a similar manner.Citation6,Citation24,Citation28 Clearly an important challenge is to obtain crystal structures of additional PCI-domain complexes to explore the precise atomic-resolution details of the interfaces between them. Several PCI-proteins appear to bind nucleic acids and, in the case of the Sac3-Thp1-Sem1 complex, some of the residues involved in this interaction have been identified using structure-based engineered mutations. However, details of the molecular recognition underlying interactions between PCI-proteins and nucleic acids need to be established to enable exploration of their functional significance.
Acknowledgments
We are most grateful to our colleagues in Melbourne and Cambridge for the helpful comments and suggestions. Supported in part by Medical Research Council Grant U105178939 (M.S.) and a Wellcome Trust Programme grant (M.S.). AME held EMBO and Marie Curie Postdoctoral fellowships.
References
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci 1998; 23:204 - 5; http://dx.doi.org/10.1016/S0968-0004(98)01217-1; PMID: 9644972
- Pick E, Hofmann K, Glickman MH. PCI complexes: Beyond the proteasome, CSN, and eIF3 Troika. Mol Cell 2009; 35:260 - 4; http://dx.doi.org/10.1016/j.molcel.2009.07.009; PMID: 19683491
- García-Oliver E, García-Molinero V, Rodríguez-Navarro S. mRNA export and gene expression: The SAGA-TREX-2 connection. Biochim Biophys Acta 2012; 1819; 555 - 65
- Rodríguez-Navarro S, Hurt E. Linking gene regulation to mRNA production and export. Curr Opin Cell Biol 2011; 23:302 - 9; http://dx.doi.org/10.1016/j.ceb.2010.12.002; PMID: 21227675
- Valkov E., Dean JC, Jani D, Kuhlmann SI, Stewart M. Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochim Biophys Acta 2012; 1819; 578 - 92
- Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Körner R, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci U S A 2012; 109:149 - 54; http://dx.doi.org/10.1073/pnas.1117648108; PMID: 22187461
- Ellisdon AM, Dimitrova L, Hurt E, Stewart M. Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nat Struct Mol Biol 2012; 19:328 - 36; http://dx.doi.org/10.1038/nsmb.2235; PMID: 22343721
- Wei Z, Zhang P, Zhou Z, Cheng Z, Wan M, Gong W. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J Biol Chem 2004; 279:34983 - 90; http://dx.doi.org/10.1074/jbc.M405158200; PMID: 15180986
- Scheel H, Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics 2005; 6:71; http://dx.doi.org/10.1186/1471-2105-6-71; PMID: 15790418
- Dessau M, Halimi Y, Erez T, Chomsky-Hecht O, Chamovitz DA, Hirsch JA. The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell 2008; 20:2815 - 34; http://dx.doi.org/10.1105/tpc.107.053801; PMID: 18854373
- Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol 2000; 10:110 - 6; http://dx.doi.org/10.1016/S0959-440X(99)00057-3; PMID: 10679470
- Enchev RI, Schreiber A, Beuron F, Morris EP. Structural insights into the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3. Structure 2010; 18:518 - 27; http://dx.doi.org/10.1016/j.str.2010.02.008; PMID: 20399188
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci U S A 1985; 82:8527 - 9; http://dx.doi.org/10.1073/pnas.82.24.8527; PMID: 3866238
- Fischer T, Rodríguez-Navarro S, Pereira G, Rácz A, Schiebel E, Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol 2004; 6:840 - 8; http://dx.doi.org/10.1038/ncb1163; PMID: 15311284
- Rodríguez-Navarro S, Fischer T, Luo MJ, Antúnez O, Brettschneider S, Lechner J, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 2004; 116:75 - 86; http://dx.doi.org/10.1016/S0092-8674(03)01025-0; PMID: 14718168
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 2006; 441:770 - 3; http://dx.doi.org/10.1038/nature04752; PMID: 16760982
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell 2008; 32:735 - 46; http://dx.doi.org/10.1016/j.molcel.2008.11.012; PMID: 19061648
- Fischer T, Strässer K, Rácz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, et al. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 2002; 21:5843 - 52; http://dx.doi.org/10.1093/emboj/cdf590; PMID: 12411502
- Jani D, Lutz S, Marshall NJ, Fischer T, Köhler A, Ellisdon AM, et al. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell 2009; 33:727 - 37; http://dx.doi.org/10.1016/j.molcel.2009.01.033; PMID: 19328066
- Kouba T, Rutkai E, Karásková M, Valášek LS. The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes. Nucleic Acids Res 2012; 40:2683 - 99; http://dx.doi.org/10.1093/nar/gkr1083; PMID: 22123745
- Chamovitz DA. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep 2009; 10:352 - 8; http://dx.doi.org/10.1038/embor.2009.33; PMID: 19305390
- Jimeno S, Tous C, García-Rubio ML, Ranes M, González-Aguilera C, Marín A, et al. New suppressors of THO mutations identify Thp3 (Ypr045c)-Csn12 as a protein complex involved in transcription elongation. Mol Cell Biol 2011; 31:674 - 85; http://dx.doi.org/10.1128/MCB.01188-10; PMID: 21149575
- Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell 2004; 16:1027 - 34; http://dx.doi.org/10.1016/j.molcel.2004.11.033; PMID: 15610744
- Lasker K, Förster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A 2012; 109:1380 - 7; http://dx.doi.org/10.1073/pnas.1120559109; PMID: 22307589
- Hoadley KA, Keck JL. Werner helicase wings DNA binding. Structure 2010; 18:149 - 51; http://dx.doi.org/10.1016/j.str.2010.01.007; PMID: 20159459
- Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure 2010; 18:177 - 87; http://dx.doi.org/10.1016/j.str.2009.12.011; PMID: 20159463
- Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. Structural basis for mRNA recognition by elongation factor SelB. Nat Struct Mol Biol 2005; 12:198 - 203; http://dx.doi.org/10.1038/nsmb890; PMID: 15665870
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012; 482:186 - 91; PMID: 22237024
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 2002; 297:1837 - 48; http://dx.doi.org/10.1126/science.297.5588.1837; PMID: 12228710