Abstract
Consolidated bioprocessing (CBP), which integrates enzyme production, saccharification and fermentation into a one step process, is a promising strategy for the effective ethanol production from cheap lignocellulosic and starchy materials. CBP requires a highly engineered microbial strain able to both hydrolyze biomass with enzymes produced on its own and convert the resulting simple sugars into high-titer ethanol. Recently, heterologous production of cellulose and starch-degrading enzymes has been achieved in yeast hosts, which has realized direct processing of biomass to ethanol. However, essentially all efforts aimed at the efficient heterologous expression of saccharolytic enzymes in yeast have involved laboratory strains and much of this work has to be transferred to industrial yeasts that provide the fermentation capacity and robustness desired for large scale bioethanol production. Specifically, the development of an industrial CBP amylolytic yeast would allow the one-step processing of low-cost starchy substrates into ethanol. This article gives insight in the current knowledge and achievements on bioethanol production from starchy materials with industrial engineered S. cerevisiae strains.
Plant biomass is an abundant and renewable substrate for the sustainable production of biofuels and chemicals. The main technological hurdling the widespread conversion of this renewable resource into fuels and other valuable products is the lack of low-cost technologies to overcome the biomass recalcitrance.Citation1-Citation6 Currently, bioethanol is being produced from sugarcane juice and starch-rich grains in Brazil and the United States of America using Saccharomyces cerevisiae strains.Citation7 Besides wheat and corn grains, abundant starchy feedstocks, such as wasted crop, cereal bran, potato peels and brewery spent grains, have been proposed as low-cost residual biomass for the production of bioethanol.Citation8-Citation10 However, since S. cerevisiae lacks the amylolytic enzymes required for starch utilization, expensive enzyme addition is needed for ethanol production from starchy biomass. Thus, the industrial process of converting starch into ethanol is a costly method with four steps: milling, starch hydrolysis into glucose, yeast fermentation and alcohol distillation. Moreover, starchy materials have to be cooked (gelatinized) at a high temperature (up to 140–180°C) to obtain a high ethanol yield. To reduce the energy cost for cooking, unmodified (raw) and low temperature cooking starch fermentation systems have been proposed.Citation11 However, the addition of large amounts of amylolytic enzymes is still required to hydrolyze raw starch into glucose.
The cost-effective conversion of raw starch demands the expression of starch-hydrolyzing enzymes in a fermenting yeast to achieve liquefaction, hydrolysis, and fermentation (Consolidated bioprocessing, CBP) by a single organism.Citation12 A CBP process for raw starch conversion to ethanol can save on the excess energy demand in heating of the starch slurry, as well as pumping or stirring of the slurry.Citation13-Citation15 The CBP of raw starch would require recombinant S. cerevisiae strains to produce sufficient quantities of raw starch degrading enzymes to ensure hydrolysis at a high substrate loading and at moderate temperatures. This has become the primary focus of several research groups in recent years and good progress toward proof of concept has been made.Citation15 However, for the efficient industrial ethanol production from starch, implementing the raw starch conversion technology into robust, industrially used yeasts is crucial. Laboratory strains are easily genetically modified and the functionality of starch hydrolyzing enzymes has been proved by various laboratories.Citation15,Citation16 Episomal plasmid vectors under selection have been mainly utilized for the overexpression of target genes in laboratory S. cerevisiae strains to ensure high copy numbers (50–200 copies) are maintained. Despite these advantages, most industrial yeasts are much more robust than laboratory strains and display higher specific ethanol productivities and ethanol yields, producing a lower amount of undesirable by-products, like glycerol.Citation5 Although industrial strains are particularly stable in a variety of manufacturing conditions including drying and long-term storage, their genetic engineering is challenging and, under non-selective pressure such as long run industrial processes, the usage of plasmids is undesirable as their maintenance depends on selectable markers.Citation17 Therefore, all genetic manipulations successfully demonstrated into laboratory strains must be transferred to industrial yeasts relying on a stable integration into the genome. Consequently, gene deletions or insertions have to be performed for all alleles to obtain a stable genotype.
