Abstract
Virotherapy on the basis of oncolytic vaccinia virus (VACV) strains is one novel approach for canine cancer therapy. In this study we described for the first time the characterization and the use of new VACV strain LIVP6.1.1 as an oncolytic agent against canine cancer in a panel of four canine cancer cell lines including: soft tissue sarcoma (STSA-1), melanoma (CHAS), osteosarcoma (D-17) and prostate carcinoma (DT08/40). Cell culture data demonstrated that LIVP6.1.1 efficiently infected and destroyed all four tested canine cancer cell lines. In two different xenograft models on the basis of the canine soft tissue sarcoma STSA-1 and the prostate carcinoma DT08/40 cell lines, a systemic administration of the LIVP6.1.1 virus was found to be safe and led to anti-tumor and immunological effects resulting in the significant reduction of tumor growth in comparison to untreated control mice.
In summary, the pre-clinical evaluation has demonstrated the efficacy of LIVP6.1.1 for canine cancer therapy. Furthermore, a clinical trial with canine cancer patients has already been started.
Introduction
Cancer is the most common cause of natural death in dogs and endemic in both developed and developing countries (http://www.wearethecure.org/). Incidence of cancer ranges from 1 to 2% in the canine population and currently accounts for about half of the deaths in dogs older than 10 years.Citation1,Citation2 The major treatment options for canine cancers include surgery, radiation therapy, chemotherapy, hyperthermia and photodynamic therapy. Despite progress in the diagnosis and treatment of advanced canine cancer, overall treatment outcome has not substantially improved in the past. Therefore, the development of new therapies for advanced canine cancer is a high priority. One of the most promising novel cancer therapies is oncolytic virotherapy. This method is based on the capacity of oncolytic viruses (OVs) to preferentially infect and lyse cancer cells without causing excessive damage to surrounding normal tissue. Several oncolytic viruses including various human and canine adenoviruses,Citation3-Citation5 canine distemper virusCitation6 and vaccinia virus strains, namely GLV-1h68, LIVP1.1.1Citation7-Citation9 and GLV-1h109Citation10 have been successfully tested for canine cancer therapy in preclinical settings (for reviews see refs. Citation11 and Citation12). However, in contrast to human studies, the clinical trials with oncolytic viruses for canine cancer patients are just at the beginning.
In this study, we have analyzed the oncolytic potential of a new vaccinia virus strain LIVP6.1.1 against different canine cancer cells in cell culture and analyzed the therapeutic effect against canine soft tissue sarcoma and prostate carcinoma in xenograft models. LIVP6.1.1 was isolated from a wild type stock of Lister strain of vaccinia virus (Lister strain, Institute of Viral Preparations, Moscow, Russia) and represents a “native” virus (no genetic manipulations were conducted). The sequence analysis revealed that the thymidine kinase (tk) gene of LIVP6.1.1 was broken into two ORFs and different mutations compared with GLV-1h68 are present (Chen et al. manuscript in preparation). In addition, we have selected LIVP6.1.1 for this study, since it was less virulent compared with other Lister strain isolates (Chen et. al. manuscript in preparation).
Here, we analyzed the oncolytic effects of LIVP6.1.1 in a panel of four different canine cancer cell lines and in canine soft tissue sarcoma and prostate carcinoma xenografts in nude mice.
Results and Discussion
Analysis of the oncolytic potential of LIVP6.1.1 virus against different canine cancer cell lines
The oncolytic effect of LIVP6.1.1 against a panel of four different canine cancer cell lines including soft tissue sarcoma STSA-1, melanoma CHAS, osteosarcoma D17 and prostate carcinoma DT08/40 cells, was examined. For this purpose, the cells were seeded three days prior to infection in 24-well plates and then were infected with LIVP6.1.1 at multiplicities of infection (MOIs) of 1.0 and 0.1. The cell viability was analyzed at 24, 48 and 72 h post-virus-infection (hpvi) by XTT-assays (). At MOI of 1.0, the LIVP6.1.1 virus was highly cytotoxic to three cell lines (STSA-1, D17 and CHAS), resulting in at last 83% cytotoxicity over 3 d. One day later similar cytotoxicity was also observed for LIVP6.1.1-infected DT08/40 cells.
