Abstract
IQGAP1 is an important cytoskeletal regulator, known to act at the plasma membrane to bundle and cap actin filaments, and to tether the cortical actin meshwork to microtubules via plus-end binding proteins. Here we describe the novel subcellular localization of IQGAP1 at the cytoplasmic face of the nuclear envelope, where it co-located with F-actin. The IQGAP1 and F-actin staining overlapped that of microtubules at the nuclear envelope, revealing a pattern strikingly similar to that observed at the plasma membrane. In detergent-extracted cells IQGAP1 was retained at cytoskeletal structures at the nuclear envelope. This finding has new implications for involvement of IQGAP1 in cell polarization and migration events and potentially in cell cycle-associated nuclear envelope assembly/disassembly.
Introduction
The nuclear envelope of eukaryotic cells separates nucleus from cytoplasm and mediates many cellular processes ranging from nuclear protein transport, cell polarization and cell migration, to chromatin regulation and cell cycle events.Citation1 The nuclear envelope consists of two phospholipid bilayers, the inner and outer nuclear membranes (INM and ONM, respectively), and is permeated with channels called nuclear pore complexes (NPCs) that facilitate selective movement of macromolecules between the two compartments.Citation2 Many of the proteins that traverse the INM and ONM have roles in chromatin regulation and cytoskeletal and nucleoskeletal tethering to the nuclear envelope.Citation3
Repositioning of the nucleus is mediated by all three cytoskeletal networks – microtubules, intermediate filaments and actin filaments.Citation4 Each cytoskeletal network associates with distinct outer nuclear envelope proteins that belong to large linker of nucleoskeleton and cytoskeleton (LINC) complexes.Citation4 These interactions can affect nuclear/microtubule organizing center (MTOC) repositioning events. LINC complexes comprise nesprin proteins that bind INM Sun proteins within the lumenal space of the nuclear envelope; Sun proteins in turn bind the nuclear lamina, which lies beneath the INM.Citation3 Several other cytoskeletal and nucleoskeletal elements are known to coordinate with LINC complexes to facilitate nuclear/MTOC repositioning. Cell polarization requires reorientation of the MTOC between the nucleus and the leading edge of the cell to drive locomotion.Citation5 Recent evidence suggests that an actin-dependent Cdc42/myosin II-driven mechanism repositions the nucleus behind a static MTOC during cell polarization of fibroblasts.Citation6 Actin-associated LINC complexes align along actin filaments adjacent to the nucleus to form TAN (transmembrane actin-associated nuclear) lines. These actin filaments exhibit retrograde flow and when coupled with LINC complexes drag the nucleus along with the forces of actin flow.Citation7
Cytoskeletal associations with the ONM are also important for nuclear envelope break down and re-assembly at the start and end of mitosis. Microtubules attached to the outer face of the ONM around centrosomes inflict invaginations in the nuclear envelope, causing mechanical tearing apart of the nuclear membranes.Citation8 The disintegration of the nuclear envelope permits the mitotic spindle access to the duplicated chromosomes enabling mitosis to proceed.Citation1
IQGAP1 orchestrates diverse cytoskeletal rearrangements, disseminating cortical cell signaling events for functions in cell:cell adhesion, cell migration, cell polarization and cell cycle.Citation9,Citation10 Reduced expression of IQGAP1 slows cell migration in several cell types.Citation11-Citation13 The role of IQGAP1 in cell migration is through direct action on filamentous actin (F-actin), either via bundlingCitation14 or capping the barbed-endsCitation15 of actin filaments. IQGAP1 also targets actin nucleating complexes to lamellipodial structures – plasma membrane regions of cell migration – thereby regulating actin polymerisation dynamics and its role in cell motility.Citation16 Silencing IQGAP1 expression also inhibits MTOC reorientation in several cell types.Citation12,Citation17 This effect is proposed to result from cortical cell polarization cues from IQGAP1-mediated tethering of peripheral actin meshworks to microtubules via plus-end proteins APC, CLIP-170 and others.Citation18 IQGAP1 at the plasma membrane also regulates stability of the adherens junction complex,Citation18 maintenance of which is vital for correct apical-basal polarity of the epithelium. Increased IQGAP1 expression augments these processes and can drive oncogenic transformation of various cancer types.Citation19,Citation20
In addition to the plasma membrane, IQGAP1 has been detected at other subcellular sites in mammalian cells including cell cycle-dependent localizations to the nucleusCitation21 and the midbody.Citation9,Citation22-Citation24 Here we describe the first observation of IQGAP1 at the cytoplasmic face of the nuclear envelope and postulate its role in regulating nucleo-cytoskeletal events.
