Abstract
Engineered zinc-finger nucleases (ZFNs) are powerful tools for creating double-stranded-breaks (DSBs) in genomic DNA in a site-specific manner. These DSBs generated by ZFNs can be repaired by homology-directed repair or nonhomologous end joining, in which the latter can be exploited to generate insertion or deletion mutants. Based on published literature, we designed a pair of zinc-finger nucleases and inactivated the GDP-fucose transporter gene (Slc35c1) in a previously reported CHO mutant that has a dysfunctional CMP-sialic acid transporter gene (Slc35a1). The resulting mutant cell line, CHO-gmt5, lacks functional GDP-fucose transporter and CMP-sialic acid transporter. As a result, these cells can only produce asialylated and afucosylated glycoproteins. It is now widely recognized that removal of the core fucose from the N-glycans attached to Asn297 of human IgG1 significantly enhances its binding to its receptor, FcγRIIIa, and thereby dramatically improves antibody-dependent cellular cytotoxicity (ADCC). Recent reports showed that removal of sialic acid from IgG1 also enhances ADCC. Therefore, CHO-gmt5 may represent a more advantageous cell line for the production of recombinant antibodies with enhanced ADCC. These cells show comparable growth rate to wild type CHO-K1 cells and uncompromised transfection efficiency, which make them desirable for use as a production line.
Recombinant human IgG1 antibodies have been successfully used as therapeutic drugs to target malignant cells in cancer patients. Upon binding to the target molecule expressed on cancer cells, the Fc region of the antibody recruits the effector cells such as natural killer (NK) cells to kill cancer cells by antibody-dependent cellular cytotoxicity (ADCC). Studies have shown that the major interaction sites of the Fc region by the FcγRIII are located in the hinge region and the CH2 domains of the antibody.Citation1,Citation2 The binding of the Fc and the Fc receptor is known to be dependent on the structures of the N-glycans attached to the conserved glycosylation site at Asn297 in each of the CH2 domains.Citation3
It has been demonstrated clearly that removal of the fucose residue from the N-glycan attached to Asn297 of human IgG1 significantly enhances its binding to FcγRIIIa and thereby dramatically improves ADCC.Citation4,Citation5 Detailed binding analyses indicated that removal of fucose enhanced binding enthalpy and increased binding constant of IgG1 for FcγRIIIa.Citation6 The molecular mechanism underlying the enhanced affinity between Fc and FcγRIIIa was further investigated. The structural differences between fucosylated and afucosylated Fc fragments of human IgG1 were compared in X-ray crystallographic and NMR spectroscopic studies. The overall conformations of the fucosylated and afucosylated Fc fragments are similar except for hydration mode around Tyr296.Citation7 The conformation of Tyr296 is more flexible for FcγRIIIa in afucosylated Fc than in fucosylated Fc.Citation7 Tyr296 has already been indicated in the interaction with FcγRIIIa.Citation2,Citation8
Enhanced ADCC was not only observed for the afucosylated antibody in in vitro assays, it was also confirmed in vivo in patients or animal models.Citation9-Citation12 These data have convincingly shown that removal of fucose from human IgG1 can be a general method for treating cancer patients with antibodies through improved ADCC.
In addition to fucosylation, sialylation of the Fc N-glycan may also affect the cell-killing activity of the antibody. Increased sialylation of the Fc N-glycan was shown to reduce the binding of Fc to FcγRIIIa and consequently, decrease ADCC.Citation13,Citation14 We have previously reported the isolation of a CHO mutant (CHO-gmt1) that produces asialylated glycoproteins due to the lack of a functional CMP-sialic acid transporter.Citation15 Using zinc-finger nuclease technology we have inactivated the GDP-fucose transporter in CHO-gmt1.Citation16 The resulting mutant, CHO-gmt5, is able to produce afucosylated and asialylated recombinant antibodies. The efficacy in ADCC by antibodies produced in CHO-gmt5 cells will be evaluated.
