Abstract
While the bacteriocin Nisin has been employed by the food industry for 60 y, it remains the only bacteriocin to be extensively employed as a food preservative. This is despite the fact that the activity of Nisin against several food spoilage and pathogenic bacteria is poor and the availability of many other bacteriocins with significant potential in this regard. An alternative route to address the deficiencies of Nisin is the application of bioengineered derivatives of the peptide which, despite differing only subtly, possess enhanced capabilities of commercial value. The career path which has taken me from learning for the first time what bacteriocins are to understanding the potential of bacteriocin bioengineering has been a hugely enjoyable experience and promises to get even more interesting in the years to come.
My Introduction to the World of “Bioengineering”
My career as a “bioengineer” of microbes took flight 11 years ago when I was recruited by my former mentors, and now collaborators, Colin Hill and Paul Ross to use genetic approaches to manipulate bacteriocin producing strains. Bacteriocins are ribosomally synthesized antimicrobial peptides produced by one bacterium that are active against other bacteria, either in the same species (narrow spectrum), or across genera (broad spectrum). Producer organisms are immune to their own bacteriocin(s), a property that is mediated by specific immunity proteins.Citation1 Bacteriocins and bacteriocin producers have attracted significant interest from a fundamental and commercial perspective over the years. My first responsibility in my new bacteriocin-related role was to adapt the tools which I’d employed when creating mutants of Listeria monocytogenes (in that instance with a view to identifying genes involved in stress resistanceCitation2,Citation3) and apply them to bacteriocin-producing lactococci instead. The idea of modifying bacteriocins produced by Gram positive bacteria was not a new one. As a consequence of the ribosomal nature of these antimicrobials and, thus, the fact that bacteriocin producing bacteria possess a gene which encodes the structural (albeit as yet inactive) peptide, it was recognized that bacteriocins were likely to be more tolerant of bioengineering than classical antibiotics, as the latter are typically generated from small building blocks through muti-enzyme complexes i.e., are non-ribosomal in nature. Site-directed approaches were first employed in bacteriocin research in the early 1990s when they were applied to the lantibiotics (a group of bacteriocins which undergo posttranslational modification and are thus members of the now expanding class I, i.e., modified, bacteriocins) nisin and subtilin by trailblazers such as Oscar Kuipers, Norm Hansen and Mike Gasson.Citation4-Citation6 Subsequently, the efforts of Cindy van Kraaij in the Kuipers laboratory were critical in the creation of nisin derivatives which contributed to the mechanism of action of the antimicrobial being elucidated.Citation7-Citation10 These developments in turn prompted parallel investigations which focused on other lantibiotics such as epidermin/gallidermin,Citation11 Pep5,Citation12 mutacin II,Citation13 lacticin 481,Citation14 mersacidinCitation15 and cinnamycin.Citation16
Despite being slower to get out of the starting blocks, the manipulation of unmodified, i.e., Class II, Gram positive bacteriocins became a hot topic after 1996, primarily as a consequence of the ground-breaking work by Gunnar Fimland and Jon Nissen Meyer who created a number of hybrid class II peptides, or class II peptides which had been subjected to more subtle changes, to provide a detailed insight into how these antimicrobials work.Citation17-Citation22 While these class II-related developments continued, the momentum that had been built up in the class I field had slowed somewhat, presumably as a consequence of the failure of bioenginering to generate lantibiotic derivatives with enhanced activity against Gram positive pathogens.
