Abstract
Gram-negative bacteria Helicobacter pylori cause gastric ulcer, duodenal cancer, and found in almost half of the world’s residents. The protein responsible for this disease is secreted through type IV secretion system (TFSS) of H. pylori. TFSS is encoded by 40-kb region of chromosomal DNA known as cag-pathogenicity island (PAI). TFSS comprises of three major components: cytoplasmic/inner membrane ATPase, transmembrane core-complex and outer membranous pilli, and associated subunits. Core complex consists of CagX, CagT, CagM, and Cag3(δ) proteins as per existing knowledge. In this study, we have characterized one of the important component of core-complex forming sub-unit protein, i.e., CagX. Complete ORF of CagX except signal peptide coding region was cloned and expressed in pET28a vector. Purification of CagX protein was performed, and polyclonal anti-sera against full-length recombinant CagX were raised in rabbit model. We obtained a very specific and high titer, CagX anti-sera that were utilized to characterize endogenous CagX. Surface localization of CagX was also seen by immunofluorescence microscopy. In short for the first time a full-length CagX was characterized, and we showed that CagX is the part of high molecular weight core complex, which is important for assembly and function of H. pylori TFSS.
Introduction
Helicobacter pylori are human pathogenic bacteria responsible for chronic activities in the stomach. Its infection causes peptic ulcer, gastric cancer, and mucosa associated lymphoid tissue lymphoma.Citation1-Citation5H. pylori infection is also associated with non-gastric diseases like cardio vascular disease, Idiopathic thrombocytopenic purpura, etc.Citation6 It is a common and important transmissible bacterial human pathogen. Its infection varies worldwide, being as low as 10% in developed countries and higher than 80% among the developing countries of the world. H. pylori infection is a major concern of developing countries like India and more than 20 million Indians are estimated to suffer from peptic ulcer.Citation7H. pylori infected persons with gastric intestinal metaplasia are also at higher risk of developing gastric cancer, which is responsible for considerable morbidity and mortality.Citation8
The virulence of H. pylori strains correlates with the intensity of the inflammatory response to the infection.Citation9 Cag-pathogenicity island (cag-PAI) is an established H. pylori virulence factor because prevalence of gastric cancer is higher in patients infected with cagPAI positive H. pylori than cagPAI negative H. pylori.Citation10 Product of CagX gene, present in cagPAI region, plays an important role in pathogen-associated virulence.Citation11 CagX is a part of core complex of H. pylori TFSS and plays an important role in CagA (an effector protein) translocation into the host cell as shown by systematic mutagenesis study.Citation12 It has partial similarity with virB9 of Agarobacterium TFSS,Citation13 and its association with pilus, the structure forms between host and H. pylori, has also been reported.Citation14 Therefore characterization, localization, and other studies are required to understand the mechanistic role of CagX in H. pylori virulence and pathogenesis. Preliminary study on CagX has been performed by few groups, but their investigations were based on the use of polyclonal antibodies raised against C-terminal part of CagX or small polypeptides only.Citation15 In this work, polyclonal antibody has been raised against whole recombinant CagX protein, and localization and existence of endogenous protein has been explored using raised antibody. We have also shown that CagX is part of a high molecular weight core complex using blue native gradient PAGE and proposed that CagX is important for the assembly and function of the H. pylori TFSS for the first time.
Results
Cloning and expression of CagX gene
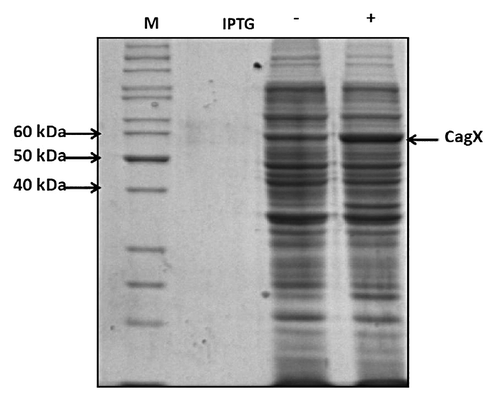
Complete ORF excluding signal peptide coding sequence (-N-terminal 30 aa) was cloned in pET28a vector at NcoI and HindIII site. The stop codon was also present in reverse primer so that there was no tag even at C-terminal of CagX protein. The clones were screened and confirmed by colony PCR and restriction digestion. Molecular weight of recombinant protein produced by CagX gene clone in pET28a (without signal sequence) is predicted to be 57 kDa. We optimized the expression conditions of CagX by using IPTG at concentrations of 1 mM. Expression was very good as shown in by arrow.
