Abstract
Papain-like cysteine proteases of malaria parasites are considered important chemotherapeutic targets or valuable models for the evaluation of drug candidates. Consequently, many of these enzymes have been cloned and expressed in Escherichia coli for their biochemical characterization. However, their expression has been problematic, showing low yield and leading to the formation of insoluble aggregates. Given that highly-productive expression systems are required for the high-throughput evaluation of inhibitors, we analyzed the existing expression systems to identify the causes of such apparent issues. We found that significant divergences in codon and nucleotide composition from host genes are the most probable cause of expression failure, and propose several strategies to overcome these limitations. Finally we predict that yeast hosts Saccharomyces cerevisiae and Pichia pastoris may be better suited than E. coli for the efficient expression of plasmodial genes, presumably leading to soluble and active products reproducing structural and functional characteristics of the natural enzymes.
Introduction
Malaria, the most prevalent protozoal infection worldwide, remains a major public health problem globally.Citation1 It is caused in humans by five species of the genus Plasmodium and causes annually the death of about 1 million people; more than 350 million new infections and 3.2 billion persons are at continuous risk of infection (http://www.dndi.org/diseases/malaria.html). Due to the lack of an effective vaccine, chemotherapy constitutes the only line of defense against malaria. However, the emergence and wide spread of drug-resistant strains reduce the effectiveness of current therapy and demand the urgent development of new drugs with novel mechanisms of action.Citation2
Over the last ten years, the study of peptidases from malaria parasites has acquired considerable importance. Some of these enzymes have been proposed to play central roles in diverse processes such as cell invasion, differentiation, cell cycle progression, catabolism of host proteins, parasite feeding and evasion of the host immune response; making them attractive targets for therapeutic intervention.Citation3 Peptidases involved in hemoglobin degradation, and particularly those belonging to Clan CA family C1 (C1A), have proven very attractive as metabolic targets, considering the experimental evidence that has arisen from both the use of specific inhibitors and gene-disruption experiments.Citation4
Papain-like cysteine hemoglobinases from human malaria parasites P. falciparum (Falcipains 2, 2´ and 3)Citation5-Citation8 and P. vivax (Vivipains 2, 3 and 4)Citation9,Citation10 have been extensively characterized. In addition, several cysteine peptidases from rodent malaria parasites have attracted some attention, given the relevance of murine models for the in vivo evaluation of inhibitors with therapeutic potential. These include Vinckepains (P. vinckei),Citation11 Berghepains (P. berghei)Citation12 and Chabaupains (P. chabaudi).Citation13 Other closely-related isoforms of these enzymes have also been identified and characterized in different plasmodial species,Citation13-Citation15 but these are not involved in hemoglobin degradation and their roles in parasite physiology are now being elucidated.
A sequence analysis of the mature domain of these enzymes shows that they are fairly typical C1A cysteine peptidases (), containing all the conserved residues important for enzymatic catalysis. However, they display several unusual features for a papain family enzyme. An N-terminal extension of the catalytic domain has been identified (or computationally predicted) in several of them, displaying a moderate degree of sequence similarity. At least for Falcipain-2, this segment has proven to be functionally relevant for refolding of mature enzyme in vitro.Citation16,Citation17 The X-ray structure of mature Falcipain-2 confirmed the presence of a short α-helix in this segment, with conserved residues making multiple side-chain interactions with the protein core.Citation7 Second, they have an unusual motif near the C terminus that is absent in Papain, which in Falcipain-2 is involved in hemoglobin binding.Citation18 Furthermore, plasmodial enzymes possess four disulfide bonds, one more than in the case of Papain, with a total of nine Cys residues. Like the rest of the enzymes of the family, they contain an inhibitory prodomain which is removed by auto-proteolysis. In contrast to other C1A enzymes, this prodomain seems to be unnecessary for the correct folding of plasmodial cysteine peptidases.Citation17
Figure 1. Multiple alignment of amino acid sequences (confirmed or predicted mature form) of several plasmodial cysteine hemoglobinases and some closely-related isoforms. Numbers of amino acids are marked at the right side of each of the alignments. Hyphens (-) represent gaps introduced to maximize alignment and asterisks (*) indicate conserved active-site residues. Clear boxes indicate the position of conserved cysteine residues implicated in disulfide bonds (based on the Falcipain-2 structure).Citation7 Shaded boxes indicate the position of a short α-helix (α1) within the N-terminal extension playing a critical role in folding of the mature protein and the C-terminal motif involved in hemoglobin binding. Amino acid sequences of chabaupain-1 (AAP43629); vinckepain-1 (AAL48319); berghepain-1 (XP_677643); yoelipain-1 (XP_729023); vivapain-1 (XP_001615807); falcipain-1 (AAA29578); falcipain-2 (AF282975); falcipain-2´ (AAX77225); falcipain-3 (AAN35746); vivapain-2 (XP_001615274); vivapain-3 (XP_001615273); vivapain-4 (XP_001615272); chabaupain-2 (AAP43630); vinckepain-2 (AAL48319); berghepain-2 (AAL48318); yoelipain-2 (XP_726900) were obtained from GeneBank database (www.ncbi.nlm.nih.gov/genbank/). Papain sequence (AAB02650) was included for comparison.