The engineering of industrial S. cerevisiae strains can be achieved through the integration of foreign genes into their chromosomes by homologous recombination to ensure mitotic stability under non-selective conditions. The reiterated DNA sequences such as δ-sequences of the Ty retrotransposon and rDNA have been efficiently proposed as target sites which results in a greater number of integrated gene copies, and therefore higher expression levels.Citation18,Citation19
This approach has been recently assessed for the first time by integrating a glucoamylase gene into an industrial S. cerevisiae yeast, resulting in a promising and improvable CBP amylolytic yeast, capable of efficiently converting raw starch into ethanol.Citation20
To achieve this purpose, several fungal strains of Aspergillus oryzae and A. awamori, screened for their efficient raw and soluble starch hydrolyzing activities, showed high amylolytic activities in liquid assays (data not shown). A. awamori CBS 115.52 was found to be a promising raw starch degrader and the cDNA copy of the glucoamylase gene GAI was amplified by PCR for expression in the laboratory strain S. cerevisiae Y294. To ensure efficient secretion of enzymes (one of the main obstacles in achieving high recombinant protein levels in engineered yeasts), the GAI gene was fused to the T. reesei β-xylanase 2 secretion signal called XYNSEC.Citation21 In order to further improve raw starch utilization in recombinant yeasts, the codon usage of the gene GAI from A. awamori as well as of the XYNSEC T. reesei β-xylanase 2 secretion signal was adapted to that of the S. cerevisiae but without changing the amino acid sequence (). The resulting optimized sGAI gene had a CAI (Codon Adaptation Index) value of 0.921 and retained conserved domains for hydrolysis of raw starch (PLWYVTVTLPA)Citation22 and the Gp-I region, which is heavily O-glycosylated. The glycosylation is required for both enzyme stability and enhanced activity on raw starch. Furthermore, the same Gp-I region was found to be crucial for correct folding of the enzyme.Citation23
Figure 1. Predicted protein sequence of the sGAI gene of A. awamori (GenBank:JX559863) expressed in S. cerevisiae Y294[ySYAG]. The XYNSEC secretion signal is indicated in bold font. The sequence identified in glucoamylases essential for raw starch hydrolysis was conserved (PL(W-597)YVTVTLPA),Citation19 as well as the second tryptophan (Trp) residue and is double underlined in gray text. The Gp-I region is indicated as text in italics bold. The Cp-I region or Starch Binding Domain is indicated in underlined text.Citation25 The general acid and base catalysts Glu-213 and Glu-434, as well as Tyr-85, Trp-87, Arg-89, Asp-90, Trp-154, Glu-214, Arg-339, Asp-343, Trp-351 sites, which play a role in substrate transition-state stabilization and/or ground-state binding, are indicated in gray text. Likely N-glycosylation sites are underlined by a broken line, although only the first and third sites were found to be glycosylated when expressed in yeast.Citation26
![Figure 1. Predicted protein sequence of the sGAI gene of A. awamori (GenBank:JX559863) expressed in S. cerevisiae Y294[ySYAG]. The XYNSEC secretion signal is indicated in bold font. The sequence identified in glucoamylases essential for raw starch hydrolysis was conserved (PL(W-597)YVTVTLPA),Citation19 as well as the second tryptophan (Trp) residue and is double underlined in gray text. The Gp-I region is indicated as text in italics bold. The Cp-I region or Starch Binding Domain is indicated in underlined text.Citation25 The general acid and base catalysts Glu-213 and Glu-434, as well as Tyr-85, Trp-87, Arg-89, Asp-90, Trp-154, Glu-214, Arg-339, Asp-343, Trp-351 sites, which play a role in substrate transition-state stabilization and/or ground-state binding, are indicated in gray text. Likely N-glycosylation sites are underlined by a broken line, although only the first and third sites were found to be glycosylated when expressed in yeast.Citation26](/cms/asset/6a4ac329-3157-4cd1-a6e7-2078b8de65d9/kbie_a_10922268_f0001.gif)
Both genes encoding the native glucoamylase from A. awamori, GAI, and the codon-optimized counterpart, sGAI, were cloned under the transcriptional control of the S. cerevisiae PGK1 promoter and terminator and expressed in S. cerevisiae Y294 laboratory strain. The ability of the glucoamylolytic strains to produce functional recombinant enzyme was visualized as cleared hydrolysis zones in raw starch agar stained with iodine (). The engineered yeasts were able to grow on raw starch as sole carbon source and, as reported in , the enzymatic assays clearly indicated that codon adaptation resulted in an improvement of the extracellular glucoamylolytic activity of the recombinant strains. S. cerevisiae Y294[ySYAG], secreting the optimized sGAI, showed a 31 and 40% increase in enzymatic activity from raw and soluble starch, respectively, compared with the enzymatic values of the native GAI, secreted by S. cerevisiae Y294[yASAG]. Both recombinant strains, but particularly S. cerevisiae Y294[ySYAG], produced limited α-amylolytic activities. Although most forms of glucoamylase can hydrolyze α-1,6-D-glucosidic bonds when the next bond in the sequence is 1,4-linked,Citation24 this finding is of great interest since the integration of a glucoamylase gene resulted in a recombinant yeast capable of exhibiting also a weak α-amylolytic phenotype.