Purification of recombinant CagX
In order to partially purify the protein and eliminate the contaminating host protein, inclusion bodies were solubilized in 0.5% sarcosine containing buffer and fractionated by different concentration of ammonium sulfate (30%, 40%, 50%, and 70%). Maximum CagX recovery was observed at 30% ammonium sulfate. Ammonium sulfate precipitated protein was dialyzed overnight against buffer and mixed with pre-equlibrated S-sepharose matrix (CagX is cationic), and bound protein(s) was purified. Profile of eluted purified protein was checked by SDS-PAGE and pure CagX band was observed (). Purification of CagX protein was further confirmed by MALDI-TOF/MS (data not shown).
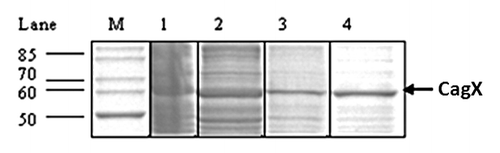
Figure 2. SDS-PAGE showing partial purification of recombinant CagX. Lane 1–4 represent 30% ammonium sulfate fraction, flow through, washing and NaCl eluted recombinant protein respectively. M indicates standard molecular size markers. Arrow indicates position of the recombinant protein. Gel was stained with Coomassie blue.
Production of polyclonal antibody against recombinant CagX in rabbit
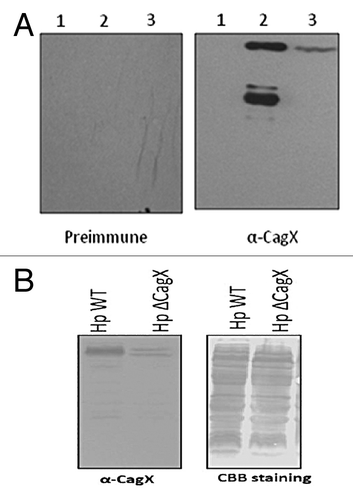
Preimmune and immune serum was tested for specificity of CagX by western blotting (), and it was specific for CagX. There is no cross reactivity with E. coli protein as a band was not seen in uninduced lane and also there was no cross reactivity with other protein of H. pylori because only one band, corresponding to CagX protein, was observed in total H. pylori cell extract. We also used H. pylori CagX null strain for further verification, and there were no bands corresponding to CagX (, right panel). We have used H. pylori WT and CagX null strain for loading control.
Figure 3. (A) western blot showing specificity of the anti-CagX antibody. Lane -1, total E. coli cell extract (uninduced), lane-2, induced E. coli extract, lane-3, total H. pylori 26695 cell extract. Blot was probed with preimmune serum (left panel) and anti-CagX serum (right panel). (B) western blot (left panel) and CBB stain (right panel) of total cell extract of H. pylori wild type and H. pylori cagX null strain to show specificity of raised CagX antisera.
CagX is cell surface exposed protein
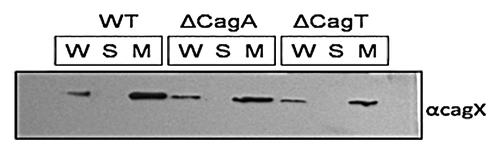
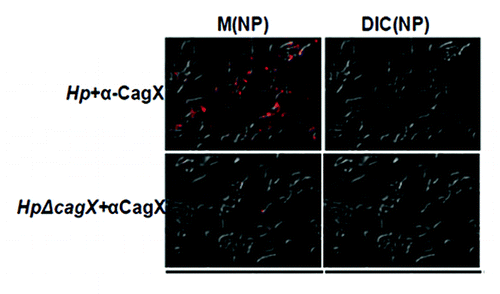
CagX is a bacterial membrane associated protein exposed partially on the cell surface. To check sub-cellular localization of CagX in wild-type (Hp26695 strain) and isogenic mutants (Hp26695ΔcagT and Hp26695ΔcagA) strains western blotting was performed with anti-CagX antibody (). CagX was detected exclusively in the membrane fraction suggesting that it is a membrane associated protein. One research group reported surface localization of both CagX and CagT by immunofluorescence microscopy.Citation15 However, another group studied the surface topology of the organism and failed to detect CagX surface localization.Citation14 We therefore, re-examined H. pylori surface for the presence of CagX by immunofluorescence microscopy. Fluorescence was observed in H. pylori wild type but not in cagX null mutant strain (), which further indicated the surface localization of CagX.