Since their early identification, it was evident that heterologous expression systems would be needed to produce sufficient amounts of these enzymes for their further characterization. However, many of these expression systems have displayed relatively low productivity. Under the rationale that highly-efficient expression systems will accelerate the high-throughput screening identification of novel inhibitors with therapeutic potential, we review in this paper the expression systems developed so far for plasmodial cysteine proteases in order to identify the possible causes of this phenomenon. We found the striking differences in codon and nucleotide composition between plasmodial and host genes to be the principal cause of expression failure, and discuss several strategies to overcome these limitations. In addition, we highlight the potentialities of non-bacterial expression systems as alternatives for the efficient expression of these particular enzymes.
Analysis of Current Heterologous Expression Systems for Cysteine Peptidases from Human and Rodent Malaria Parasites
The ideal expression system for these targets should meet several requirements. Primarily, it should provide abundant amounts of soluble and active enzymes, with structural and functional characteristics resembling those of the natural molecule. In addition, the system should allow easy downstream purification of the recombinant product, being operationally simple and economically efficient. reviews some characteristics of the expression systems described to date for expression of these enzymes.
Table 1. Heterologous expression systems of Papain-like cysteine proteases from human and rodent malaria parasites
A rapid analysis of this table highlights relevant similarities. First, E. coli has been the most popular expression host used for the recombinant production of these enzymes, accounting for the vast majority of them. This selection is based on well-known advantages it displays over other expression systems, such as (1) high productivity, (2) fast growth at a high cell density in inexpensive media, (3) well-characterized genetics and (4) the availability of a large number of cloning vectors and mutant host strains.
Second, almost all the constructs have been based on native plasmodial genes. Only one study was performed using a codon-optimized synthetic gene for the efficient expression in the selected host.Citation19 In addition, a variety of expression vectors have been used for cloning all the genes, mainly under the regulation of strong IPTG-inducible promoters. In most cases, the recombinant proteins have been N-terminal fused to purification tags such as 6xHis, Maltose-Binding Protein (MBP) or Glutathione-S transferase (GST). For expression, almost all the studies employed a conventional induction protocol, using different IPTG concentrations to regulate gene expression. Despite these apparent similarities in methodologies, the resultant expression levels varied greatly from one study to another, ranging from non-detectable expression to reasonable levels. Low-to-moderate expression levels were the most common results even after optimization of expression conditions,Citation6,Citation9,Citation11,Citation12,Citation15,Citation20,Citation21 with some cases displaying abundant expression.Citation5,Citation10,Citation14,Citation19,Citation22,Citation23
Finally, the great majority of the systems developed have led to the formation of insoluble inclusion bodies, with the exception of those where the protein was fused to MBP as a solubility-enhancing partner. However, these fusion proteins have typically showed limited recovery after proteolytic removal of the tag.Citation22,Citation24 On the other hand, the rapid-dilution methodology invariably used for the refolding of solubilized inclusion bodies failed to renaturate these proteins efficiently to their active conformation. Despite the fact that in many cases the refolding step was performed under previously optimized conditions, a poor recovery of about 10–15% of the protein was typically obtained, except in the case of Falcipain-3, where a rather high renaturation yield of ~43% was reported.Citation6
Strategies to Enhance Productivity of Existing Expression Systems
Increasing the expression level in E. coli
As can be observed, the pre-existing expression systems are far from the expected efficiency level. The first item to be solved is the low expression levels observed for plasmodial genes in E. coli expression hosts. There is consensus that expression of these enzymes in heterologous systems is problematic and unpredictable.Citation14,Citation15,Citation20,Citation22 In this context, it is prudent to select E. coli as the first choice to try the expression of these enzymes, given all the advantages shown by this organism as an expression host. However, the existing evidence indicates that multiple factors must also be considered if a highly-productive expression system is desired.