Figure 1. Viability of soft tissue sarcoma STSA-1 (A), melanoma CHAS (B), osteosarcoma D17 (C) and prostate carcinoma DT08/40 (D). cells after LIVP6.1.1 infection at MOIs of 1.0 and 0.1. Viable cells after infections with LIVP6.1.1virus at MOIs of 0.1 and 1.0 were detected using a XTT assay (Cell Proliferation Kit II, Roche Diagnostics, Mannheim, Germany)(Sigma, Taufkirchen, Germany). Mean values (n = 3) and standard deviations are shown as percentages of respective controls. The data represent three independent experiments
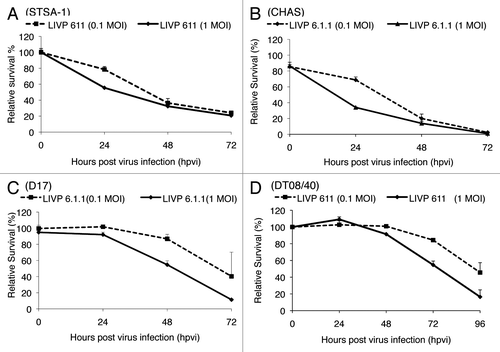
These results indicated that the LIVP6.1.1 virus infection led to efficient killing of all four canine cancer cell lines in these experimental settings.
Efficiency of LIVP6.1.1-replication in STSA-1 and DT08/40 canine cancer cells
The oncolytic potential of OVs is dependent on their ability to efficiently infect and replicate in cancer cells. In order to test the efficiency of virus replication, STSA-1 and DT08/40 cells were infected with either LIVP6.1.1 or GLV-1h68 at an MOI of 0.1. In these experimental setting GLV-1h68 was used as a control. Standard plaque assays were performed for all samples to determine the viral titers at different time points during the course of infection (). Efficient LIVP6.1.1 viral replication (> 100-fold titer increase at 48 or 96 hpvi) was observed in both cell lines. The maximum viral titers were determined for LIVP6.1.1 (5.34 × 106 pfu/well) and for GLV-1h68 (2.98 × 106 pfu/well) in STSA-1 at 48 hpvi (). Interestingly, the highest virus titers in virus-infected DT08/40 cells were identified for LIVP6.1.1 (8.24 × 106 pfu/well) and for GLV-1h68 (1.53 × 106 pfu/well) at 96 hpvi ().
Figure 2. Replication capacity of the vaccinia virus strains LIVP6.1.1 and GLV-1h68 in different canine cancer cells. For the viral replication assay, STSA-1 (A) and DT08/40 (B) cells grown in 24-well plates were infected with either LIVP6.1.1 or GLV-1h68 at an MOI of 0.1. Cells and supernatants were collected for the determination of virus titers at various time points. Viral titers were determined as pfu per ml in triplicates by standard plaque assay in CV-1 cell monolayers. Averages plus standard deviation are plotted. The data represent three independent experiments.
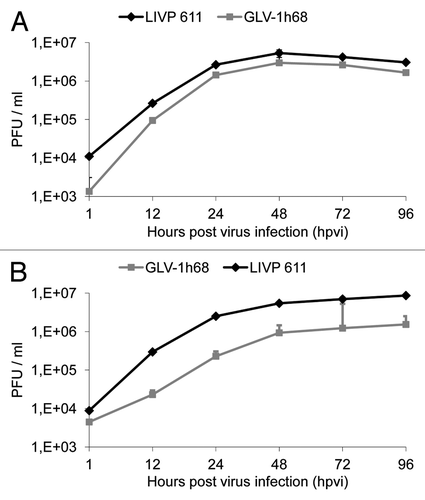
These data demonstrated that the virus replication efficiency is dependent on the infection time point and tumor types. Under these experimental conditions, LIVP6.1.1 can replicate more efficiently than GLV-1h68 in both cancer cell lines.
Oncolytic effect of a single systemic application of LIVP6.1.1 on STSA-1 and DT08/40 cell xenografts
In this study the therapeutic effect of LIVP6.1.1 on the progression of soft tissue sarcoma STSA-1 subcutaneous xenografts was evaluated in vivo by measuring tumor volumes at various time points. Tumors were generated by implanting 1 × 106 STSA-1 canine soft tissue sarcoma cells subcutaneously into the right hind leg of 6- to 8-week-old female nude mice (NCI/Hsd/Athymic Nude-Foxn1nu). Five weeks post implantation, all mice developed tumors with volumes of 600 to 1000 mm3. We have chosen mice with larger tumors for virus injection since this late stage of the tumor development is more representative and interesting for the clinical praxis. Animals were separated into two groups (n = 6/group) and were injected either with a single dose of LIPV6.1.1 (5 × 106 pfu) or PBS intravenously into the lateral tail vein. As shown in , the virus treatment led to a significant decrease in STSA-1 tumor growth in all virus-treated mice compared with PBS control mice. Due to excessive tumor burden (> 3000 mm3), all animals in the control PBS group were euthanized after 14 dpi.