Native CagX forms high molecular mass complex
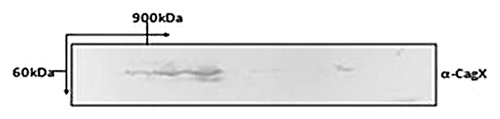
Being a VirB9 homolog of A. tumifaciens, CagX is expected to interact with other components of H. pylori TFSS and also expected to form homo-multimer. DDM (n-Dodecyl-β-D-Maltoside: a non-ionic detergent) solubilized H. pylori extract was subjected to blue-native gradient PAGE in first dimension followed by SDS-PAGE in the second dimension. Proteins separated through SDS-PAGE were transferred onto PVDF membrane and probed with anti-CagX antibody. CagX specific bands were detected in high range molecular mass (> 900 kDa) suggesting its interaction and ability of these proteins to form complex ().
CagX forms oligomer
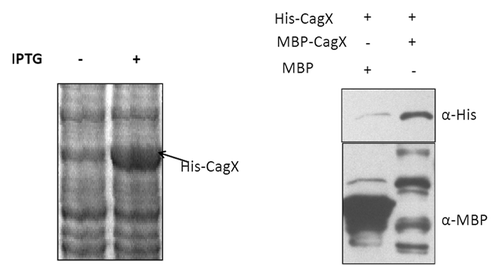
Blue native PAGE analysis indicated that native CagX forms high molecular mass complex. In order to test whether CagX could form homo-multimer, we cloned and expressed CagX with His-6 and MBP sequence tags in E. coli. To test homo-multimer formation these differentially tagged CagX were mixed and subjected to MBP-pull down analysis. It is worthwhile mentioning that while MBP-CagX was in soluble form, CagX-His-6 was not. CagX-His-6 was further solubilized in sarcosine and MBP-CagX efficiently pulled down CagX-His-6, which further indicated the CagX-CagX self-interaction or multimerization (). Some degradation of MBP-CagX complex was seen, which was unavoidable. It was not only found in this result but also observed earlier by Busler et al. (2006).Citation16
Figure 7. SDS PAGE showing expression profile of recombinant CagX protein conjugated with 6-His at C-terminal (left). Western blot (right panel) of MBP pull-down sample showing CagX oligomerization. Recombinant MBP tagged CagX and poly-histidine tagged CagX were mixed and subjected to pull-down analysis. MBP was used as negative control. Anti- MBP and Anti-His antibody was used to probe the pull down sample. MBP and histidine tagged proteins are indicated along with antibodies used.
Discussion
Presently the structure and assembly of cagPAI encoded TFSS is scarcely known in H. pylori. The characterization of full-length CagX proteins (as described in this study), would be helpful in the investigations of its architecture, role in virulence, pathogenesis, etc. During expression, most of the recombinant protein was insoluble and formed inclusion bodies. This phenomenon was not unusual because membrane protein typically comes in inclusion bodies, when expressed in a heterologous host. We solubilized the inclusion bodies in N-lauryle sarcosine which is frequently used to solubilize membrane protein. Sometimes sarcosine denatures protein, but our purpose was only to raise polyclonal antibodies without any tag. The purpose of using untagged protein was to get polyclonal antibody (anti-sera) without contaminating them with antibody against tag protein. Raised CagX polyclonal antibodies were very specific, and the titer was also very high (picks band even at 1:50 K dilution) without any cross reactivity with either H. pylori or E. coli protein (). CagX expressions were also seen in the membrane fraction of wild type as well as mutant constructs (). Rhode et al. (2003), used anti-CagX antibodies but had not mentioned the source of antibodies.Citation14 Previously reported antibodies by Tanaka et al. (2003), were not against the whole CagX protein.Citation15 They raised CagX antibody against 20 amino acid polypeptide of C-terminal. Characterization and localization of CagX in earlier reported studies was not based on antibody raised against whole CagX protein. Kutter et al. (2008), raised antibody against whole CagX,Citation17 but their titer value was very low (1:3K dilution) in comparison to ours (1:50K dilution). As per our knowledge, we raised polyclonal antibody against the whole CagX protein of H. pylori, unlike previous studies with a high titer value and more specific than any available anti-CagX antisera to date. The antibody raised against small portion/peptide that may or may not bind with other portions of protein, but an antibody raised against the whole protein will be accessible to any part of protein. Sensitivity of detection also increases using antiserum against whole protein because IgG would be available against more epitopes of protein than small peptides.Citation18 Our antibody recognizes the surface exposed CagX without any modification as observed by simple fluorescence microscopy (). We used these CagX antibodies to understand CagX interaction with other cag-PAI protein clearly. We showed high molecular weight complex of CagX using total H. pylori lysate through 2-D electrophoresis (BNP) () for the first time, which further indicated that CagX is an important component of the core complex of TFSS and providing direct evidence for CagX association with other core-complex protein of cag-PAI.