Standing on the Shoulder of Giants
The aforementioned frustrations did not dampen our enthusiasm with respect to becoming engaged in the bacteriocin bioengineering field as our initial goal was to apply bioengineering to the research of the lantibiotic lacticin 3147 in a manner similar to that which had been employed by the aforementioned "giants" in the field for nisin and other bacteriocins in the past. Lacticin 3147 had been identified by our joint Teagasc-University College Cork research team (also known as the Cork Bacteriocin Group) in 1995 [Aside – Teagasc is a Gaelic word that translates to "teaching" or "instruction" and is the name given to the agriculture and food development authority in Ireland]. Lacticin 3147 initially attracted attention by virtue of its activity at neutral pH and the fact that it possesses greater activity than nisin against several targets.Citation23,Citation24 However, from a fundamental perspective, lacticin 3147 is also interesting by virtue of being a two peptide lantibiotic and the fact that it possesses three D-alanine residues (introduced through post-translational modification) across the two peptides (Ltnα and Ltnβ).Citation25 It was the latter characteristic that first prompted us to become interested in the utilization of bioengineering based strategies. D-amino acids are exceedingly rare in ribosomally synthesized peptides, with lacticin 3147, another lantibiotic lactocin SCitation26 and a handful of eukaryotic peptides being notable exceptions. In the case of the two lantibiotics, the means via which these D-amino acids are incorporated is particularly unusual in that it involves a post-translational modification which changes both the identity and chirality of the corresponding residue in the unmodified peptide, i.e., from L-serine to D-alanine. To facilitate this, we modulated a strategy developed by Leenhouts et al.,Citation27 to facilitate the creation of bioengineered lacticin 3147 peptides through a "food-grade" approach.Citation28 This technique took advantage of the temperature-sensitive RepA+ plasmid pVE6007 and the RepA− vector pORI280 and relies crucially on the temporary integration of pORI280 into the target plasmid (facilitated by using a pORI280 derivative containing an insert bearing homology to the target plasmid). This cointegrate is stable in the presence of an antibiotic marker, but its resolution can be readily detected by screening for the loss of β-galactosidase activity or the erythromycin resistance phenotype associated with pORI280 when the selective pressure is removed.Citation28 Armed with this strategy, we set about changing the relevant serine codons in the corresponding genes to codons for glycine, L-alanine, L-valine and L-threonine, respectively. This analysis revealed that the natural d-alanines were required for optimal activity and that replacement of these with L-alanine and L-valine had extremely negative consequences. Notably, however, the incorporation of residues that lacked chirality, i.e., glycine or dehydrobutyrine (the latter was incorporated as a consequence of the post-translational modification of L-threonine), was better tolerated with respect to production levels and activity.Citation29
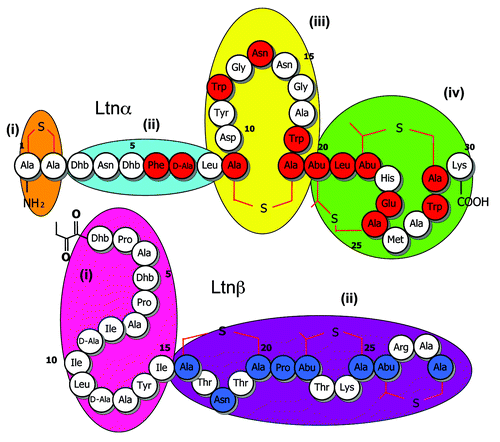
The aforementioned bioengineering-based strategy was attractive in that it facilitated the creation of lantibiotic producing derivatives which changed from the corresponding parental strain with respect to one codon, and in some cases one nucleotide, alone. This was potentially of great value in situations where the producing strains were food-grade bacteria such as lactococci and there was a desire to preserve their food-grade or non-genetically modified (non-GM) status. It also ensured that the changes made were stable. The downside was that the creation of each mutant took a minimum of 2 mo. It was thus, with some hesitation, that Lucy Deegan, a PhD student at the time, Elaine Lawton (research assistant) and myself took on the daunting task of applying this technology to carry out alanine scanning mutagenesis of all 59 amino acids across the lacticin 3147 peptides i.e., to create 59 mutants in which each amino acid (Ltnα 30 residues, Ltnβ 29 residues) was in turn converted to alanine. I recall the look of incredulity on the faces of Hans-Georg Sahl and Imke Wiedemann when we informed them of our plans at the Gordon Research Conference in Barga, Italy in 2003 with the primary question being, to paraphrase, “Why bother?” Colin, Paul and I explained that the logic was that such an approach would provide a valuable insight into the tolerance or intolerance of different regions of the peptides to change, thereby highlighting important functional domains and identifying regions that may accommodate further bioengineering in the future. We estimated that, were we to have created these derivatives one at a time, the process would have taken 118 mo (or almost 10 y) but through the creation of multiple derivatives simultaneously and the significant efforts of Elaine and Lucy, we crossed the finishing line in under 2 y. To facilitate the rapid investigation of the consequences of alanine incorporation, we assessed the "bioactivity" of these strains. "Bioactivity" based assays are those which assess the antimicrobial activity of bacteriocin-producing strains and make no effort to discriminate between changes in activity that are due to altered production levels or the altered specific antimicrobial activity of the peptide. These assays revealed that several residues appeared intolerant of change in that conversion to alanine resulted in the elimination of bioactivity. This included several residues within a proposed receptor (lipid II) binding domain and others apparently which were proposed to be involved in peptide-peptide interactionsCitation30 ().