It is well documented that significant differences in codon usage and A+T content between the target gene and the selected host have a great impact on the resultant expression level of foreign genes in heterologous systems.Citation25,Citation26 These divergences often cause problems during expression, such as decreased mRNA stability and slow translation rates; early termination of transcription and translation; occurrence of insertions, deletions and frame-shifts; and finally inhibition of protein synthesis or even arrest of cell growth. The relative abundance of codon-specific tRNAs in different host systems explains partially why some genes are better expressed than others in a selected system. The Codon Adaptation Index (CAI) parameter is commonly used to describe how well the codons of the target gene match the codon usage preference of the host organism.Citation27 Consequently, the maximum value of CAI of 1.0 indicates a perfect adaptation; but in practice CAI values > 0.9 are considered as very good predictors of high expression.
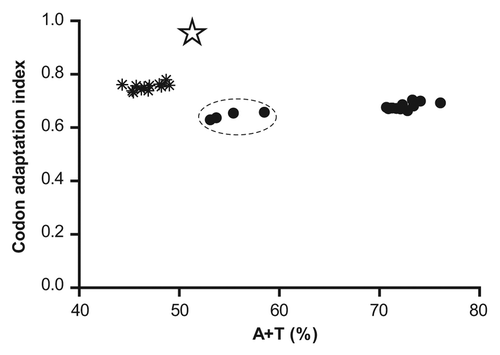
shows that plasmodial genes for cysteine peptidases are distant from highly expressed genes in E. coli. First, they show CAI values of ~0.6 whereas those for reference genes are around 0.75. However, the most striking difference is found in the nucleotide composition of plasmodial genes, which are among the A/T richest genomes sequenced so far. The average A+T content of reference E. coli genes is near 45%, while it is ~70% in plasmodial ones. Interestingly, among them, the genes for Vivipains seem to show a lower A+T content, suggesting a less problematic expression in this host. The divergences in nucleotide composition observed for the rest of the genes are mainly focused in the first (62.1% vs. 38.7%) and third (82.6% vs. 41.9%) codon positions. Consequently, it is frequent to find in these plasmodial genes, codons that are rarely used in E. coli and vice versa (see for more detail); leading to low expression levels, in particular when rare codons are present at the 5′-end of the mRNA or in clusters along the sequence.
Figure 2. Comparison of plasmodial genes for cysteine proteases (●) with highly expressed Escherichia coli genes (٭) corresponding to glycolytic and tricarboxylic acid cycle enzymes. The cluster corresponding to vivipain genes is indicated with dashed lines. The optimized falcipain-2 gene is also indicated with a star. CAI values and global A+T content were calculated using the CAIcal server (http://genomes.urv.es/CAIcal/).Citation27
However, the expression levels of these “problematic” cysteine peptidase genes can be significantly enhanced by means of genetic strategies. One possible approach consists of expanding the tRNA stock of the E. coli host for these rare codons by co-transformation with RIG (AGA/AGG/ATA/GGA), RIL (AGA/AGG/ATA/CTA), RP (AGA/AGG/CCC) or pRARE (ATA/AGG/AGA/CTA/CCC/GGA/CGG) plasmids. However, as can be observed in , these codons account only for a minor part of the actual rare codons, and in some cases not even for the most divergent ones. Moreover, the addition of a highly-expressed fusion partner at the N-terminus of the heterologous protein often results in high level expression of the fusion protein. MBP has been the most used with these enzymes, resulting not only in abundant expression (probably by the effect of harmonized codon usage at the 5′ end of the construct) but also in improved solubility of the fusion protein.