Figure 2. Glucoamylases production by wild type and recombinant S. cerevisiae strains. (A) Hydrolysis of raw starch appears as clear zones around S. cerevisiae colonies secreting functional glucoamylases; strain (a) Y294 (reference), (b) Y294[yASAG] secreting the native GAI, (c) Y294[ySYAG] secreting the codon-optimized sGAI, were grown for 4 d on agar containing raw starch and then stained with iodine solution. (B) Glucoamylase and α-amylase activities of the strains Y294[yASAG] and Y294[ySYAG]; glucoamylase activity, determined at pH 5.4 and 30°C, was reported as nanokatals per gram dry cell weight (nKat/g DCW), which is the enzyme activity needed to produce 1 nmol of glucose per second per gram dry cell weight. α-amylase activity, detected at pH 5.4 and 50°C, was expressed as Ceralpha Units per gram dry cell weight (CU/g DCW), which is the enzyme activity required to release 1 micromol p-nitrophenyl per min per gram dry cell weight.
![Figure 2. Glucoamylases production by wild type and recombinant S. cerevisiae strains. (A) Hydrolysis of raw starch appears as clear zones around S. cerevisiae colonies secreting functional glucoamylases; strain (a) Y294 (reference), (b) Y294[yASAG] secreting the native GAI, (c) Y294[ySYAG] secreting the codon-optimized sGAI, were grown for 4 d on agar containing raw starch and then stained with iodine solution. (B) Glucoamylase and α-amylase activities of the strains Y294[yASAG] and Y294[ySYAG]; glucoamylase activity, determined at pH 5.4 and 30°C, was reported as nanokatals per gram dry cell weight (nKat/g DCW), which is the enzyme activity needed to produce 1 nmol of glucose per second per gram dry cell weight. α-amylase activity, detected at pH 5.4 and 50°C, was expressed as Ceralpha Units per gram dry cell weight (CU/g DCW), which is the enzyme activity required to release 1 micromol p-nitrophenyl per min per gram dry cell weight.](/cms/asset/71027212-b28d-4601-a3c4-971c60564fb5/kbie_a_10922268_f0002.gif)
Upon functional expression of both the native and synthetic glucoamylase genes into the laboratory S. cerevisiae Y294 strain, the sGAI expression cassette was integrated into the industrial S. cerevisiae 27P at multiple δ-sites. The resulting recombinant strains were tested for their mitotic stability using the method described in Favaro et al.Citation13 and the mitotically stable integrants F2 and F6 were selected as the most efficient hydrolysing yeasts on the basis of their raw starch degradation halos (data not shown).
The glucoamylolytic activity of sGAI, tested in liquid enzymatic assays at pH values of 4.0, 4.5, 5.0, 5.5 and 6.0, was found to be optimal at pH 4.5. The raw and soluble starch hydrolysing activity were assessed at different temperature values: 30, 37 and 60°C (). When raw corn starch was used as carbon source, the recombinant strains produced about 48% of the enzymatic activity obtained on soluble starch (). S. cerevisiae F2 displayed higher soluble and raw starch hydrolysing activities than S. cerevisiae F6. This finding could be attributed to either higher number copies of sGAI genes integrated into the S. cerevisiae F2 genome compared with S. cerevisiae F6, or integration in regions of the genome that promotes higher transcription levels in S. cerevisiae F2 than in S. cerevisiae F6. However, further genetic studies are in progress to confirm both hypotheses. The highest enzymatic activity was detected at 60°C, while at 30°C, the growth temperature preferred by S. cerevisiae, the glucoamylolytic activity notably decreased to 23%. Increasing the enzymatic assay temperature up to 37°C resulted in 15% higher enzymatic values for both recombinant strains compared with 30°C.