Results
IQGAP1 is generally regarded as a plasma membrane-associated scaffolding protein as it is most commonly detected at cell:cell membrane junctions and membrane ruffles.Citation19 We previously confirmed the specificity of IQGAP1 polyclonal antibody H-109 (Santa Cruz Biotech, CA) by siRNA-mediated knockdown, and showed that this antibody detects similar endogenous IQGAP1 staining patterns to that of ectopically-expressed GFP-tagged IQGAP1.Citation21 A careful analysis of IQGAP1 subcellular localization with this antibody has revealed a ‘nuclear halo’ pattern of IQGAP1 by immunofluorescence microscopy, reminiscent of that previously reported for actin in interphase NIH 3T3 cells.Citation25 IQGAP1 is inextricably linked to F-actin,Citation14 therefore we co-stained for F-actin with FITC-conjugated phalloidin to determine the degree of co-localization. Human epithelial-derived breast (MCF-7) and colon (HT29) cancer cells, as well as immortalized non-tumorigenic mouse fibroblasts (NIH 3T3), each showed IQGAP1 nuclear halos that co-localized with F-actin (). The nuclear halo staining was also detected in SW480 (colon cancer), U2OS (osteosarcoma) and HeLa (cervical) cancer cells, but not in HCT116 colon cancer cells, and the staining pattern was abolished by IQGAP1 knockdown (data not shown).
Figure 1. Novel nuclear envelope localization of IQGAP1. (A) Nuclear halo staining of IQGAP1 occurs in several cell lines. Deconvolution fluorescence cell images of human epithelial-derived MCF-7 and HT29 cancer cells and mouse fibroblast NIH 3T3 cells. Cells were immunolabelled and stained for IQGAP1 (H-109, Santa-Cruz; red), F-actin (phalloidin-FITC; green) and DNA (Hoechst; blue). (B) IQGAP1 locates to the cytoplasmic face of the outer nuclear membrane. Deconvolution microscopy fluorescence cell images of MCF-7 cells stained with IQGAP1 (red) and FxFG-repeat nucleoporins (mAb414; green). The graph depicts pixel intensity of IQGAP1 (red) and nucleoporin (green) staining from the indicated cross-section in the enlarged micrograph (toward cytoplasm on right). (C) High resolution scoring of IQGAP1 co-localization with nuclear envelope in MCF-7 cells (n = 62). (D) Electron micrographs of ultrathin cryosections of MCF-7 cells immunolabelled with IQGAP1 pAb. Cells with no primary antibody showed no specific staining pattern (not shown). Thin closed arrow indicates nuclear rim; broad open arrow indicates immunogold-labeling of IQGAP1. White bar, 200 nm.
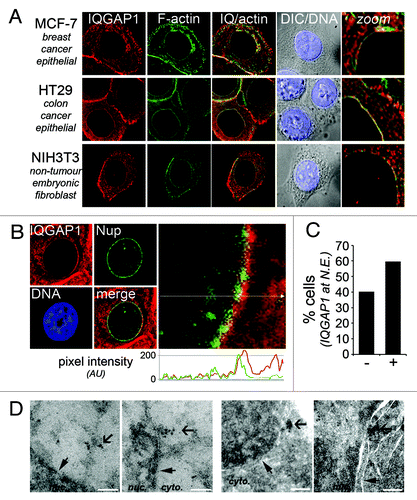
To determine whether the IQGAP1 nuclear halo correlates with INM or ONM localization we co-stained MCF-7 cells with IQGAP1 and with the nucleoporin (Nup) monoclonal antibody (mAb414) to highlight the nuclear pore complex. Confocal imaging of transverse-sections through the cell nucleus showed that IQGAP1 coats the outside of the nuclear envelope, locating at the cytoplasmic face of the nuclear membrane (), in ~60% of asynchronous MCF-7 cells (). To confirm this finding and better define the ultrastructural localization of IQGAP1 we analyzed cells by transmission electron microscopy. Electron micrographs of ultrathin cryosections of MCF-7 cells labeled with IQGAP1 H-109 pAb show distinct immunogold labeling at the nuclear envelope (). The EM staining of IQGAP1 was occasionally seen at nuclear pores but more often at electron dense structures emanating from the nuclear envelope.