The best strategy to eliminate differences in codon usage is the recodonization of specific plasmodial genes, by replacing codons that are rarely found in highly expressed E. coli genes with more favorable codons throughout the whole sequence. This strategy allows not only the optimization of CAI value for the selected host, but also the modification of the global A+T content and its distribution along the sequence (particularly important to reduce secondary structure in RNA at the translation initiation region), to achieve maximal resemblance with highly expressed host genes. This strategy was successfully used by us to enhance the expression of Falcipain-2.Citation19 As can be observed in , the optimized construct showed an almost perfect match to the codon usage preferences of the host (CAI = 0.96) and a base composition (%A+T = 51.3) very close to that of the host reference genes. Consequently, the construct was expressed to very high-level in several E. coli strains, with the recombinant enzyme representing 35–55% of the total proteins (depending on the host). These expression levels were 3.5–5.5 fold higher than those obtained with the native P. falciparum gene under similar conditions,Citation20 exemplifying the positive effect that codon harmonization could exert in the expression of these enzymes.
Although not strictly a genetic strategy, we also introduced an auto-induction medium to achieve high-cell-density E. coli cultures as an additional step to further enhance productivity.Citation19 As a result, saturation OD600nm in auto-inducing medium was considerably higher than that obtained in LB under similar culture conditions, bringing a ~1.5 fold increase in the volumetric productivity of the system. In addition to higher productivity, the system proved to be more economic and operationally simpler than classical IPTG-induction, with a high degree of adaptability to other related targets.
Soluble and active expression
In addition to the afore-mentioned approaches, the optimization of expression conditions can also influence the global yield of active enzymes. The rationale of this strategy is not based on the augmentation of expression levels, but on increasing the proportion of properly folded product by modifying the expression conditions. Consequently, the main goal would be the identification of induction conditions to achieve the expression of soluble and active enzymes, bypassing solubilization, renaturation and repurification of active enzyme from insoluble/inactive aggregates. Despite sporadic and fortuitous successes,Citation13,Citation19 many classical approaches have repeatedly failed, including changes in culture/induction temperature and time, promoter strength, inducer type and concentration, growth media, E. coli host strain, cellular density at which induction began and addition of folding enhancers. As mentioned above, the most reproducible condition to achieve soluble and active plasmodial cysteine proteases has been its fusion to MBP, with the inconvenience of low efficiency removal of the fusion partner.Citation22
The formation of insoluble inclusion bodies has been the most common and important productive limitation of the expression systems developed so far for plasmodial cysteine proteases. Although the compositions of refolding buffers have been optimized with high-throughput technology, the current refolding methodology is so inefficient that it constitutes the limiting-step in the production of these enzymes. The eight cysteine residues forming disulphide bonds and the remaining catalytic Cys residue represent a huge structural challenge to recover the native conformation of these enzymes.Citation28 Any improvement in the existing refolding methodologies will constitute an actual progress, with great impact on final productivity.
Non-Bacterial Expression Systems
Non-prokaryotic expression systems have been used only twice for the expression of plasmodial cysteine proteases: the methylotrophic yeast Pichia pastoris for Falcipain-2 and Trichoplusia ni insect cells for Falcipain-1. In both cases the results were unsatisfactory, showing no detectable expression and limited recovery of the active protein, respectively.Citation14,Citation22 However, eukaryotic systems might be predicted to be better hosts than E. coli for the expression of these enzymes; as they share more compatible translational machinery and are capable to perform many post-translational modifications (disulfide bond formation, glycosylation, proteolytic processing, etc) critical for the folding and activity of C1A cysteine proteases.
shows the distribution of CAI values of these plasmodial genes for several eukaryotic systems, commonly used for the heterologous expression of Papain-like cysteine proteases. As expected, all the genes showed higher CAI values for eukaryotic hosts than for E. coli, indicating better adaptation to the codon usage preferences of these organisms, with the highest values for Saccharomyces cerevisiae. Interestingly, again vivipain genes deviate from the general behavior of the group; clearly indicating species-specific differences that must be considered for the design of efficient expression systems.