CHO-gmt1 Cells
CHO-gmt1 cells (originally called MAR-11) were isolated from CHO-K1 cells treated with Maackia amurensis agglutinin (MAA) which is specific for Neu5Acα2,3Gal structure. The cells have a dysfunctional CMP-sialic acid transporter (CMP-SAT).Citation15 A point mutation in the CMP-SAT gene in CHO-gmt1 cells results in a premature stop codon. As a result, these cells express a truncated version of CMP-SAT which contains only 100 amino acids, rather than the normal CMP-SAT which consists of 336 amino acids. As a result, glycoproteins produced by this cell line are completely free of sialic acid. These cells have been used to study the structure-function relationships of CMP-SAT.Citation17
A Simplified “Modular Assembly” Strategy to Design Zinc-Finger Nucleases (ZFNs) Based on Publically Available Information
Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to the cleavage domain of restriction enzyme FokI. The zinc finger DNA-binding domain of ZFNs consists of three or four zinc finger units. Each of these recognizes a 3-bp motif in the chromosomal DNA. The specificity of the ZFNs is determined by 7 amino acids within each zinc-finger unit that interact with the DNA. In order to allow the two FokI cleavage domains to dimerize and cleave DNA, the two ZFNs must bind opposite strands of DNA and the two binding sites have to be separated by 5–7 bps. The double-stranded-breaks (DSBs) in genomic DNA created by ZFNs can be repaired by nonhomologous end joining (NHEJ). During NHEJ, cells often create insertion or deletion mutations.
ZFNs generated by the combinatorial selection methods may have high DNA-binding affinity and low toxicity.Citation18 Sangamo Biosciences has used its own proprietary information to create highly specific ZFNs.Citation19-Citation21 However, most laboratories do not have the randomized libraries or the selection expertise to do so. Alternatively, a modular assembly strategy can be used based on publically available information in the literature.Citation22,Citation23 We also used the modular assembly strategy to design ZFNs to interrupt the GDP-fucose transporter gene in CHO cells. To target a gene with ZFNs, the first step is to identify an ideal target site in the gene of interest. The open reading frame of the cDNA can be analyzed by the web-based ZiFiT program provided by the Zinc Finger Consortium (ZiFiT: software for engineering zinc finger proteins (V3.0)) at: http://bindr.gdcb.iastate.edu/ZiFiT/.Citation24 The ZiFiT output will suggest a few potential target sites. The fingers that bind the 5′-GNN-3′ sequences are the best studied and strongest DNA-binding fingers.Citation25-Citation27 Two binding sites for ZFNs should be separated by 5–7 bps which is the optimal distance for the two FokI domains to dimerize and cleave the targeted site. Therefore, an ideal target sequence for two 4-fingered ZFNs should be: 5′-NNCNNCNNCNNCxxxxxxGNNGNNGNNGNN-3′. This sequence ensures each zinc finger binds a 5′-GNN-3′ sequence. The 5′-GNG-3′ sequences are better binding sites compared with other 5′-GNN-3′ sequences. In addition to the 5′-GNN-3′ sequences, other sequences have also be successfully used in the literature. These include CTG, TGG, AAG and AAA triplets. It is generally believed that 3-fingered ZFNs should work as well as the 4-fingered ZFNs. In the event that there is no ideal site in the open reading frame, one can try to find two ideal sites that flank an exon in the open reading frame of the targeted gene. ZFNs designed to target two different sites can introduce two concurrent DNA double-strand breaks in the chromosome and create deletions of the genomic segment between the two sites.Citation28 In this situation, cells will be transfected with two pairs of ZFNs simultaneously in order to generate targeted deletions of genomic segments. A simplified “modular assembly” strategy to design zinc-finger nucleases (ZFNs) based on publically available information is outlined in .
Figure 1. Outline for the interruption of a target gene using zinc-finger nucleases designed by the “modular assembly” strategy.
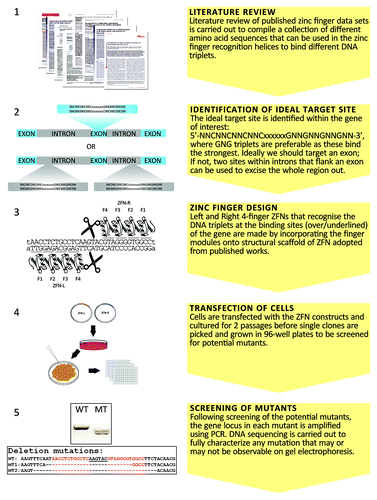
The structural scaffold for the ZFNs can be adopted from previous publications.Citation19,Citation20 To eliminate unwanted homodimerization of FokI cleavage domain, the high-fidelity FokI-KK and FokI-EL variants can be used.Citation29 The amino acid sequences of the DNA-binding domains in the ZFNs are assembled using an archive of zinc-finger motifs collected from previous publicationsCitation25-Citation27 and many other related publications which are not listed here.