Figure 1. Insights revealed from alanine scanning mutagenesis of the lacticin 3147 peptides and subsequent bioactivity based analyses. Residues that are apparently intolerant of change, on the basis of the elimination of bioactivity following alanine conversion, are depicted in red and blue in Ltnα and Ltnβ, respectively. Apparently distinct functional domains in Ltnα (i-iv) and Ltnβ (i and ii) are grouped according to oval shapes of different color.
Undeterred by the mental scarring inflicted by alanine scanning, Lucy subsequently took the lead in a study aiming at a closer inspection of the consequences to manipulating the charged residues in lacticin 3147 in different ways. This baton was taken up, in 2007, by a new PhD student, Srinivas Suda. The combined efforts of Lucy and Srinivas resulted in the generation of 16 additional derivatives and allowed us to confirm the importance of LtnαE24, reveal the requirement for positively charged residues in Ltnβ when targeting cells with reduced levels of cell envelope-associated D-alanylation or lysinylation and resulted in, for the first time, the creation of a derivative of a lacticin 3147 peptide (LtnβR27A) which displays enhanced specific activity, albeit only against Lactococcus lactis. Notably, however, this enhancement was not evident when this peptide was combined with its partner, Ltnα, peptide.Citation31,Citation32 Prior to the completion of his studies, Srinivas also investigated the tolerance of the lanthionine structures in the two peptides to change. He noted that switching lanthionine and β-methyllanthionine bridges in the peptides had variable consequences. Notably, it was also apparent that although the N-terminal lanthionine bridge in Ltnα is unusual in that its removal does not eliminate antimicrobial activity, the presence of this structure does confer Ltnα with enhanced resistance to thermal and proteolytic degradation.Citation33
Random Thoughts
In 2003 we were fortunate enough to recruit a new PhD student, Des Field, who, among other things, was charged with developing systems to facilitate the random mutagenesis of lacticin 3147. Based on the blueprint that had been developed through site-directed mutagenesis, we were now becoming more ambitious and suspected that the "Holy Grail," i.e., the generation of lantibiotic derivatives with enhanced activity against Gram positive pathogens, may be achievable. However, given the lack of success in this regard in the past, we didn’t want to rely exclusively on our ability to predict which changes might allow this goal to be realized. Thus we decided to implement a parallel strategy whereby random bioengineering was employed. The logic in this case was that if we generated a large enough bank, coupled with a system that would allow random screening thereof, we could identify bioengineered peptides of interest. However, as is evident from the above, the creation of a large bank of bioengineered peptides using existing strategy wasn’t feasible and so an alternative approach was required. For this purpose we used a two plasmid system consisting of one vector containing the Ltnα and β-encoding genes (ltnA1 and ltnA2) and their corresponding promoter and a second containing all of the other genes required for biosynthesis of and immunity to lacticin 3147. Once it was established that this approach facilitated the production of lacticin 3147, the next step was to employ error-prone PCR to randomly generate errors in ltnA1A2 and express them using our new system. This system did not yield "food-grade" lantibiotic producers but, were peptides of interest to emerge, the option was available to re-create the peptides of greatest interest using the more laborious, "food-grade" approach. An initial screen of a small bank of bioengineered peptide-producers revealed the success of the strategy and validated the findings of the alanine scanning study.Citation34 However, at this time we got distracted by what we anticipated would be a short summer project that took on a life of its own.