Figure 3. Comparison of several eukaryotic systems (hosts) for the expression of plasmodial genes for cysteine proteases (P. pastoris: Pichia pastoris; S. cerevisiae: Saccharomyces cerevisiae; T. ni: Trichoplusia ni; S. frugiperda: Spodoptera frugiperda; C. griseus: Cricetulus griseus; H. sapiens: Homo sapiens). The mean for each group is indicated by a horizontal line. The cluster corresponding to vivipain genes is indicated with dashed lines.
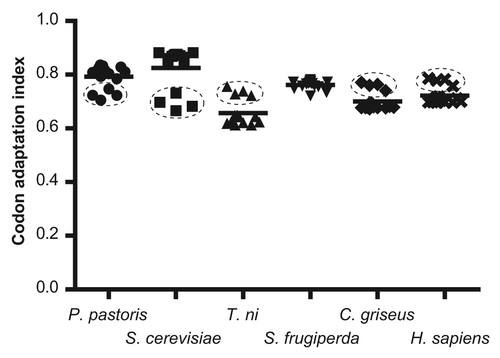
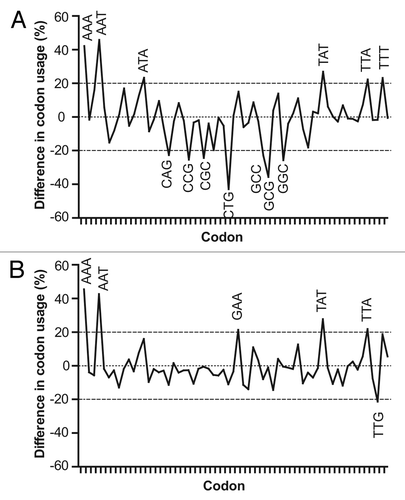
Figure 4. Comparative profile of relative codon frequencies in plasmodial cysteine protease, E. coli (A) and P. pastoris (B) genes. Differences in frequency (per thousand) of codon usage (FCUPLASMODIUM – FCUHOST) were calculated for each codon, using codon composition from plasmodial cysteine protease genes and information contained in codon usage tables for E. coli and P. pastoris, obtained from Codon Usage Database.Citation30 Those codons showing modular differences ≥ 20% are indicated.
Among these organisms, yeast hosts clearly constitute the best alternative to support highly-efficient expression systems for the target enzymes, considering their productivity, easy handling and cost-effective cultivation. In particular, the methylotrophic yeast Pichia pastoris is among the most used expression platforms for biomedical and biotechnological applications, mainly due to: (1) the presence of a strong, tightly regulated, and easily manipulated promoter derived from the P. pastoris alcohol oxidase I gene (AOX1); (2) the existence of an efficient secretory pathway to export recombinant products; (3) the low costs of fermentation and (4) the low risk of carrying human pathogenic agents. An additional advantage of this system is the presence of a single (non-secreted) endogenous C1A cysteine protease (in contrast with insect or mammalian cells), which facilitates the analysis and subsequent purification of the heterologous enzyme from culture supernatant. Furthermore, this organism can grow in a wide pH range, both in complex or chemically defined media and using a great variety of carbon and nitrogen sources, allowing optimization of expression conditions to expedite downstream processing of recombinant product. All these combined, can lead to the cost-effective production of correctly folded, safe and fully active recombinant proteins in milligram-to-gram quantities.Citation29
compares the nucleotide composition of E. coli and P. pastoris genes (codon usage tables)Citation30 with those from plasmodial cysteine protease genes. Interestingly, significant differences can be observed with E. coli genes in all the parameters compared, while only moderate divergences were found for P. pastoris. Apparently, both plasmodial and P. pastoris genes show a similar A/T bias, making them more alike. In contrast, E. coli genes display a modest C/G bias, accounting for bigger differences in nucleotide composition. When compared with E. coli, P. pastoris also showed a more convergent codon usage in relation to plasmodial genes (). Although in both cases a similar number of unmatched A/T-rich codons were found, the profile obtained for P. pastoris is more flat, representing lesser differences in the relative frequency of codon usage. Several C/G-rich codons, almost absent in plasmodial genes but frequently used in host genes, were also identified; notably, seven of them in E. coli and only one in P. pastoris, emphasizing the divergences previously described. All together, these elements explain the higher CAI values obtained for plasmodial genes in P. pastoris over the E. coli host.