Due to the co-localization of IQGAP1 with F-actin (), we sought to determine whether IQGAP1 was anchored by cytoskeletal structures at the nuclear rim. MCF-7 cells were transfected with plasmid DNA encoding GFP-tagged emerin, an insoluble INM protein that anchors lamina and chromatin to the INM,Citation26 and the cells then permeabilized with Triton X-100 to wash out all soluble material. Emerin-GFP expressing cells were either fixed-first (-CSK buffer) or detergent-extracted (+CSK buffer; soluble material washed out) then fixed and stained for IQGAP1 (). After mild CSK extraction, IQGAP1 remained clearly visible at the nuclear rim by confocal microscopy (), where it overlapped emerin staining in punctuate clusters at the envelope but predominantly was anchored at structures on the cytoplasmic side of the pores.
Figure 2. IQGAP1 is associated with cytoskeletal architecture at the nuclear envelope. (A and B) IQGAP1 is anchored at the nuclear rim. MCF-7 cells transiently expressing emerin-GFP were either (A) fixed-first (-CSK) and detergent permeabilized or (B) CSK-extracted (+CSK) and fixed, then stained for IQGAP1 (red). (C) Confocal fluorescence microscopy images of CSK-extracted MCF-7 cells tri-labeled for IQGAP1 (a; red), F-actin (b; green) and β-tubulin (c; blue). Bottom panel and enlarged image on right show merged fluorescence micrographs. (D) Schematic proposing the roles of IQGAP1 at the cytoplasmic face of the nuclear rim. a, At plasma membrane sites involved in cell migration IQGAP1 tethers microtubule networks to the cortical actin mesh via the MT +end protein APC for cell polarization cues. b, Tethering for polarization cues? At the cytoplasmic face of the NE, IQGAP1 overlays with MTs and actin and may tether these cytoskeletal networks via APC. c, Nuclear repositioning? TAN lines in mesenchymal cells assist in nuclear repositioning during cell polarization and cell migration. d, Orchestrating actin polymerisation and rearrangements? IQGAP1 targets numerous actin-associated proteins to subcellular sites. IQGAP1 anchors actin-branching and nucleating proteins to membrane ruffles and to yeast cytokinetic ringsCitation9 to orchestrate actin rearrangements. IQGAP1 may target other proteins to the nuclear envelope during cell migration or cell cycle events such as during nuclear envelope breakdown.
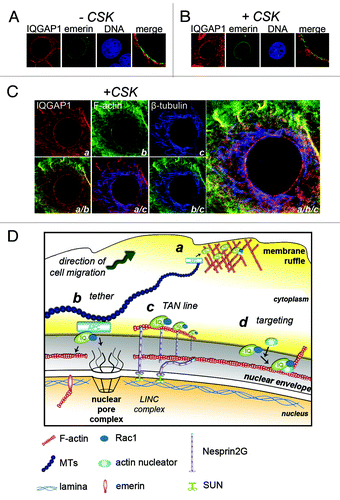
At the plasma membrane IQGAP1 often co-localizes with both F-actin and microtubules.Citation18 In CSK-extracted MCF-7 cells tri-labeled with IQGAP1, F-actin and tubulin, IQGAP1 staining was also found to overlap with both F-actin and microtubules at the ONM (), implicating a cytoskeletal tethering role analogous to that at plasma membrane sites.