Literature indicated that CagX interact with other Cag-TFSS component as well,Citation15-Citation17 and we have also found that other proteins of cag-PAI are interacting with CagX (data not shown). Using MBP pull down assay, we observed that CagX forms an oligomer (), and the occurrence of high molecular weight may be due to the homoologomerization of CagX. CagX homolog in other system also forms oligomer.Citation19 This research communication is focused on CagX characterization only and speculates that homooligomerization is one of the reasons of the high molecular weight complex as seen in . We have also investigated the reason of this high molecular weight complex, whether it is a heteromer of CagX and other Cag-TFSS component or only CagX forms homooligomer or both. It has been shown that CagX forms heteromer with CagT and CagM,Citation15 and in a separate study, CagX directly interacted with CagY and CagZ.Citation16 So this high molecular weight complex may also comprise these proteins along with CagX oligomer. CagX, CagY, and CagT homolog form a supra molecular complex having size 1.1 megadalton in other TFSS.Citation19,Citation20 We had molecular weight markers up to 900 kDa, which was a limitation of this study. Therefore, it is difficult to assess the exact molecular weight of this complex. Our study has contributed to building more insight into complex architecture profiling of the clinically important cag-PAI encoded TFSS of H. pylori. Further studies are necessary to understand the exact role of CagX in assembly of cag-TFSS and translocation of the effector protein, CagA. In the future, if it proves conclusively that CagA translocation is dependent upon presence of CagX, then our full-length specific antibody can be used as a therapeutic tool, and purified CagX recombinant protein may further open the doors to understand TFSS structure and function in depth.
Materials and Methods
Genomic DNA isolation from H. pylori
Genomic DNA was isolated from H. pylori as described by Simor et al. (1990)Citation21 with minor modification. Briefly, H. pylori cells were scraped from BHI-agar plate’s and centrifuged. The pellet was washed first with PBS then with TE. The washed pellet was re-suspended in 160 μl/10 mg weight cells of TE (50 mM-TRIS-HCl, 5 mM EDTA) and lysozyme (500 μg/ml) for 30 min at room temperature. One percent SDS and 0.05 mg/ml of RNase-A was added and incubated again for 60 min at 37 °C. Proteinase K was added (final concentration 0.5 mg/ml) and incubated over-night at RT. Equal volume of Phenol:chloroform:isoamyl (25:24:1) was added, and centrifuged at 12K for 10 min. Aqueous layer was separated and DNA was precipitated by adding 0.1 volume of sodium acetate and 2.1 volume of ethanol. Precipitated DNA was dissolved in TE buffer.
PCR amplification of CagX
PCR was performed in 50 μl reaction volume containing 50–100 ng templates of genomic DNA, 10 pmoles each of appropriate forward and reverse primer (F528ΔN30N/R528H for without His tag cloning and F528ΔN30N/R528X pair for His tag at –C’ terminal cloning) (), 200 µM dNTPs and 1.25 unit of Pfu 1X reaction buffer (20 mM TRIS-HCl (pH 8.8), 10 mM (NH4)2SO4, 10 mM KCl, 0.1%(V/V) TritonX-100, 0.1 mg/ml BSA, 2 mM MgSO4 or 20 mM tris-HCl, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% (V/V) TritonX-100 (pH 8.8). Generally 30 cycles of following PCR steps were used for amplification of desired DNA templates. Denaturation at 94 °C for 1 min, annealing at 50–55 °C for 45 s, extension at 72 °C for appropriate time and final extension step was for 7 min.