Figure 3. Effects of systemic LIVP6.1.1 virus injection on tumor growth (A, B) and the body weights (C, D) of STSA-1 or DT08/40 xenografted mice. Two groups each of (A, C) STSA-1 tumor-bearing nude mice (n = 6) and (B, D) DT08/40 tumor bearing mice (n = 6) were either treated with a single dose of 5x106 pfu LIVP6.1.1 or with PBS (mock control) intravenously (iv). The statistical significance was confirmed by Student’s t-test where * and ** indicate p < 0.05 and 0.01 respectively.
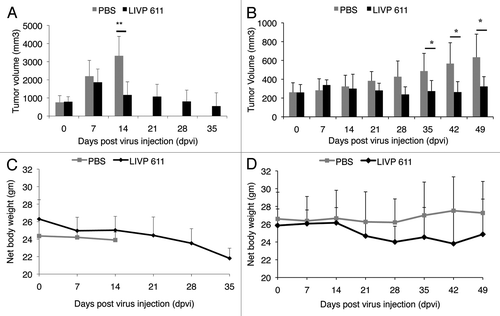
The therapeutic effect of LIVP6.1.1 was also evaluated on the progression of the slow growing canine prostate carcinoma DT08/40 tumors in nude mice by measuring the tumor volume at various time points. Data demonstrated again that a single injection with LIVP6.1.1 vaccinia virus led to significant inhibition of the tumor growth (*p < 0.05) of all virus-treated mice compared with the control PBS animals on 35, 42 and 49 dpvi ().
Finally, the toxicity of the LIVP6.1.1 virus was determined by monitoring the relative weight change of mice over time (). All LIVP6.1.1 treated mice showed relatively stable mean weight over the course of studies. There were no signs of virus-mediated toxicity.
In summary, therapy with the vaccinia strain LIVP6.1.1 demonstrated anti-tumor activity in canine soft tissue sarcoma and canine prostate xenograft models. Therefore we propose that vaccinia virus strain LIVP6.1.1 may be useful for the treatment of these cancer types in dogs.
Biodistribution and persistence of LIVP6.1.1 in STSA-1 tumor-bearing nude mice
At late time points after virus treatment, we analyzed the virus distribution and persistence in LIVP6.1.1-treated STSA-1 xenografted mice. summarizes the virus distribution data at 35 dpvi. In all virus-treated mice the highest viral titers were identified in primary tumors. In addition, low copies of LIVP6.1.1 virus particles were also detected in liver, lung, spleen and kidney of the treated animals (). The later presence of LIVP6.1.1 in these organs could be due to leakiness of blood vessels in solid tumors, circulating virus-infected tumor cells or cell particles may end up in healthy tissues such as the lung, liver, spleen and kidney. However, LIVP6.1.1 seems to be tumor-selective, as we found about 104–105 fold more virus particles in solid tumors compared with healthy tissues of the treated animals.
Table 1. Biodistribution of LIVP6.1.1 in virus-treated STSA-1 xenografted mice at 35 d post virus injection (dpvi)
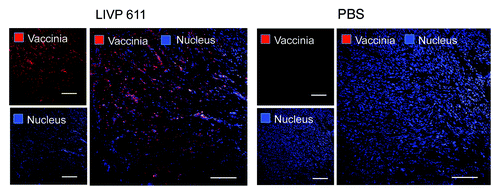
The virus biodistribution in the primary STSA-1 tumors was also analyzed by immunohistochemical staining at 35 dpvi (). The data revealed that the tumors in all treated mice were completely infected with vaccinia virus, which led to oncolysis and destruction of tumor tissues.
Figure 4. Immunohistochemical staining of infected and uninfected STSA-1 xenograft tumors at 35 dpvi. Tumor-bearing mice were either mock treated (PBS) or infected with LIVP6.1.1. Tumor sections were labeled with anti-vaccinia virus antibodies (red) and nuclei were stained with Hoechst (blue). Scale bars, 2.5 mm.