Nisincredible
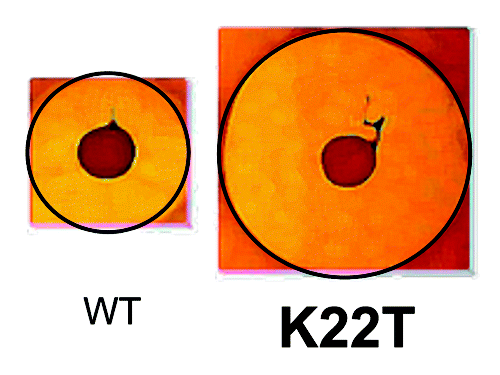
In the summer of 2006, we hosted an undergraduate student who was keen to learn some molecular microbiology. Not being sure how the student, Evelyn Molloy, would work out, we thought it best to develop a dedicated project that was distinct from our ongoing lacticin 3147 studies. Thus we thought it might be interesting to engage in the first nisin bioengineering studies in our group. In hindsight it is probably surprising that it has taken us so long to engage in nisin research given that it is the prototypical lantibiotic, having been first marketed in England in 1953 and has since been approved for use in over 50 countries. Nisin has been assessed to be safe for food use by the Joint Food and Agriculture Organization/World Heath Organization Expert Committee on Food Additives in 1969. In 1983, this bacteriocin was added to the European food additive list as number E234 (indeed it is the only natural antibacterial to have been approved for as a food preservative by the EU) and, in 1988, it was approved by the US Food and Drug Agency (FDA) for use in pasteurized, processed cheese spreads and is currently used in a wide variety of foods across the world.Citation35 As noted earlier, nisin had previously been the focus of several bioengineering based studies but, notably, prior to our entry into the field, random mutagenesis of nisin had only been performed on one previous occasion and in that instance was on a relatively small scale.Citation36 We postulated that if we created a considerably larger bank of nisin derivative producers and developed an efficient means of collecting and screening these, there would be a greater chance of finding elusive "enhanced" derivatives. There had been some exceptional instances of success in that some nisin derivatives with enhanced activity against Gram positive indicator strains had been identifiedCitation37-Citation40 and some nisin derivatives with enhanced activity against Gram negative targets had also been identified.Citation41 However, derivatives with activity against Gram positive pathogens, i.e., the targets that nisin is usually employed to control, remained elusive. Aided by our lacticin 3147-associated bioengineering experience, a bank of 8000 producers was rapidly generated and put to good use long after the end of Evelyn’s summer project. As a consequence of the availability of this resource, Des became increasingly involved in this new nisin-related research, especially so when it appeared that the long sought-after "holy grail" had been uncovered i.e., a producer displaying an enhanced zone of inhibition against a Gram positive pathogen (Streptococcus agalactiae ATCC13813) ().Citation42 The enhanced strain was found to produce a nisin that possessed a lysine to threonine change at position 22 (K22T) of the nisin peptide, within a region of the peptide known as the "hinge" i.e., a three amino acid stretch (consisting of Asn-Met-Lys) which links the receptor-binding N-terminal domain and C-terminal pore-forming domains. Notably, other changes within this hinge region were responsible for the enhanced activity of other derivatives, referred to above, against Gram negative targets. A more targeted bioengineering of hinge-associated residues which involved site directed mutagenesis and site saturation mutagenesis followed. The latter was prompted by the success of Rick Rink, Gert Moll and others at BioMade who had used saturation mutagenesis to elegantly investigate the consequences of changing residues within the N-terminal domain of Nisin.Citation38 Our targeted manipulation of the hinge uncovered several additional derivatives of interest with those containing N20P, M21V, K22S or the aforementioned K22T being selected for closer inspection as a consequence of the impressive bioactivity of the associated producers. Through the purification of these peptides and the completion of minimum inhibitory concentration-based assays, it was apparent that these "lead" peptides possessed enhanced specific activity but that this activity could be target specific e.g., Nisin N20P exhibited enhanced activity against Staphylococcus aureus but activity against Streptococcus agalactiae was reduced.Citation42 The producers of the most interesting Nisin derivatives were re-created using a "food-grade" approach corresponding to that described for lacticin 3147 above and further investigations with M21V and K22T, which were renamed Nisin V and Nisin T, respectively, established that both peptides exhibited enhanced activity relative to Nisin A against a broad spectrum of targets. More specifically, broth-based minimum inhibitory concentration assays revealed that Nisin T was enhanced against S. agalactiae, Streptococus mutans, Clostridium difficile, several S. aureus strains, L. lactis and a variety of mycobacteria while Nisin V was enhanced against this same selection but differed by virtue of also exhibiting enhanced activity against Listeria monocytogenes, Enterococcus faecium and Bacillus cereus.Citation43,Citation44 These enhanced potencies were apparent when growth and kill curve type assays were employed or when Nisin V was added to frankfurter meat in order to control a lux-tagged L. monocytogenes.Citation43 A number of other Nisin derivatives with enhanced specific activity have been identified and corresponding manuscripts have recently been submitted. Finally, some Nisin hinge-derivatives seem to instead be specifically enhanced in complex matrices such as those containing agar or carrageenan. In two instances, i.e., peptides containing hinges consisting of Ser-Val-Ala or Asn-Ala-Lys, this translated to enhanced activity against L. monocytogenes in a carrageenan-containing chocolate milk despite the fact that neither peptide exhibited enhanced activity against this pathogen when assessed using broth-based MIC assays.Citation45