Using ZFNs to Inactivate GDP-Fucose Transporter Gene in CHO Cells and Fluorescence-Activated Cell Sorting to Rapidly Isolate Mutant Cells
Analysis of the open reading frame of Chinese hamster GDP-fucose transporter gene using the ZiFiT programCitation24 identified one potential target site in the first exon of the GDP-fucose transporter coding region (5′-tAACCTCTGCCTCAAGTACGTAGGGGTGGCCt-3′). As discussed earlier, this sequence allows each zinc finger in the left ZFN and the right ZFN to bind to a 5′-GNN-3′ DNA triplet.
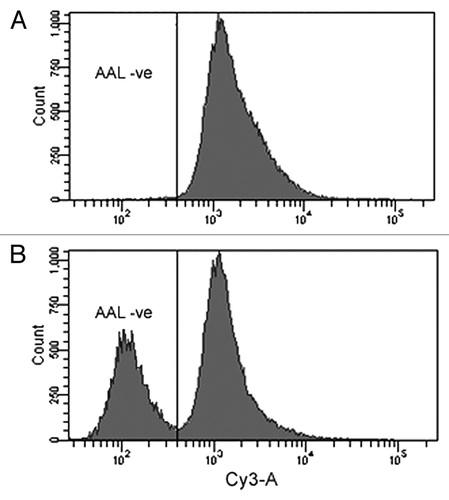
CHO cells were transiently transfected with plasmids expressing the left and right zinc-finger nucleases as described previously.Citation16 In our previous report, two days after transfection, single cells were seeded in a 96-well plate for clone isolation. Genomic DNA from each single clone was isolated and the targeted GDP-fucose transporter locus was amplified by two specific PCR primers flanking the targeted site in the gene. The PCR products were sequenced in order to identify the mutation. However, this is a labor intensive and time-consuming process and we identified only one mutant.Citation16 To increase the selection efficiency, a fucose-specific lectin combined with fluorescence-activated cell sorting (FACS) strategy was employed. After transfection with the ZFN constructs, the cells were cultured for several generations to allow the daughter cells of successful knockout mutants to express afucosylated glycoproteins on their surface. Mutant and the wild type CHO cells in the transfected pool were stained with biotinylated Aleuria Aurantia lectin (AAL) and Cy3-conjugated streptavidin. Stained cells were sorted by FACS using a Becton Dickenson FACSAria IIu SORP cell sorter. The negatively stained cells (AAL-ve) were pooled and cultured (). When this population was subsequently stained with FITC and analyzed by FACS, a significant number of the cells were AAL-negative (). Following two more cycles of this lectin-based cell sorting essentially all the cells in the population became AAL negative cells (data not shown).
Figure 2. Using FACS to rapidly isolate cells with inactivated GDP-fucose transporter. Cells transfected with constructs expressing the ZFNs were cultured to near confluence and subcultured at 1:6 ratio for 2 passages. Ten million of the resulting cells were labeled with biotinylated AAL and Cy3-conjugated streptavidin. Stained cells were sorted on a Becton Dickenson FACSAria IIu SORP cell sorter. (A) FACS histogram for transfected cells at the first round of sorting. The sorting gate for AAL-negative (AAL-ve) cells was set to collect the lowest 0.5% of AAL-stained cells. Approximately 12,000 cells were collected from a total of 3.5 million cells sorted. These cells were cultured for 2 weeks before being subjected to a second round of sorting. (B) Second round of FACS shows that more than 30% of the cells are AAL-ve cells after the first round of sorting. Single AAL-ve cells were isolated from this pool for further characterization.
In order to confirm that the phenotypic change observed in the flow cytometry was indeed due to the disruption of the GDP-fucose transporter gene, the cells were transfected with a construct that expressed wild type GDP-fucose transporter in a rescue assay. The conversion of the AAL-negative phenotype to AAL-positive following this assay is indicative of a mutated GDP-fucose transporter. The gene locus in the mutants was then amplified by PCR and sequenced. The results further confirmed the genetic mutation at the targeted site. Taken together, the fucose-specific lectin in combination with FACS is a successful method to rapidly isolate mutants.