Increased presence of immune cells in tumors of STSA-1 xenografted mice after LIVP6.1.1 treatment
To investigate the immunological aspect of oncolytic therapy, we have analyzed the effect of virus infection on host immune cells in tumors of STSA1-tumor-bearing mice. Our flow cytometry data demonstrated a significant increase of Gr-1highCD11b+ (MDSCs, granulocytes), Gr-1intCD11b+ (MDSCs, monocytes), F4/80+CD45+ (macrophages) and MHCII+CD45+ cells in the LIVP6.1.1 infected tumors compared with PBS-treated tumors (). In addition, we used the Gr-1highCD11b+ cells as markers for monitoring viral infection on a systemic level. For this purpose, a parallel flow cytometric analysis of Gr-1highCD11b+ cells in the peripheral blood was performed. There was no significant difference in the number of Gr-1highCD11b+cells in the peripheral blood between virus and PBS-treated STSA-1 xenografted mice (). These data suggest that the changes in granulocytic MDSCs were not systemic, but rather due to a change in recruitment and/or persistence within virus treated tumors.
Table 2. Presence of immune cells in tumors and peripheral blood of STSA-1 xenografted mice 7 d after LIVP6.1.1 or PBS-treatments
On the basis of these data, we assume that the strong anti-tumor effect of LIVP6.1.1 in STSA-1 tumor bearing mice could be at least in part dependent on the increased number of these host immune cells in the tumor bed. Moreover, we and others have reported that virotherapy induces massive tumoral infiltration of MDSCs resembling neutrophils and macrophages, which may be part of virotherapy-mediated antitumor mechanism.Citation13-Citation15
Taken together, therapy with the vaccinia strain LIVP6.1.1 demonstrated outstanding anti-tumor activity in several canine cancer cell lines and in two xenograft models. Our findings suggest that virotherapy-mediated anti-tumor mechanism in STSA-1 xenografts could be a combination of the direct viral oncolysis of tumor cells and the virus-dependent infiltration of tumor-associated host immune cells.
In conclusion, we propose that the LIVP6.1.1 vaccinia virus strain may be useful for the treatment of canine cancer patients. First clinical trial with this virus has already started.
Materials and Methods
Ethics statement
All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Explora Biolabs (San Diego, CA, USA; protocol number: EB11–025) and/or the government of Unterfranken, Germany (permit number: 55.2–2531.01–17/08 and 55.2–2531.01–24/12).
Cell culture
African green monkey kidney fibroblasts (CV-1) were obtained from the American Type Culture Collection (ATCC). STSA-1 cells were derived from a canine patient with a low grade II soft tissue sarcoma.Citation15 DT08/40 was diagnosed as canine prostate carcinoma.Citation16 The canine melanoma CHAS cell line was provided by Dr. Ogilvie (Angel Care Cancer Center, Carlsbad, USA), and canine osteosarcoma D17 was obtained from ATCC.
Cells were cultured in DMEM supplemented with antibiotic-solution (100 U/ml penicillin G, 100 units/ml streptomycin) and 10% fetal bovine serum (FBS; Invitrogen GmbH, Karlsruhe, Germany) for CV-1 and 20% FBS for D17 and DT08/40 at 37°C under 5% CO2.
STSA-1 cells were cultivated in minimum essential medium (MEM) with Earle’s salts supplemented with 2 mM glutamine, 50 U/mL penicillin G, 50 µg/mL streptomycin, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids (MEM-C) and 10% FBS.
CHAS cells were cultured in DMEM/F-12 50/50 media (Cellgro) supplemented with antibiotic-solution, HuMEC supplement (Gibco) and 10% FBS (Cellgro), at 37°C under 5% CO2.
Virus strains
Vaccinia virus strain LIVP6.1.1 was derived from LIVP (Lister strain, Institute of Viral Preparations, Moscow, Russia). The sequence analysis of LIVP6.1.1 revealed the presence of different mutations in several genes (Chen et al., manuscript in preparation). In addition, LIVP6.1.1 demonstrated different plaque morphology in comparison to GLV-1h68 in CV-1 cells. GLV-1h68 is an oncolytic virus strain designed to colonize and destroy cancer cells without harming healthy tissues or organs.Citation17
Cell viability assay
1 × 104 cells/well were seeded in 96-well plates (Nunc). After 24 h in culture, cells were infected with vaccinia virus strains using multiplicities of infection (MOI) of 0.1 and 1.0. The cells were incubated at 37°C for 1 h, then the infection medium was removed and subsequently the cells were incubated in fresh growth medium.