Legislation
It has been suggested that we are entering into a ‘Golden era’ of bacteriocin engineering.Citation46 Comprehensive bioengineering based strategies corresponding to those described above have also been employed to study and/or enhance other lantibiotics such as actagardine, mersacidin and nukacin ISK-1.Citation47-Citation50 A number of other strategies have been successfully employed to facilitate the expression of lantibiotic gene clusters in quite different hosts, such as the expression of a S. pneumoniae encoded cluster in L. lactisCitation51 or of the B. licheniformis associated lichenicidin in Escherichia coli.Citation32 The other option available to bioengineers of bacteriocins is to utilize bacteriocin-associated modification proteins in vitro which provides even greater flexibility in terms of the changes that can be made. The group of Wilfred van der Donk at the University of Illinois at Urbana-Champaign have been trailblazers in this area.Citation52 While all bioengineering based strategies are valid should the ultimate goal be the investigation of the peptides from a fundamental perspective or the generation of antimicrobials for pharma-based applications (provided sufficient quantities can be produced), the application of bioengineered bacteriocin peptides as food preservatives is a bigger obstacle, particularly in the EU. For the latter to be an option, we anticipated that the producer of the peptide would need to be a derivative of a generally regarded as safe (GRAS) strain, such as a L. lactis, that was altered in a manner that did not lead to the strain being regarded as a genetically modified organism (GMO). Notably, as reported Sybesma et al.,Citation53 self cloning of non-pathogenic microorganisms is not considered to lead to a GMO as long as containment of the organism is guaranteed (directive 90/219/EC). According to Council Directive 98/81/EC (amending Directive 90/219/EC), "self-cloning" means the removal of nucleic acid from a cell or organism, followed by the re-insertion of all or part of that nucleic acid—with or without further enzymatic, chemical or mechanical steps—into the same cell type (or cell-line) or into a phylogenetically closely related species which can naturally exchange genetic material with the donor species. Accordingly, the temporary introduction of plasmids, the deletion of specific DNA sequences, or introduction of DNA from another microorganism belonging to the same species fall within the definition of self-cloning. Thus, microorganisms such as the Nisin V producer, when generated using our aforementioned "food-grade" approach fall outside the remit of the Contained Use legislation and therefore are not regulated as GMMs.
In the period of time since our initial identification of enhanced Nisin derivatives, I have moved from one wing of the Cork Bacteriocin Group (University College Cork) to another (Teagasc) to take up a new position but have continued to enjoy working with Colin, Paul R., Des and the many other excellent members of the group (including Mary Rea, Sheila Morgan, Paula O’Connor, Lorraine Draper, Karen Daly, Brian Healy, Alan Marsh, Dan Burke and Harsh Mathur) and those who have recently departed (Evelyn Molloy, Alleson Dobson, Sarah Norberg). I am particularly optimistic about the future for bacteriocin research and see the use of bioengineered bacteriocins as a genuine means of increasing the safety and quality of our foods. While bacteriocins other than Nisin have not been extensively employed as food preservatives by the food industry in the past, it may be that Nisin derivatives will be more rapidly accepted and provide a more direct route to market than has been the case for other bacteriocins.
About Dr Paul Cotter
Dr Paul Cotter graduated from University College Cork (UCC), Ireland with a PhD in Microbiology in 2001. He continued to carry out research and lecture at UCC before being appointed as a Principal Research Officer at Teagasc Food Research Centre at Fermoy, Cork in 2009 where he also manages the high throughput DNA sequencing platform and is a Principal Investigator within the Alimentary Pharmabiotic Centre (APC). His research focuses on the study and application of bacteriocins and the use of high throughput sequencing to study the microbiology of food and the gastrointestinal tract. Paul was appointed to Faculty of 1000 in 2006 and has received awards ESCMID-FEMS (2007), SfAM (W.H. Pierce prize 2008) and Teagasc (inaugural Excellence in Research award 2012). .
Acknowledgments
In addition to all current and past members of the Cork Bacteriocin Groups, I would like to thanks funding agencies such as Science Foundation Ireland, the Irish Department for Agriculture, Food and the Marine, Enterprise Ireland and the Health Research Board.