Table 2. Nucleotide composition of plasmodial genes in comparison to E. coli and P. pastoris genes
In addition, yeast-based systems have been successfully used for the expression of numerous functionally active papain-like cysteine proteases from diverse organisms, including some protozoan parasites.Citation28 Remarkably, all these successful attempts included the whole inhibitory prodomain of the enzymes in the expressed construct, in contrast with the failed expression of the mature domain of Falcipain-2 in P. pastoris.Citation22 The absence of the prodomain might be one of the plausible causes of this failure, given that significant divergence in codon usage seems unlikely. Although it was postulated that the prodomain is not strictly necessary for the folding of the enzyme, it is unclear whether its inclusion could exert a positive influence in the expression levels of the entire protein in this host. As previously described for E. coli, the optimization of codon composition and A+T content of the target gene would also be beneficial to improve the resultant expression level further. Other strategies to increase productivity, such as the evaluation of different secretion signals and mutated AOX1 promoter libraries, the generation/selection of multi-copy transformant clones, optimization of expression conditions (pH, media composition, feed regime, etc.) and the use of protease inhibitors during expression, can be used as part of the vast repertory of possibilities offered by this versatile expression platform.
Conclusions
Thanks to the development of bioengineered organisms, the design of highly-efficient expression systems for cysteine proteases of malaria parasites is not only possible, but closer than ever before. The confluence of: (1) rational gene design and synthesis, (2) availability of highly-expressed fusion tags to enhance expression levels, solubility and purification of problematic target genes, (3) tools for the fine genetic and metabolic regulation of protein expression and (4) a variety of bacterial and eukaryotic hosts with significant expression capabilities, provides the basis for the development of highly efficient expression systems for these structurally-complex chemotherapeutic targets. The development of such systems will definitely fuel the identification of novel and potent antimalarial drugs lacking parasite resistance.
| Abbreviations: | ||
| CAI | = | codon adaptation index |
| GST | = | glutathione S-transferase |
| FCU | = | frequency (per thousand) of codon usage |
| 6xHis-tag | = | a stretch containing six consecutive histidine residues tag |
| IPTG | = | isopropyl-1-thio-β-D-galactopyranoside |
| LB | = | Luria-Bertani medium |
| MBP | = | maltose-binding protein |
| OD600nm | = | Optical density at 600 nm |
Acknowledgments
We would like to thank Drs. Manuel Mansur, Valentina Cattaneo, Martín Blasco and Colin Berry for helpful discussion and critical reading of the manuscript. This material is based on work partially supported by Grants F/4081–1 and F/4081–2 from Internacional Foundation for Science (Sweden) and fellowships from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina), Red Latinoamericana de Ciencias Biológicas (RELAB), Universidad de las Naciones Unidas/Biotecnología para América Latina y el Caribe (UNU-BIOLAC) and Programa Pablo Neruda/Red Iberoamericana de Biotecnología Isla Negra (RIABIN).