The amount of viable cells after infection was measured using 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide inner salt (XTT) assay (Cell Proliferation Kit II, Roche Diagnostics), according to the manufacturer’s protocol at 24, 48, or 72 h after virus infection. Quantification of cell viability was performed in an ELISA plate reader (Tecan Sunrise, Tecan Trading AG) at 490 nm with a reference wavelength of 690 nm. The relative number of viable cells was expressed as percent cell viability. Uninfected cells were used as reference and were considered as 100% viable.
Viral replication
For the viral replication assay, different canine cancer cells were infected with LIVP6.1.1 or GLV-1h68 at an MOI of 0.1. After one hour of incubation at 37°C with gentle agitation every 20 min, the infection medium was removed and replaced by a fresh growth medium. After 1, 12, 24, 48, 72 and 96 h, the cells and supernatants were harvested. Following three freeze-thaw cycles, serial dilutions of the supernatants and lysates were titered by standard plaque assays on CV-1 cells. All samples were measured in triplicate.
Vaccinia virus-mediated therapy of STSA-1 and DT08/40 xenografts
Tumors were generated by implanting either 1 × 106 canine soft tissue sarcoma STSA-1 cells or 5 × 106 canine prostate DT08/40 cells subcutaneously into the right hind leg of 6- to 8-week-old female nude mice [Hsd:Athymic Nude-Foxn1nu; Harlan, Holland]. Tumor growth was monitored weekly in two dimensions using a digital caliper. Tumor volume was calculated as [(length × widthCitation2)/2]. When tumor volume reached approximately 600–1000 mm3 (STSA-1) and 200–400 mm3 (DT08/40), groups of mice (n = 6) were injected either with 5 × 106 pfu of LIVP6.1.1 virus or PBS (control) into the tail vein intravenously (i.v.). The significance of the results was calculated by Student’s t-test. Results are displayed as means ± standard deviation (SD). P values of < 0.05 were considered significant.
Mice were also monitored for change in body weight and signs of toxicity.
Histology and microscopy
For histological studies, tumors were excised and snap-frozen in liquid nitrogen, followed by fixation in 4% paraformaldehyde/PBS at pH 7.4 for 16 h at 4°C. After dehydration in 10% and 30% sucrose (Carl Roth) specimens were embedded in Tissue-Tek® O.C.T. (Sakura Finetek Europe B.V.). Tissue samples were sectioned (10 µm thickness) with the cryostat 2800 Frigocut (Leica Microsystems GmbH). Labeling of tissue sections was performed as described in detail elsewhere.Citation18,Citation19 LIVP6.1.1 was labeled using polyclonal rabbit anti vaccinia virus (anti-VACV) antibody (Abcam), which was stained using Cy3-conjugated donkey anti rabbit secondary antibodies obtained from Jackson ImmunoResearch. Hoechst 33342 was used to label nuclei in tissue sections.
The fluorescence-labeled preparations were examined using the Leica TCS SP2 AOBS confocal laser microscope equipped with argon, helium-neon and UV laser and the LCS 2.16 software (1024 × 1024 pixel RGB-color images). Digital images were processed with Photoshop 7.0 (Adobe Systems).
Flow cytometric (FACS) analysis
For flow cytometric analysis, three mice from each group were sacrificed by CO2 inhalation and tumors were removed. The preparation of single cell suspensions derived from LIVP6.1.1- or PBS- treated tumors was performed as described by Gentschev et al.Citation9
To block non-specific staining, single cells were preincubated with 0.5 µg of anti-mouse CD16/32 antibody (clone 93, Biolegend) per one million cells for 20 min on ice. After that, the cells were incubated at 4°C for 15 min in PBS with 2% FBS in the presence of appropriate dilutions of labeled monoclonal antibodies: anti-mouse MHCII-PE (Clone M5 114.15.2, eBioscience), anti-CD11b-PerCPCy5.5 (Clone M1/70, eBioscience), anti-F4/80-APC (Clone BM8, eBioscience), anti-Gr-1-APC (Ly-6G, Clone RB6–8C5, eBioscience). The Anti-Gr-1 mAb (RB6–8C5) has long been used to stain MDSCs and allows the distinction of at least two subsets of granulocytes (Gr-1highCD11b+) and monocytic cells (Gr-1intCD11b+).Citation20
Stained cells were subsequently analyzed, using Accuri C6 Cytometer and FACS analysis software CFlow Version 1.0.227.4 (Accuri Cytometers, Inc.).