Discussion
We provide here the first reported detection of IQGAP1 at the outer nuclear envelope. We previously showed that IQGAP1 accumulates in the nucleus of cells but only when cells are arrested in late G1/early S phase, implicating a nucleoplasmic role of IQGAP1 in the DNA replication stress response.Citation21 The observed co-staining with emerin at punctuate clusters in the nuclear envelope is interesting in light of a recent study that linked emerin with IQGAP1. Emerin is an INM protein occasionally also detected at the ONM, and is thought to stabilize the nuclear lamina through its association with lamin A and chromatin-binding proteins.Citation27 A proteomics study identified IQGAP1 within two distinct emerin-containing multi-protein fractions from HeLa cell nuclei.Citation28 Interestingly, emerin also binds and stimulates actin polymerisation through a pointed-end capping mechanism.Citation29
At the plasma membrane, IQGAP1 regulates actin filaments at dynamic membrane ruffles – through bundling, cross-linking and barbed-end capping of actin filaments – for roles in cell migration and macropinocytosis.Citation14,Citation18,Citation30 IQGAP1 functions as a cross-linking agent at the plasma membrane, as it also tethers microtubules through its interaction with plus-end binding proteins APC and CLIP-170 for cortical cell polarization cues ().Citation18 Our combined confocal and ultrastructural EM data strongly suggest a similar cytoskeleton scaffolding role for IQGAP1 at the nuclear envelope (). Indeed, the IQGAP1 partner, APC, was reported to bind the nucleoporin Nup153 to promote nuclear envelope anchorage of proximal microtubules emanating from the centrosome.Citation31 Perinuclear actin has been shown to polymerise at the cytoplasmic face of the nuclear envelope.Citation32 We thus speculate that one role of IQGAP1 might be to tether microtubules to perinuclear actin to regulate MTOC and nuclear positioning for cell polarization during cell migration.
In migrating fibroblasts, repositioning of the nucleus can be more important than that of the MTOC in cell polarization.Citation6 Retrograde flow of actin cables coupled to TAN lines provide the physical forces to move the nucleus into an appropriate nuclear-centrosome axis during polarized cell migration ().Citation5,Citation7 Silencing IQGAP1 expression in Vero fibroblasts inhibited MTOC reorientation.Citation12 This effect may not only have been due to a deficiency in IQGAP1 cortical polarization cues, signaled from IQGAP1-APC-microtubule complexes, as suggested, but perhaps also attributable to defects in actin dynamics at the perinuclear zone. IQGAP1 targets several actin regulating proteins to membrane ruffles, including APC,Citation12 Diaphanous1,Citation33 and N-WASp.Citation34,Citation35 By analogy, we therefore predict that IQGAP1 can determine ONM localization of specific actin-associated binding partners for cytoplasmic actin rearrangements at the perinuclear zone (). Moreover, a cytoskeletal cross-linking role of IQGAP1 could contribute to regulation of nuclear envelope breakdown or reassembly, a mitosis-timed process involving both actin and microtubule motor proteins.Citation8 Future proteomics experiments aimed at defining the composition of ONM IQGAP1 protein complexes will help resolve its functional roles, and determine whether this protein might coordinate actin/microtubule rearrangements at the nucleus and plasma membrane.