References
- Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 2005; 3:777 - 88; http://dx.doi.org/10.1038/nrmicro1273; PMID: 16205711
- Cotter PD, Emerson N, Gahan CG, Hill C. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J Bacteriol 1999; 181:6840 - 3; PMID: 10542190
- Cotter PD, Gahan CG, Hill C. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol 2001; 40:465 - 75; http://dx.doi.org/10.1046/j.1365-2958.2001.02398.x; PMID: 11309128
- Liu W, Hansen JN. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J Biol Chem 1992; 267:25078 - 85; PMID: 1460009
- Kuipers OP, Rollema HS, Yap WM, Boot HJ, Siezen RJ, de Vos WM. Engineering dehydrated amino acid residues in the antimicrobial peptide nisin. J Biol Chem 1992; 267:24340 - 6; PMID: 1447185
- Dodd HM, Horn N, Gasson MJ. A cassette vector for protein engineering the lantibiotic nisin. Gene 1995; 162:163 - 4; http://dx.doi.org/10.1016/0378-1119(95)00342-4; PMID: 7557409
- Breukink E, van Kraaij C, Demel RA, Siezen RJ, Kuipers OP, de Kruijff B. The C-terminal region of nisin is responsible for the initial interaction of nisin with the target membrane. Biochemistry 1997; 36:6968 - 76; http://dx.doi.org/10.1021/bi970008u; PMID: 9188693
- Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 1999; 286:2361 - 4; http://dx.doi.org/10.1126/science.286.5448.2361; PMID: 10600751
- Van Kraaij C, Breukink E, Rollema HS, Siezen RJ, Demel RA, De Kruijff B, et al. Influence of charge differences in the C-terminal part of nisin on antimicrobial activity and signaling capacity. Eur J Biochem 1997; 247:114 - 20; http://dx.doi.org/10.1111/j.1432-1033.1997.00114.x; PMID: 9249016
- Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, et al. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 2001; 276:1772 - 9; PMID: 11038353
- Ottenwälder B, Kupke T, Brecht S, Gnau V, Metzger J, Jung G, et al. Isolation and characterization of genetically engineered gallidermin and epidermin analogs. Appl Environ Microbiol 1995; 61:3894 - 903; PMID: 8526502
- Bierbaum G, Szekat C, Josten M, Heidrich C, Kempter C, Jung G, et al. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl Environ Microbiol 1996; 62:385 - 92; PMID: 8593044
- Chen P, Novak J, Kirk M, Barnes S, Qi F, Caufield PW. Structure-activity study of the lantibiotic mutacin II from Streptococcus mutans T8 by a gene replacement strategy. Appl Environ Microbiol 1998; 64:2335 - 40; PMID: 9647795
- Hindré T, Didelot S, Le Pennec JP, Haras D, Dufour A, Vallée-Réhel K. Bacteriocin detection from whole bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol 2003; 69:1051 - 8; http://dx.doi.org/10.1128/AEM.69.2.1051-1058.2003; PMID: 12571028
- Szekat C, Jack RW, Skutlarek D, Färber H, Bierbaum G. Construction of an expression system for site-directed mutagenesis of the lantibiotic mersacidin. Appl Environ Microbiol 2003; 69:3777 - 83; http://dx.doi.org/10.1128/AEM.69.7.3777-3783.2003; PMID: 12839744
- Widdick DA, Dodd HM, Barraille P, White J, Stein TH, Chater KF, et al. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proc Natl Acad Sci U S A 2003; 100:4316 - 21; http://dx.doi.org/10.1073/pnas.0230516100; PMID: 12642677
- Fimland G, Blingsmo OR, Sletten K, Jung G, Nes IF, Nissen-Meyer J. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol 1996; 62:3313 - 8; PMID: 8795220
- Fimland G, Eijsink VG, Nissen-Meyer J. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 2002; 41:9508 - 15; http://dx.doi.org/10.1021/bi025856q; PMID: 12135373
- Johnsen L, Fimland G, Eijsink V, Nissen-Meyer J. Engineering increased stability in the antimicrobial peptide pediocin PA-1. Appl Environ Microbiol 2000; 66:4798 - 802; http://dx.doi.org/10.1128/AEM.66.11.4798-4802.2000; PMID: 11055926
- Johnsen L, Fimland G, Nissen-Meyer J. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Biol Chem 2005; 280:9243 - 50; http://dx.doi.org/10.1074/jbc.M412712200; PMID: 15611086
- Kaewsrichan J, Douglas CW, Nissen-Meyer J, Fimland G, Teanpaisan R. Characterization of a bacteriocin produced by Prevotella nigrescens ATCC 25261. Lett Appl Microbiol 2004; 39:451 - 8; http://dx.doi.org/10.1111/j.1472-765X.2004.01608.x; PMID: 15482437
- Kazazic M, Nissen-Meyer J, Fimland G. Mutational analysis of the role of charged residues in target-cell binding, potency and specificity of the pediocin-like bacteriocin sakacin P. Microbiology 2002; 148:2019 - 27; PMID: 12101290
- Piper C, Draper LA, Cotter PD, Ross RP, Hill C. A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species. J Antimicrob Chemother 2009; 64:546 - 51; http://dx.doi.org/10.1093/jac/dkp221; PMID: 19561147
- Iancu C, Grainger A, Field D, Cotter PD, Hill C, Ross RP. Comparison of the Potency of the Lipid II Targeting Antimicrobials Nisin, Lacticin 3147 and Vancomycin Against Gram-Positive Bacteria. Probiotics Antimicrob Prot 2012.