CHO-gmt5 Cells
The GDP-fucose transporter gene was successfully inactivated by ZFNs in CHO-gmt1 cells.Citation16 The resultant CHO-gmt5 cells lack functional CMP-sialic acid transporter and GDP-fucose transporter. They can be used to produce afucosylated and asialylated recombinant antibodies such as Rituximab. The ability of fucose-free and sialic acid-free antibodies to enhance ADCC will be assessed. We have also shown that CHO-gmt5 cells have comparable growth rate to wild type CHO-K1 upon adaptation to suspension culture in chemically defined medium, as well as uncompromised transfection efficiency when transfected with GFP and recombinant Rituximab constructs.
Perspectives
This work demonstrated that the “modular assembly” strategy can be successfully utilized by labs without special expertise in zinc finger design. Utilizing the archive of zinc-finger motifs collected from all the zinc finger work published in the literature, researchers will be able to design better zinc fingers.
FACS is a simple method to rapidly isolate mutant cells provided the mutant cell can be identified by a fluorescent tag. In our case, fucosylated proteins on the wild type cell surface can be positively stained by AAL, which consequently excludes the mutant cells. Based on this feature we were able to enrich and isolate the cells with inactivated GDP-fucose transporter gene. FACS can also rapidly enrich and isolate mutant cells in which the dihydrofolate reductase (DHFR) gene is interrupted by ZFNs. Wild type cells that express DHFR bind fluorescent methotrexate.Citation30 Mutants that are no longer able to bind fluorescent methotrexate can be isolated by FACS. We have successfully used methotrexate to isolate DHFR negative CHO cells using previously reported ZFNs.Citation21 Thus, the FACS strategy can be applied to the isolation of a variety of mutants, including mutations in the genes that encode cell surface proteins, provided antibodies specific for the proteins are available.
CHO-gmt5 cells lack functional CMP-sialic acid transporter and GDP-fucose transporter. These cells can be used to study structure-function relationships of these two receptors.Citation16,Citation17 They can also be used to produce afucosylated and asialylated recombinant antibodies. It is of great interest for us to investigate whether these antibodies will have enhanced ADCC against the target cells.
Acknowledgments
The authors would like to thank Ms. Natasha A. Pereira for critically reviewing the manuscript. This work was funded by the Agency for Science, Technology and Research (A*STAR), Singapore.
References
- Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature 2000; 406:267 - 73; http://dx.doi.org/10.1038/35018508; PMID: 10917521
- Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The structure of a human type III Fcgamma receptor in complex with Fc. J Biol Chem 2001; 276:16469 - 77; http://dx.doi.org/10.1074/jbc.M100350200; PMID: 11297532
- Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol 2003; 325:979 - 89; http://dx.doi.org/10.1016/S0022-2836(02)01250-0; PMID: 12527303
- Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem 2002; 277:26733 - 40; http://dx.doi.org/10.1074/jbc.M202069200; PMID: 11986321
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem 2003; 278:3466 - 73; http://dx.doi.org/10.1074/jbc.M210665200; PMID: 12427744
- Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, et al. Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcgammaRIIIa. J Mol Biol 2004; 336:1239 - 49; http://dx.doi.org/10.1016/j.jmb.2004.01.007; PMID: 15037082
- Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, et al. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J Mol Biol 2007; 368:767 - 79; http://dx.doi.org/10.1016/j.jmb.2007.02.034; PMID: 17368483
- Radaev S, Sun P. Recognition of immunoglobulins by Fcgamma receptors. Mol Immunol 2002; 38:1073 - 83; http://dx.doi.org/10.1016/S0161-5890(02)00036-6; PMID: 11955599