Materials and Methods
Cell culture, reagents and transfection
MCF-7, HT29 and NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics (penicillin and streptomycin) at 37°C in 5% CO2 humidified atmosphere. Cells were grown on glass coverslips in 6-well dishes (Nunc) for immunofluorescence microscopy analysis. Cells were transfected with plasmid DNA as previously described.Citation30
Immunocytochemistry, antibodies and plasmids
Cells were washed, fixed and immuno-stained as previously described,Citation30 then visualized by fluorescence microscopy. The following antibodies and dilutions were used for immunofluorescence (IF): IQGAP1 polyclonal antibody (1:150; Santa Cruz H-109), FxFG-repeat nucleoporin monoclonal antibody (1:2000 by MeOH fixation; 1:500 by formalin fixation; Covance mAb414 (MMS-120P)) and β-tubulin monoclonal antibody (IF 1:2000; Sigma #T0198). FITC-phalloidin (0.5 μg/ml) was used to label F-actin and Hoechst dye was used to stain chromatin; each were purchased from Sigma. Secondary antibodies used were anti-mouse Alexa-Fluor-405 (1:50), anti-mouse or anti-rabbit Alexa-Fluor-488 (1:500) and anti-mouse or anti-rabbit AlexaFlour-594 (1:1500) (Molecular Probes). Emerin-GFP plasmid DNA was a kind gift from Ewa Markiewicz (Durham University).Citation26
In situ retention assay
To determine the extent of nuclear retention in situ, a detergent extraction assay was used to remove soluble proteins from cells prior to fixation. Cells were grown on poly-l-lysine (Sigma) coated coverslips. For CSK-extraction (+CSK), cells were incubated in CSK extraction buffer (10 mM Pipes (pH 6.8); 300 mM sucrose; 5 mM MgCl2; 100 mM NaCl; 0.1% Triton X-100) containing protease inhibitor cocktail (Roche) for 1 min on ice. The processed cells were fixed with 3.7% formaldehyde/PBS for 15 min at RT and processed according to the immuno-staining protocol. For control cells (-CSK), the cells were prepared in a similar way except that they were first fixed with 3.7% formaldehyde before they were processed with CSK extraction buffer. Immunofluorescence images were taken at equal exposures in each condition.
Immunofluorescence image acquisition
For basic fluorescence analysis for subcellular localization studies samples were observed under an Olympus BL51 fluorescence microscope. For advanced fluorescence image analysis, cells were visualized through a 60 x oil immersion lens using Olympus FV1000 confocal laser scanning microscope with images processed using Fluoview Version 1.6a software, or cells were visualized through a 100 x 1.4 numerical aperture oil immersion lens with an inverted Olympus IX-70 microscope (DeltaVision Image Restoration Microscope; Applied Precision/Olympus) and a photometrics CoolSnap QE camera. We acquired 10–20 serial optical sections of 0.2–0.5 μm. Then the images were deconvolved and generated volume projections of the entire z-series using DeltaVision SoftWoRx software (version 3.4.4.) The images were compiled in Adobe Photoshop CS5.
Electron microscopy and image acquisition
MCF-7 cells were grown in DMEM to 70% confluence. On day of harvest, cells were rinsed in serum-free media for 30 min at 37°C. Media was replaced with 4% formaldehyde [freshly prepared from paraformaldehyde] (PF)/Sorensens phosphate buffer (SPB) + 0.1% EM grade glutaraldehyde at RT for 30 min. Fixative replaced with fresh 4% PF/SPB for a further 30 min. Cells were then resuspended in 12% gelatin. Cell/gelatin pellet was infiltrated in 2.3 M sucrose for ~3–5 d at 4°C with over-end mixing and then frozen in LN2.
70–90 nm sections were cut at –80 to –100°C in a Leica Ultracut S/FCS cryo- ultramicrotome using a Diatome 35° cryo diamond knife. Sections were collected on Formvar/Pioloform-coated nickel grids (GCu200tbh, PST), then incubated in 50 mM glycine/PBS for ~15 min at RT and blocked in BSA for 30 min. Sections were immunolabelled with IQGAP1 polyclonal antibody (1:10; Santa Cruz H-109) overnight at 4°C. Secondary immunolabelling with 10 nm gold-conjugated goat anti-rabbit IgG (ProSciTech; JB15726) was performed at RT using a Leica EM IGL immunolabelling machine. Sections were embedded in methylcellulose (2%) and uranyl acetate (0.3%) for ~15 min on ice.