- Ryan MP, Jack RW, Josten M, Sahl HG, Jung G, Ross RP, et al. Extensive post-translational modification, including serine to D-alanine conversion, in the two-component lantibiotic, lacticin 3147. J Biol Chem 1999; 274:37544 - 50; http://dx.doi.org/10.1074/jbc.274.53.37544; PMID: 10608807
- Skaugen M, Nissen-Meyer J, Jung G, Stevanovic S, Sletten K, Inger C, et al. In vivo conversion of L-serine to D-alanine in a ribosomally synthesized polypeptide. J Biol Chem 1994; 269:27183 - 5; PMID: 7961627
- Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, et al. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 1996; 253:217 - 24; http://dx.doi.org/10.1007/s004380050315; PMID: 9003306
- Cotter PD, Hill C, Ross RP. A food-grade approach for functional analysis and modification of native plasmids in Lactococcus lactis. Appl Environ Microbiol 2003; 69:702 - 6; http://dx.doi.org/10.1128/AEM.69.1.702-706.2003; PMID: 12514066
- Cotter PD, O’Connor PM, Draper LA, Lawton EM, Deegan LH, Hill C, et al. Posttranslational conversion of L-serines to D-alanines is vital for optimal production and activity of the lantibiotic lacticin 3147. Proc Natl Acad Sci U S A 2005; 102:18584 - 9; http://dx.doi.org/10.1073/pnas.0509371102; PMID: 16339304
- Cotter PD, Deegan LH, Lawton EM, Draper LA, O’Connor PM, Hill C, et al. Complete alanine scanning of the two-component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol Microbiol 2006; 62:735 - 47; http://dx.doi.org/10.1111/j.1365-2958.2006.05398.x; PMID: 17076667
- Deegan LH, Suda S, Lawton EM, Draper LA, Hugenholtz F, Peschel A, et al. Manipulation of charged residues within the two-peptide lantibiotic lacticin 3147. Microb Biotechnol 2010; 3:222 - 34; http://dx.doi.org/10.1111/j.1751-7915.2009.00145.x; PMID: 21255322
- Suda S, Hill C, Cotter PD, Ross RP. Investigating the importance of charged residues in lantibiotics. Bioeng Bugs 2010; 1:345 - 51; http://dx.doi.org/10.4161/bbug.1.5.12353; PMID: 21326835
- Suda S, Westerbeek A, O’Connor PM, Ross RP, Hill C, Cotter PD. Effect of bioengineering lacticin 3147 lanthionine bridges on specific activity and resistance to heat and proteases. Chem Biol 2010; 17:1151 - 60; http://dx.doi.org/10.1016/j.chembiol.2010.08.011; PMID: 21035738
- Field D, Collins B, Cotter PD, Hill C, Ross RP. A system for the random mutagenesis of the two-peptide lantibiotic lacticin 3147: analysis of mutants producing reduced antibacterial activities. J Mol Microbiol Biotechnol 2007; 13:226 - 34; http://dx.doi.org/10.1159/000104747; PMID: 17827973
- Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 1996; 69:193 - 202; http://dx.doi.org/10.1007/BF00399424; PMID: 8775979
- Rollema HS, Kuipers OP, Both P, de Vos WM, Siezen RJ. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl Environ Microbiol 1995; 61:2873 - 8; PMID: 7487019
- Kuipers OP, Bierbaum G, Ottenwälder B, Dodd HM, Horn N, Metzger J, et al. Protein engineering of lantibiotics. Antonie Van Leeuwenhoek 1996; 69:161 - 9; http://dx.doi.org/10.1007/BF00399421; PMID: 8775976
- Rink R, Wierenga J, Kuipers A, Kluskens LD, Driessen AJ, Kuipers OP, et al. Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C-terminal truncation. Appl Environ Microbiol 2007; 73:5809 - 16; http://dx.doi.org/10.1128/AEM.01104-07; PMID: 17660303