- Niwa R, Shoji-Hosaka E, Sakurada M, Shinkawa T, Uchida K, Nakamura K, et al. Defucosylated chimeric anti-CC chemokine receptor 4 IgG1 with enhanced antibody-dependent cellular cytotoxicity shows potent therapeutic activity to T-cell leukemia and lymphoma. Cancer Res 2004; 64:2127 - 33; http://dx.doi.org/10.1158/0008-5472.CAN-03-2068; PMID: 15026353
- Suzuki E, Niwa R, Saji S, Muta M, Hirose M, Iida S, et al. A nonfucosylated anti-HER2 antibody augments antibody-dependent cellular cytotoxicity in breast cancer patients. Clin Cancer Res 2007; 13:1875 - 82; http://dx.doi.org/10.1158/1078-0432.CCR-06-1335; PMID: 17363544
- Cardarelli PM, Moldovan-Loomis MC, Preston B, Black A, Passmore D, Chen TH, et al. In vitro and in vivo characterization of MDX-1401 for therapy of malignant lymphoma. Clin Cancer Res 2009; 15:3376 - 83; http://dx.doi.org/10.1158/1078-0432.CCR-08-3222; PMID: 19401346
- Junttila TT, Parsons K, Olsson C, Lu Y, Xin Y, Theriault J, et al. Superior in vivo efficacy of afucosylated trastuzumab in the treatment of HER2-amplified breast cancer. Cancer Res 2010; 70:4481 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-09-3704; PMID: 20484044
- Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science 2008; 320:373 - 6; http://dx.doi.org/10.1126/science.1154315; PMID: 18420934
- Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science 2006; 313:670 - 3; http://dx.doi.org/10.1126/science.1129594; PMID: 16888140
- Lim SF, Lee MM, Zhang P, Song Z. The Golgi CMP-sialic acid transporter: A new CHO mutant provides functional insights. Glycobiology 2008; 18:851 - 60; http://dx.doi.org/10.1093/glycob/cwn080; PMID: 18713811
- Zhang P, Haryadi R, Chan KF, Teo G, Goh J, Pereira NA, et al. Identification of functional elements of the GDP-fucose transporter SLC35C1 using a novel Chinese hamster ovary mutant. Glycobiology 2012; 22:897 - 911; http://dx.doi.org/10.1093/glycob/cws064; PMID: 22492235
- Chan KF, Zhang P, Song Z. Identification of essential amino acid residues in the hydrophilic loop regions of the CMP-sialic acid transporter and UDP-galactose transporter. Glycobiology 2010; 20:689 - 701; http://dx.doi.org/10.1093/glycob/cwq016; PMID: 20181793
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 2008; 31:294 - 301; http://dx.doi.org/10.1016/j.molcel.2008.06.016; PMID: 18657511
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005; 435:646 - 51; http://dx.doi.org/10.1038/nature03556; PMID: 15806097
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 2008; 26:702 - 8; http://dx.doi.org/10.1038/nbt1409; PMID: 18500334
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A 2008; 105:5809 - 14; http://dx.doi.org/10.1073/pnas.0800940105; PMID: 18359850
- Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc 2006; 1:1329 - 41; http://dx.doi.org/10.1038/nprot.2006.231; PMID: 17406419
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 2009; 19:1279 - 88; http://dx.doi.org/10.1101/gr.089417.108; PMID: 19470664
- Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res 2007; 35:Web Server issue W599-605; http://dx.doi.org/10.1093/nar/gkm349; PMID: 17526515
- Dreier B, Segal DJ, Barbas CF 3rd. Insights into the molecular recognition of the 5′-GNN-3′ family of DNA sequences by zinc finger domains. J Mol Biol 2000; 303:489 - 502; http://dx.doi.org/10.1006/jmbi.2000.4133; PMID: 11054286
- Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J Biol Chem 2002; 277:3850 - 6; http://dx.doi.org/10.1074/jbc.M110669200; PMID: 11726671
- Segal DJ, Dreier B, Beerli RR, Barbas CF 3rd. Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci U S A 1999; 96:2758 - 63; http://dx.doi.org/10.1073/pnas.96.6.2758; PMID: 10077584
- Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 2010; 20:81 - 9; http://dx.doi.org/10.1101/gr.099747.109; PMID: 19952142
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 2007; 25:778 - 85; http://dx.doi.org/10.1038/nbt1319; PMID: 17603475
- Gaudray P, Trotter J, Wahl GM. Fluorescent methotrexate labeling and flow cytometric analysis of cells containing low levels of dihydrofolate reductase. J Biol Chem 1986; 261:6285 - 92; PMID: 3700393