Samples were imaged using a Philips CM10 Transmission Electron Microscope operated at 80kV. Images were recorded using an SIS Megaview G2 digital camera and iTEM software. Post processing of images for cropping and contrast / brightness adjustment was performed using Adobe Photoshop CS5.
| Abbreviations: | ||
| APC | = | adenomatous polyposis coli |
| CLIP-170 | = | cytoplasmic linker protein 170 |
| CSK | = | cytoskeletal |
| F-actin | = | filamentous actin |
| INM | = | inner nuclear membrane |
| IQGAP1 | = | IQ-domain GTPase-activating protein 1 |
| LINC | = | linker of nucleoskeleton and cytoskeleton |
| MTOC | = | microtubule organising centre |
| N-WASp | = | neural-Wiskott–Aldrich syndrome protein |
| ONM | = | outer nuclear membrane |
| TAN line | = | transmembrane actin-associated nuclear line |
Acknowledgments
The authors wish to thank members of the Henderson lab for helpful comments and discussions, and give particular appreciation to Emma Kettle and Ross Boadle (Institute for Clinical Pathology and Medical Research, Westmead, Sydney) for their assistance with electron microscopy. This work was funded by grants to B.R.H. from the National Health and Medical Research Council (NH&MRC) of Australia and the Cancer Council of New South Wales. B.R.H. is a Senior Research Fellow of the NH&MRC.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000539; http://dx.doi.org/10.1101/cshperspect.a000539; PMID: 20300205
- Görlich D, Kutay U. Transport between the cell nucleus and the cyotplasm. Annu Rev Cell Dev Biol 1999; 15:607-60; 10611974 doi:10.1146/annurev.cellbio.15.1.607.
- Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci 2010; 123:1973 - 8; http://dx.doi.org/10.1242/jcs.019042; PMID: 20519579
- Starr DA. Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Mol Biosyst 2007; 3:583 - 9; http://dx.doi.org/10.1039/b703878j; PMID: 17700857
- Luxton GWG, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol 2011; 23:579 - 88; http://dx.doi.org/10.1016/j.ceb.2011.08.001; PMID: 21885270
- Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 2005; 121:451 - 63; http://dx.doi.org/10.1016/j.cell.2005.02.022; PMID: 15882626
- Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 2010; 329:956 - 9; http://dx.doi.org/10.1126/science.1189072; PMID: 20724637
- Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 2009; 10:178 - 91; http://dx.doi.org/10.1038/nrm2641; PMID: 19234477
- Shannon KB. IQGAP Family Members in Yeast, Dictyostelium, and Mammalian Cells. Int J Cell Biol 2012; 2012:894817; http://dx.doi.org/10.1155/2012/894817; PMID: 22505937
- White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cell Signal 2012; 24:826 - 34; http://dx.doi.org/10.1016/j.cellsig.2011.12.005; PMID: 22182509
- Mataraza JM, Briggs MW, Li Z, Frank R, Sacks DB. Identification and characterization of the Cdc42-binding site of IQGAP1. Biochem Biophys Res Commun 2003; 305:315 - 21; http://dx.doi.org/10.1016/S0006-291X(03)00759-9; PMID: 12745076
- Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, et al. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell 2004; 7:871 - 83; http://dx.doi.org/10.1016/j.devcel.2004.10.017; PMID: 15572129
- Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L, Dikalov SI, Chen YE, Tojo T, et al. IQGAP1, a novel vascular endothelial growth factor receptor binding protein, is involved in reactive oxygen species--dependent endothelial migration and proliferation. Circ Res 2004; 95:276 - 83; http://dx.doi.org/10.1161/01.RES.0000136522.58649.60; PMID: 15217908
- Mateer SC, Wang N, Bloom GS. IQGAPs: integrators of the cytoskeleton, cell adhesion machinery, and signaling networks. Cell Motil Cytoskeleton 2003; 55:147 - 55; http://dx.doi.org/10.1002/cm.10118; PMID: 12789660