Table 1. Glucoamylolytic activity (nKat/DCW) of the engineered S. cerevisiae F2 and F6 strains once grown in YPD broth for 72 h
Starch conversion was subsequently evaluated at 37°C and aerobic growth on soluble starch were more rapid than that observed at 30°C (). Both engineered yeasts were able to grow faster at 37°C than at 30°C, confirming that the higher incubation temperature positively affected the enzymatic activity of the recombinant glucoamylase (). As a result, at 37°C, the starch to glucose conversion rate was enhanced (data not shown) and the recombinant strains reached the stationary phase earlier than at 30°C.
Figure 3. Effect of the temperature on the aerobic growth of S. cerevisiae 27P (♦), F2 (□) and F6 (Δ) incubated at 30°C (A) and 37°C (B) in soluble starch (20 g/L) medium.
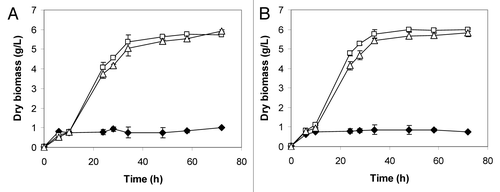
The recombinant S. cerevisiae F2 and F6 were evaluated for their ability to ferment glucose, soluble and raw starch under oxygen-limited conditions (). From glucose, the growth rate and the fermentative performance were comparable for all the strains. Both S. cerevisiae F2 and F6 showed biomass yield and ethanol production level from glucose similar to those of the parental yeast S. cerevisiae 27P, indicating that multiple gene integrations did not significantly affect the yeast fermentative abilities (). From soluble starch, the S. cerevisiae F2 and F6 produced high ethanol levels and their fermentative abilities were similar to those of previously engineered strains.Citation16,Citation27,Citation28 The conversion rate of starch to ethanol was found to be much more efficient in the case of S. cerevisiae F2 (data not shown). However, both recombinant yeasts were not able to metabolize all the starch available and the same result was observed from raw starch where the recombinants produced limited ethanol concentrations (). Their volumetric productivity, about 0.02 g/L/h, was much lower than those determined for previously generated strains, 0.31–0.46 g/L/h,Citation28 but the latter yeasts were grown in batch fermentations with higher carbon source and 25-fold larger inoculum. However, S. cerevisiae F2 and F6 showed the interesting ethanol yield of 75% of the theoretical, which is similar to those reported by Yamada et al.Citation28 As a consequence, although S. cerevisiae F2 and F6 secreted only the glucoamylase, their raw starch-to-ethanol conversion capability should be considered promising since the yeasts described in Yamada et al.Citation28 were constructed by mating two integrated haploid strains, each expressing either α-amylase or glucoamylase gene.
Table 2. Ethanol production by S. cerevisiae strains engineered for the multiple integration of amylolytic genes.
This study showed that codon optimization is an interesting tool for enhancing heterologous expression of genes into industrial S. cerevisiae strains. However, the choice of a properly selected yeast having the traits tailored for the industrial scale bioethanol process revealed to be crucial toward the design of efficient CBP amylolytic yeasts. The constructing strategy proved effective and will contribute to the high expression levels of other heterologous sequences into industrial CBP yeasts.
Acknowledgments
This work was supported by the BiotechIIbis and BiotechIII projects (Regione Veneto, Italy), the Progetto di Ateneo 2010 N. CPDA102570 (University of Padova, Italy), the bilateral joint research project N. ZA11MO2 entitled “Development of robust yeast strains for bioethanol production from starchy and cellulosic plant biomass” and by the National Research Foundation (South Africa). Dr. Lorenzo Favaro is recipient of “Assegno di ricerca Junior” grant (University of Padova). Stefania Zannoni and Federico Fontana (University of Padova) are acknowledged for technical analysis.