| Abbreviations: | ||
| ATCC | = | American Type Culture Collection |
| DMEM | = | Dulbecco's Modified Eagle's Medium |
| FBS | = | Fetal Bovine Serum |
| hpvi | = | hours post-virus-infection |
| IACUC | = | Institutional Animal Care and Use Committee |
| LIVP | = | a Lister strain of vaccinia virus |
| MDSCs | = | Myeloid-derived suppressor cells |
| MOI | = | Multiplicity of Infection |
| ORF | = | Open Reading Frame |
| OV | = | oncolytic virus |
| pfu | = | plaque-forming unit |
| tk | = | thymidine kinase |
| VACV | = | vaccinia virus |
| XTT | = | 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide inner salt |
Acknowledgments
We thank Mrs J. Langbein-Laugwitz, Mr J. Aguilar, Mr T. Trevino and Mrs I. Smirnow for technical support, Dr I. Nolte and Dr A. Macneill for providing of canine cell lines used in this study and Dr Z. Sokolovic for critical reading of the manuscript.
Disclosure of Potential Conflicts of Interest
This work was supported by Genelux Corporation, San Diego, USA, and a Service Grant to the University of Wuerzburg, Germany also funded by Genelux Corporation. I.G., U.G., A.F., N.G.C., Y.A.Y., Q.Z. and A.A.S. are employees and shareholders of Genelux. M.A. and S.W. were supported by grants of Genelux Corporation. S.S.P. is a graduate fellow and supported by a grant of the German Excellence Initiative to the Graduate School of Life Sciences, University of Wuerzburg. No additional external funding was received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Notes
† These authors contributed equally to this work.
References
- Merlo DF, Rossi L, Pellegrino C, Ceppi M, Cardellino U, Capurro C, et al. Cancer incidence in pet dogs: findings of the Animal Tumor Registry of Genoa, Italy. J Vet Intern Med 2008; 22:976 - 84; http://dx.doi.org/10.1111/j.1939-1676.2008.0133.x; PMID: 18564221
- Kelsey JL, Moore AS, Glickman LT. Epidemiologic studies of risk factors for cancer in pet dogs. Epidemiol Rev 1998; 20:204 - 17; http://dx.doi.org/10.1093/oxfordjournals.epirev.a017981; PMID: 9919439
- Ternovoi VV, Le LP, Belousova N, Smith BF, Siegal GP, Curiel DT. Productive replication of human adenovirus type 5 in canine cells. J Virol 2005; 79:1308 - 11; http://dx.doi.org/10.1128/JVI.79.2.1308-1311.2005; PMID: 15613357
- Smith BF, Curiel DT, Ternovoi VV, Borovjagin AV, Baker HJ, Cox N, et al. Administration of a conditionally replicative oncolytic canine adenovirus in normal dogs. Cancer Biother Radiopharm 2006; 21:601 - 6; http://dx.doi.org/10.1089/cbr.2006.21.601; PMID: 17257075
- Le LP, Rivera AA, Glasgow JN, Ternovoi VV, Wu H, Wang M, et al. Infectivity enhancement for adenoviral transduction of canine osteosarcoma cells. Gene Ther 2006; 13:389 - 99; http://dx.doi.org/10.1038/sj.gt.3302674; PMID: 16292351
- Suter SE, Chein MB, von Messling V, Yip B, Cattaneo R, Vernau W, et al. In vitro canine distemper virus infection of canine lymphoid cells: a prelude to oncolytic therapy for lymphoma. Clin Cancer Res 2005; 11:1579 - 87; http://dx.doi.org/10.1158/1078-0432.CCR-04-1944; PMID: 15746063
- Gentschev I, Ehrig K, Donat U, Hess M, Rudolph S, Chen N, et al. Significant Growth Inhibition of Canine Mammary Carcinoma Xenografts following Treatment with Oncolytic Vaccinia Virus GLV-1h68. J Oncol 2010; 2010:736907; http://dx.doi.org/10.1155/2010/736907; PMID: 20631910