References
- Mathers CD, Ezzati M, Lopez AD. Measuring the burden of neglected tropical diseases: the global burden of disease framework. PLoS Negl Trop Dis 2007; 1:e114; http://dx.doi.org/10.1371/journal.pntd.0000114; PMID: 18060077
- Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int 2009; 58:201 - 9; http://dx.doi.org/10.1016/j.parint.2009.04.004; PMID: 19393762
- Wegscheid-Gerlach C, Gerber HD, Diederich WE. Proteases of Plasmodium falciparum as potential drug targets and inhibitors thereof. Curr Top Med Chem 2010; 10:346 - 67; http://dx.doi.org/10.2174/156802610790725461; PMID: 20166950
- Rosenthal PJ. Falcipains and other cysteine proteases of malaria parasites. Adv Exp Med Biol 2011; 712:30 - 48; http://dx.doi.org/10.1007/978-1-4419-8414-2_3; PMID: 21660657
- Shenai BR, Sijwali PS, Singh A, Rosenthal PJ. Characterization of native and recombinant falcipain-2, a principal trophozoite cysteine protease and essential hemoglobinase of Plasmodium falciparum. J Biol Chem 2000; 275:29000 - 10; http://dx.doi.org/10.1074/jbc.M004459200; PMID: 10887194
- Sijwali PS, Shenai BR, Gut J, Singh A, Rosenthal PJ. Expression and characterization of the Plasmodium falciparum haemoglobinase falcipain-3. Biochem J 2001; 360:481 - 9; http://dx.doi.org/10.1042/0264-6021:3600481; PMID: 11716777
- Hogg T, Nagarajan K, Herzberg S, Chen L, Shen X, Jiang H, et al. Structural and functional characterization of Falcipain-2, a hemoglobinase from the malarial parasite Plasmodium falciparum. J Biol Chem 2006; 281:25425 - 37; http://dx.doi.org/10.1074/jbc.M603776200; PMID: 16777845
- Singh N, Sijwali PS, Pandey KC, Rosenthal PJ. Plasmodium falciparum: biochemical characterization of the cysteine protease falcipain-2′. Exp Parasitol 2006; 112:187 - 92; http://dx.doi.org/10.1016/j.exppara.2005.10.007; PMID: 16337629
- Na BK, Shenai BR, Sijwali PS, Choe Y, Pandey KC, Singh A, et al. Identification and biochemical characterization of vivapains, cysteine proteases of the malaria parasite Plasmodium vivax. Biochem J 2004; 378:529 - 38; http://dx.doi.org/10.1042/BJ20031487; PMID: 14629194
- Na BK, Bae YA, Zo YG, Choe Y, Kim SH, Desai PV, et al. Biochemical properties of a novel cysteine protease of Plasmodium vivax, vivapain-4. PLoS Negl Trop Dis 2010; 4:e849; http://dx.doi.org/10.1371/journal.pntd.0000849; PMID: 20967286
- Singh A, Shenai BR, Choe Y, Gut J, Sijwali PS, Craik CS, et al. Critical role of amino acid 23 in mediating activity and specificity of vinckepain-2, a papain-family cysteine protease of rodent malaria parasites. Biochem J 2002; 368:273 - 81; http://dx.doi.org/10.1042/BJ20020753; PMID: 12169096
- Singh A, Walker KJ, Sijwali PS, Lau AL, Rosenthal PJ. A chimeric cysteine protease of Plasmodium berghei engineered to resemble the Plasmodium falciparum protease falcipain-2. Protein Eng Des Sel 2007; 20:171 - 7; http://dx.doi.org/10.1093/protein/gzm009; PMID: 17430972
- Caldeira RL, Gonçalves LM, Martins TM, Silveira H, Novo C, Rosário V, et al. Plasmodium chabaudi: expression of active recombinant chabaupain-1 and localization studies in Anopheles sp. Exp Parasitol 2009; 122:97 - 105; http://dx.doi.org/10.1016/j.exppara.2009.03.003; PMID: 19292986
- Salas F, Fichmann J, Lee GK, Scott MD, Rosenthal PJ. Functional expression of falcipain, a Plasmodium falciparum cysteine proteinase, supports its role as a malarial hemoglobinase. Infect Immun 1995; 63:2120 - 5; PMID: 7768590
- Kumar A, Kumar K, Korde R, Puri SK, Malhotra P, Singh Chauhan V. Falcipain-1, a Plasmodium falciparum cysteine protease with vaccine potential. Infect Immun 2007; 75:2026 - 34; http://dx.doi.org/10.1128/IAI.01533-06; PMID: 17242063