References
- Hamelinck CN, Van Hooijdonk G, Faaij APC. Ethanol from lignocellulosic biomass: techno economic performance in short-, middle- and long-term. Biomass Bioenergy 2005; 28:384 - 410; http://dx.doi.org/10.1016/j.biombioe.2004.09.002
- Balat M, Balat H. Recent trends in global production and utilization of bioethanol fuel. Appl Energy 2009; 86:2273 - 82; http://dx.doi.org/10.1016/j.apenergy.2009.03.015
- Rumbold K, van Buijsen HJJ, Gray VM, van Groenestijn JW, Overkamp KM, Slomp RS, et al. Microbial renewable feedstock utilization: a substrate-oriented approach. Bioeng Bugs 2010; 1:359 - 66; http://dx.doi.org/10.4161/bbug.1.5.12389; PMID: 21326838
- Weber C, Farwick A, Benisch F, Brat D, Dietz H, Subtil T, et al. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl Microbiol Biotechnol 2010; 87:1303 - 15; http://dx.doi.org/10.1007/s00253-010-2707-z; PMID: 20535464
- Oreb M, Dietz H, Farwick A, Boles E. Novel strategies to improve co-fermentation of pentoses with D-glucose by recombinant yeast strains in lignocellulosic hydrolysates. Bioengineered 2012; 3; http://dx.doi.org/10.4161/bioe.21444; PMID: 22892590
- Saha BC, Cotta MA. Ethanol production from lignocellulosic biomass by recombinant Escherichia coli strain FBR5. Bioengineered 2012; 3; http://dx.doi.org/10.4161/bbug.19874; PMID: 22705843
- Bothast RJ, Schlicher MA. Biotechnological processes for conversion of corn into ethanol. Appl Microbiol Biotechnol 2005; 67:19 - 25; http://dx.doi.org/10.1007/s00253-004-1819-8; PMID: 15599517
- Kim S, Dale BE. Global potential bioethanol production from wasted crops and crop residues. Biomass Bioenergy 2004; 26:361 - 75; http://dx.doi.org/10.1016/j.biombioe.2003.08.002
- Favaro L, Basaglia M, van Zyl WH, Casella S. Using an efficient fermenting yeast enhances ethanol production from unfiltered wheat bran hydrolysates. Appl Energy 2012; http://dx.doi.org/10.1016/j.apenergy.2012.05.059
- Favaro L, Basaglia M, Casella S. Processing wheat bran into ethanol using mild treatments and highly fermentative yeasts. Biomass Bioenergy 2012; http://dx.doi.org/10.1016/j.biombioe.2012.07.001
- Fukuda H, Kondo A, Tamalampudi S. Bioenergy: sustainable fuels from biomass by yeast and fungal whole-cell biocatalysts. Biochem Eng J 2009; 44:2 - 12; http://dx.doi.org/10.1016/j.bej.2008.11.016
- van Zyl WH, Lynd LR, den Haan R, McBride JE. Consolidated bioprocessing for bioethanol production using Saccharomyces cerevisiae.. Adv Biochem Eng Biotechnol 2007; 108:205 - 35; http://dx.doi.org/10.1007/10_2007_061; PMID: 17846725
- Favaro L, Basaglia M, Saayman M, Rose SH, van Zyl WH, Casella S. Engineering amylolytic yeasts for industrial bioethanol production. Chemical Engineering Transactions 2010; 20:97 - 102; http://dx.doi.org/10.3303/CET1020017
- Favaro L, Basaglia M, Trento A, Saayman M, Rose SH, van Zyl WH, et al. Development of raw starch hydrolysing yeasts for industrial bioethanol production. J Biotechnol 2010; 150:Supplement 1 142; http://dx.doi.org/10.1016/j.jbiotec.2010.08.371
- van Zyl WH, Bloom M, Viktor MJ. Engineering yeasts for raw starch conversion. Appl Microbiol Biotechnol 2012; 95:1377 - 88; http://dx.doi.org/10.1007/s00253-012-4248-0; PMID: 22797599