- Pelikan-Conchaudron A, Le Clainche C, Didry D, Carlier M-F. The IQGAP1 protein is a calmodulin-regulated barbed end capper of actin filaments: possible implications in its function in cell migration. J Biol Chem 2011; 286:35119 - 28; http://dx.doi.org/10.1074/jbc.M111.258772; PMID: 21730051
- Brandt DT, Grosse R. Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep 2007; 8:1019 - 23; http://dx.doi.org/10.1038/sj.embor.7401089; PMID: 17972901
- Kanwar N, Wilkins JA. IQGAP1 involvement in MTOC and granule polarization in NK-cell cytotoxicity. Eur J Immunol 2011; 41:2763 - 73; http://dx.doi.org/10.1002/eji.201040444; PMID: 21681737
- Noritake J, Watanabe T, Sato K, Wang S, Kaibuchi K. IQGAP1: a key regulator of adhesion and migration. J Cell Sci 2005; 118:2085 - 92; http://dx.doi.org/10.1242/jcs.02379; PMID: 15890984
- Johnson M, Sharma M, Henderson BR. IQGAP1 regulation and roles in cancer. Cell Signal 2009; 21:1471 - 8; http://dx.doi.org/10.1016/j.cellsig.2009.02.023; PMID: 19269319
- White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett 2009; 583:1817 - 24; http://dx.doi.org/10.1016/j.febslet.2009.05.007; PMID: 19433088
- Johnson M, Sharma M, Brocardo MG, Henderson BR. IQGAP1 translocates to the nucleus in early S-phase and contributes to cell cycle progression after DNA replication arrest. Int J Biochem Cell Biol 2011; 43:65 - 73; http://dx.doi.org/10.1016/j.biocel.2010.09.014; PMID: 20883816
- Morita E, Sandrin V, Chung H-Y, Morham SG, Gygi SP, Rodesch CK, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 2007; 26:4215 - 27; http://dx.doi.org/10.1038/sj.emboj.7601850; PMID: 17853893
- Skop AR, Liu H, Yates J 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 2004; 305:61 - 6; http://dx.doi.org/10.1126/science.1097931; PMID: 15166316
- Tekletsadik YK, Sonn R, Osman MA. A conserved role of IQGAP1 in regulating TOR complex 1. J Cell Sci 2012; 125:2041 - 52; http://dx.doi.org/10.1242/jcs.098947; PMID: 22328503
- Clubb BH, Locke M. Peripheral nuclear matrix actin forms perinuclear shells. J Cell Biochem 1998; 70:240 - 51; http://dx.doi.org/10.1002/(SICI)1097-4644(19980801)70:2<240::AID-JCB10>3.0.CO;2-R; PMID: 9671230
- Markiewicz E, Tilgner K, Barker N, van de Wetering M, Clevers H, Dorobek M, et al. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J 2006; 25:3275 - 85; http://dx.doi.org/10.1038/sj.emboj.7601230; PMID: 16858403
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol 2005; 6:21 - 31; http://dx.doi.org/10.1038/nrm1550; PMID: 15688064
- Holaska JM, Wilson KL. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry 2007; 46:8897 - 908; http://dx.doi.org/10.1021/bi602636m; PMID: 17620012
- Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol 2004; 2:E231; http://dx.doi.org/10.1371/journal.pbio.0020231; PMID: 15328537
- Sharma M, Henderson BR. IQ-domain GTPase-activating protein 1 regulates beta-catenin at membrane ruffles and its role in macropinocytosis of N-cadherin and adenomatous polyposis coli. J Biol Chem 2007; 282:8545 - 56; http://dx.doi.org/10.1074/jbc.M610272200; PMID: 17255093
- Collin L, Schlessinger K, Hall A. APC nuclear membrane association and microtubule polarity. Biol Cell 2008; 100:243 - 52; http://dx.doi.org/10.1042/BC20070123; PMID: 18042042
- Münter S, Enninga J, Vazquez-Martinez R, Delbarre E, David-Watine B, Nehrbass U, et al. Actin polymerisation at the cytoplasmic face of eukaryotic nuclei. BMC Cell Biol 2006; 7:23; http://dx.doi.org/10.1186/1471-2121-7-23; PMID: 16719903
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol 2007; 178:193 - 200; http://dx.doi.org/10.1083/jcb.200612071; PMID: 17620407
- Benseñor LB, Kan H-M, Wang N, Wallrabe H, Davidson LA, Cai Y, et al. IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J Cell Sci 2007; 120:658 - 69; http://dx.doi.org/10.1242/jcs.03376; PMID: 17264147
- Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, et al. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem 2007; 282:426 - 35; http://dx.doi.org/10.1074/jbc.M607711200; PMID: 17085436