- Gentschev I, Stritzker J, Hofmann E, Weibel S, Yu YA, Chen N, et al. Use of an oncolytic vaccinia virus for the treatment of canine breast cancer in nude mice: preclinical development of a therapeutic agent. Cancer Gene Ther 2009; 16:320 - 8; http://dx.doi.org/10.1038/cgt.2008.87; PMID: 18949014
- Gentschev I, Müller M, Adelfinger M, Weibel S, Grummt F, Zimmermann M, et al. Efficient colonization and therapy of human hepatocellular carcinoma (HCC) using the oncolytic vaccinia virus strain GLV-1h68. PLoS One 2011; 6:e22069; http://dx.doi.org/10.1371/journal.pone.0022069; PMID: 21779374
- Patil SS, Gentschev I, Adelfinger M, Donat U, Hess M, Weibel S, et al. Virotherapy of canine tumors with oncolytic vaccinia virus GLV-1h109 expressing an anti-VEGF single-chain antibody. PLoS One 2012; e47472
- Arendt M, Nasir L, Morgan IM. Oncolytic gene therapy for canine cancers: teaching old dog viruses new tricks. Vet Comp Oncol 2009; 7:153 - 61; http://dx.doi.org/10.1111/j.1476-5829.2009.00187.x; PMID: 19691644
- Patil SS, Gentschev I, Nolte I, Ogilvie G, Szalay AA. Oncolytic virotherapy in veterinary medicine: current status and future prospects for canine patients. J Transl Med 2012; 10:3; http://dx.doi.org/10.1186/1479-5876-10-3; PMID: 22216938
- Gil M, Bieniasz M, Seshadri M, Fisher D, Ciesielski MJ, Chen Y, et al. Photodynamic therapy augments the efficacy of oncolytic vaccinia virus against primary and metastatic tumours in mice. Br J Cancer 2011; 105:1512 - 21; http://dx.doi.org/10.1038/bjc.2011.429; PMID: 21989183
- Breitbach CJ, Paterson JM, Lemay CG, Falls TJ, McGuire A, Parato KA, et al. Targeted inflammation during oncolytic virus therapy severely compromises tumor blood flow. Mol Ther 2007; 15:1686 - 93; http://dx.doi.org/10.1038/sj.mt.6300215; PMID: 17579581
- Gentschev I, Adelfinger M, Josupeit R, Rudolph S, Ehrig K, Donat U, et al. Preclinical evaluation of oncolytic vaccinia virus for therapy of canine soft tissue sarcoma. PLoS One 2012; 7:e37239; http://dx.doi.org/10.1371/journal.pone.0037239; PMID: 22615950
- Reimann-Berg N, Willenbrock S, Murua Escobar H, Eberle N, Gerhauser I, Mischke R, et al. Two new cases of polysomy 13 in canine prostate cancer. Cytogenet Genome Res 2011; 132:16 - 21; http://dx.doi.org/10.1159/000317077; PMID: 20668368
- Zhang Q, Yu YA, Wang E, Chen N, Danner RL, Munson PJ, et al. Eradication of solid human breast tumors in nude mice with an intravenously injected light-emitting oncolytic vaccinia virus. Cancer Res 2007; 67:10038 - 46; http://dx.doi.org/10.1158/0008-5472.CAN-07-0146; PMID: 17942938
- Gentschev I, Donat U, Hofmann E, Weibel S, Adelfinger M, Raab V, et al. Regression of human prostate tumors and metastases in nude mice following treatment with the recombinant oncolytic vaccinia virus GLV-1h68. J Biomed Biotechnol 2010; 2010:489759; http://dx.doi.org/10.1155/2010/489759; PMID: 20379368
- Weibel S, Stritzker J, Eck M, Goebel W, Szalay AA. Colonization of experimental murine breast tumours by Escherichia coli K-12 significantly alters the tumour microenvironment. Cell Microbiol 2008; 10:1235 - 48; http://dx.doi.org/10.1111/j.1462-5822.2008.01122.x; PMID: 18208564
- Peranzoni E, Zilio S, Marigo I, Dolcetti L, Zanovello P, Mandruzzato S, et al. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol 2010; 22:238 - 44; http://dx.doi.org/10.1016/j.coi.2010.01.021; PMID: 20171075