- Pandey KC, Sijwali PS, Singh A, Na BK, Rosenthal PJ. Independent intramolecular mediators of folding, activity, and inhibition for the Plasmodium falciparum cysteine protease falcipain-2. J Biol Chem 2004; 279:3484 - 91; http://dx.doi.org/10.1074/jbc.M310536200; PMID: 14625277
- Sijwali PS, Shenai BR, Rosenthal PJ. Folding of the Plasmodium falciparum cysteine protease falcipain-2 is mediated by a chaperone-like peptide and not the prodomain. J Biol Chem 2002; 277:14910 - 5; http://dx.doi.org/10.1074/jbc.M109680200; PMID: 11827964
- Pandey KC, Wang SX, Sijwali PS, Lau AL, McKerrow JH, Rosenthal PJ. The Plasmodium falciparum cysteine protease falcipain-2 captures its substrate, hemoglobin, via a unique motif. Proc Natl Acad Sci U S A 2005; 102:9138 - 43; http://dx.doi.org/10.1073/pnas.0502368102; PMID: 15964982
- Sarduy ES, Muñoz AC, Trejo SA, de los A Chavéz Planes M. High-level expression of Falcipain-2 in Escherichia coli by codon optimization and auto-induction. Protein Expr Purif 2012; 83:59 - 69; http://dx.doi.org/10.1016/j.pep.2012.03.008; PMID: 22450163
- Sijwali PS, Brinen LS, Rosenthal PJ. Systematic optimization of expression and refolding of the Plasmodium falciparum cysteine protease falcipain-2. Protein Expr Purif 2001; 22:128 - 34; http://dx.doi.org/10.1006/prep.2001.1416; PMID: 11388810
- Goh SL, Goh LL, Sim TS. Cysteine protease falcipain 1 in Plasmodium falciparum is biochemically distinct from its isozymes. Parasitol Res 2005; 97:295 - 301; http://dx.doi.org/10.1007/s00436-005-1430-7; PMID: 16041608
- Goh LL, Loke P, Singh M, Sim TS. Soluble expression of a functionally active Plasmodium falciparum falcipain-2 fused to maltose-binding protein in Escherichia coli. Protein Expr Purif 2003; 32:194 - 201; http://dx.doi.org/10.1016/S1046-5928(03)00225-0; PMID: 14965764
- Jeong JJ, Kumar A, Hanada T, Seo PS, Li X, Hanspal M, et al. Cloning and characterization of Plasmodium falciparum cysteine protease, falcipain-2B. Blood Cells Mol Dis 2006; 36:429 - 35; http://dx.doi.org/10.1016/j.bcmd.2006.02.003; PMID: 16595182
- Goh LL, Sim TS. Characterization of amino acid variation at strategic positions in parasite and human proteases for selective inhibition of falcipains in Plasmodium falciparum. Biochem Biophys Res Commun 2005; 335:762 - 70; http://dx.doi.org/10.1016/j.bbrc.2005.07.147; PMID: 16095562
- Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol 1995; 6:494 - 500; http://dx.doi.org/10.1016/0958-1669(95)80082-4; PMID: 7579660
- Yadava A, Ockenhouse CF. Effect of codon optimization on expression levels of a functionally folded malaria vaccine candidate in prokaryotic and eukaryotic expression systems. Infect Immun 2003; 71:4961 - 9; http://dx.doi.org/10.1128/IAI.71.9.4961-4969.2003; PMID: 12933838
- Puigbò P, Bravo IG, Garcia-Vallvé SE-CAI. E-CAI: a novel server to estimate an expected value of Codon Adaptation Index (eCAI). BMC Bioinformatics 2008; 9:65; http://dx.doi.org/10.1186/1471-2105-9-65; PMID: 18230160
- Brömme D, Nallaseth FS, Turk B. Production and activation of recombinant papain-like cysteine proteases. Methods 2004; 32:199 - 206; http://dx.doi.org/10.1016/S1046-2023(03)00212-3; PMID: 14698633
- Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast 2005; 22:249 - 70; http://dx.doi.org/10.1002/yea.1208; PMID: 15704221
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res 2000; 28:292; http://dx.doi.org/10.1093/nar/28.1.292; PMID: 10592250