- Khaw TS, Katakura Y, Koh J, Kondo A, Ueda M, Shioya S. Evaluation of performance of different surface-engineered yeast strains for direct ethanol production from raw starch. Appl Microbiol Biotechnol 2006; 70:573 - 9; http://dx.doi.org/10.1007/s00253-005-0101-z; PMID: 16133340
- Romanos MA, Scorer CA, Clare JJ. Foreign gene expression in yeast: a review. Yeast 1992; 8:423 - 88; http://dx.doi.org/10.1002/yea.320080602; PMID: 1502852
- Lee FWF, Da Silva NA. Improved efficiency and stability of multiple cloned gene insertions at the δ sequences of Saccharomyces cerevisiae.. Appl Microbiol Biotechnol 1997; 48:339 - 45; http://dx.doi.org/10.1007/s002530051059; PMID: 9352677
- Favaro L, Basaglia M, Rose SH, Trento A, Saayman M, van Zyl WH, et al. δ-Integration technique and efficient heterologous expression in yeasts tailored for bioethanol production. Yeast 2011; 28:S36; http://dx.doi.org/10.1002/yea.1863
- Favaro L, Jooste T, Basaglia M, Rose SH, Saayman M, Görgens JF, et al. Codon-optimized glucoamylase sGAI of Aspergillus awamori improves starch utilization in an industrial yeast. Appl Microbiol Biotechnol 2012; 95:957 - 68; http://dx.doi.org/10.1007/s00253-012-4001-8; PMID: 22450569
- Den Haan R, McBride JE, La Grange DC, Lynd LR, van Zyl WH. Functional expression of cellobiohydrolases in Saccharomyces cerevisiae towards one-step conversion of cellulose to ethanol. Enzyme Microb Technol 2007; 40:1291 - 9; http://dx.doi.org/10.1016/j.enzmictec.2006.09.022
- Goto M, Semimaru T, Furukawa K, Hayashida S. Analysis of the raw starch-binding domain by mutation of a glucoamylase from Aspergillus awamori var. kawachi expressed in Saccharomyces cerevisiae.. Appl Environ Microbiol 1994; 60:3926 - 30; PMID: 7993082
- Goto M, Shinoda N, Oka T, Sameshima Y, Ekino K, Furukawa K. Thr/Ser-rich domain of Aspergillus glucoamylase is essential for secretion. Biosci Biotechnol Biochem 2004; 68:961 - 3; http://dx.doi.org/10.1271/bbb.68.961; PMID: 15118335
- Fierobe HP, Clarke AJ, Tull D, Svensson B. Enzymatic properties of the cysteinesulfinic acid derivative of the catalytic-base mutant Glu400-->Cys of glucoamylase from Aspergillus awamori. Biochemistry 1998; 37:3753 - 9; http://dx.doi.org/10.1021/bi972232p; PMID: 9521694
- Belshaw NJ, Williamson G. Specificity of the binding domain of glucoamylase 1. Eur J Biochem 1993; 211:717 - 24; http://dx.doi.org/10.1111/j.1432-1033.1993.tb17601.x; PMID: 7679638
- Chen HM, Ford C, Reilly PJ. Substitution of asparagine residues in Aspergillus awamori glucoamylase by site-directed mutagenesis to eliminate N-glycosylation and inactivation by deamidation. Biochem J 1994; 301:275 - 81; PMID: 8037681
- Nakamura Y, Kobayashi F, Ohnaga M, Sawada T. Alcohol fermentation of starch by a genetic recombinant yeast having glucoamylase activity. Biotechnol Bioeng 1997; 53:21 - 5; http://dx.doi.org/10.1002/(SICI)1097-0290(19970105)53:1<21::AID-BIT4>3.0.CO;2-0; PMID: 18629955
- Yamada R, Bito Y, Adachi T, Tanaka T, Ogino C, Fukuda H, et al. Efficient production of ethanol from raw starch by a mated diploid Saccharomyces cerevisiae with integrated α-amylase and glucoamylase genes. Enzyme Microb Technol 2009; 44:344 - 9; http://dx.doi.org/10.1016/j.enzmictec.2009.01.001