- Field D, Hill C, Cotter PD, Ross RP. The dawning of a "Golden era" in lantibiotic bioengineering. Mol Microbiol 2010; 78:1077 - 87; http://dx.doi.org/10.1111/j.1365-2958.2010.07406.x; PMID: 21091497
- Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci 2008; 65:455 - 76; http://dx.doi.org/10.1007/s00018-007-7171-2; PMID: 17965835
- Yuan J, Zhang ZZ, Chen XZ, Yang W, Huan LD. Site-directed mutagenesis of the hinge region of nisinZ and properties of nisinZ mutants. Appl Microbiol Biotechnol 2004; 64:806 - 15; http://dx.doi.org/10.1007/s00253-004-1599-1; PMID: 15048591
- Field D, Connor PM, Cotter PD, Hill C, Ross RP. The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol Microbiol 2008; 69:218 - 30; http://dx.doi.org/10.1111/j.1365-2958.2008.06279.x; PMID: 18485077
- Murphy K, O’Sullivan O, Rea MC, Cotter PD, Ross RP, Hill C. Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters. PLoS One 2011; 6:e20852; http://dx.doi.org/10.1371/journal.pone.0020852; PMID: 21760885
- Carroll J, Field D, O’Connor PM, Cotter PD, Coffey A, Hill C, et al. Gene encoded antimicrobial peptides, a template for the design of novel anti-mycobacterial drugs. Bioeng Bugs 2010; 1:408 - 12; http://dx.doi.org/10.4161/bbug.1.6.13642; PMID: 21468208
- Rouse S, Field D, Daly KM, O’Connor PM, Cotter PD, Hill C, et al. Bioengineered nisin derivatives with enhanced activity in complex matrices. Microb Biotechnol 2012; 5:501 - 8; http://dx.doi.org/10.1111/j.1751-7915.2011.00324.x; PMID: 22260415
- DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol 2011; 8:523 - 31; http://dx.doi.org/10.1038/nrgastro.2011.133; PMID: 21844910
- Boakes S, Cortés J, Appleyard AN, Rudd BA, Dawson MJ. Organization of the genes encoding the biosynthesis of actagardine and engineering of a variant generation system. Mol Microbiol 2009; 72:1126 - 36; http://dx.doi.org/10.1111/j.1365-2958.2009.06708.x; PMID: 19400806
- Appleyard AN, Choi S, Read DM, Lightfoot A, Boakes S, Hoffmann A, et al. Dissecting structural and functional diversity of the lantibiotic mersacidin. Chem Biol 2009; 16:490 - 8; http://dx.doi.org/10.1016/j.chembiol.2009.03.011; PMID: 19477413
- Islam MR, Shioya K, Nagao J, Nishie M, Jikuya H, Zendo T, et al. Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol Microbiol 2009; 72:1438 - 47; http://dx.doi.org/10.1111/j.1365-2958.2009.06733.x; PMID: 19432794
- Boakes S, Ayala T, Herman M, Appleyard AN, Dawson MJ, Cortés J. Generation of an actagardine A variant library through saturation mutagenesis. Appl Microbiol Biotechnol 2012; http://dx.doi.org/10.1007/s00253-012-4041-0; PMID: 22526797
- Kuipers OP, Rollema HS, Beerthuyzen MM, Siezen RJ, de Vos WM. Protein engineering and biosynthesis of nisin and regulation of transcription of the structural nisA gene. Int Dairy J 1995; 5:785 - 95; http://dx.doi.org/10.1016/0958-6946(95)00032-1
- Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem 2012; 81:479 - 505; http://dx.doi.org/10.1146/annurev-biochem-060110-113521; PMID: 22404629
- Sybesma W, Hugenholtz J, de Vos WM, Smid EJ. Safe use of genetically modified lactic acid bacteria in food. Bridging the gap between consumers, green groups, and industry. Electron J Biotechnol 2006; 9:1 - 25; http://dx.doi.org/10.2225/vol9-issue4-fulltext-12