Table 1. Primers used in this study
Gel electrophoresis, digestion, and elution
Amplified PCR products were subjected to agarose gel (1%) electrophoresis, and the desired band was cut out of the gel and purified using Qiagene gel elution kit. Purified DNA fragment was digested with the desired restriction enzymes, extracted, and finally precipitated with ethanol in presence of 0.3 M sodium acetate. Precipitated DNA was washed twice with 70% ethanol and finally dissolved in TE and stored at –20 °C for further use. Desired plasmid DNA (vector) pET28a, pMALc-2X, plasmid was digested with suitable restriction enzymes for 3 h at 37 °C and 1 h treatment with CIP (calf intestine phosphatase, NEB) to remove 5' phosphate groups from the linearized plasmid DNA. Dephosphorylated DNA fragment was subjected to agarose gel electrophoresis, and desired DNA fragment was cut out of the gel and purified using Qiagen gel elution kit according to the manufacturer’s protocol.
Cloning of CagX
Linearized vector DNA and processed amplicons were mixed in 1:3 molar ratios and ligated using “Quick ligase” ligation kit (NEB) or “Fast Link™ DNA ligation kit” (Epicenter Biotechnologies) according to the manufacturer’s protocol. Ligated product was used for transformation of DH10β or DH5α competent E. coli cells. Transformed E. coli cells were platted on antibiotic containing LB agar plate. Colonies were picked from selective plate for screening. Positive clones were selected by double digestion and PCR.
Recombinant CagX protein expression
E. coli strains DH10β/DH5α were used for propagation of all types of E. coli cloning and expression vectors, while E. coli strain BL21(DE3) was used for expression of plasmid having pET-28a vector back bone. When desired constructs were confirmed, freshly transformed E. coli colonies were inoculated into 5 ml of LB broth containing 100 μg/ml ampicillin, or 50 μg/ml kanamycine and grown overnight at 37 °C for expression of recombinant protein. This primary culture was then used to inoculate 100 ml of fresh LB broth containing the same antibiotics. The culture was allowed to grow at 37 °C till OD600 reaches 0.6. At this stage, culture was induced with 1 mM isopropyl β -D-thiogalactoside (IPTG) for 3 h. Protein expression was checked by lysis of a small aliquot of pelleted cells subjecting to SDS-PAGE. Solubility of each protein was assessed in native lysate of desired bacterial cells followed by SDS-PAGE.
Purification of recombinant CagXΔN30
Overnight culture derived from a single E. coli BL21(DE3) colony, harboring plasmid pET-528A (without tag) was used to inoculate 100 ml of LB, containing 50 μg/ml kanamycin. Culture was allowed to grow at 37 °C till OD600 reaches 0.6. Thereafter, 1 mM IPTG was added to the media to induce the expression of recombinant CagX and incubated further at 37 °C for 3 h. Cells were then collected by centrifugation and washed with 50 mM Tris-HCl, pH 8.0 containing 150 mM NaCl. It was further resuspended in lysis buffer (50 mM Tris-HCl, pH-8.0, 150 mM NaCl, 1% Triton X-100, 10 mM βME, and 1 mM PMSF), sonicated and centrifuged at 10 K for 20 min in cold. Major portion of proteins were recovered in pellet as inclusion bodies. Protein was extracted from the inclusion body with 0.5% sarcosine containing lysis buffer. The extract was subjected to ammonium sulfate precipitation at different concentration followed by centrifugation at 10 K for 20 min at 4 °C. Dialyzed ammonium sulfate fractionated protein was partially purified by ion exchanger chromatography in S-sepharose and Q-sepharose column.
SDS-PAGE
SDS-PAGE was carried with purified recombinant CagX protein according to Laemmli’s method (1970).Citation22 A wide range molecular weight marker was used to identify and assess the molecular weights of protein bands observed in the CBB stained gel.
MALDI-TOF/MS analysis
Bands of interest were excised from gels and de-stained with 50% acetonitrile (AcCN), 25 mM ammonium bicarbonate three times. The final de-stained gel was digested with trypsin and the derivatized tryptic digests of peptide were identified by MALDI-TOF/MS (Bruker) according to Kumar et al. (2010).Citation23
Production of polyclonal antibodies against CagX
The purified CagX was separated on 10% SDS-PAGE, visualized by incubating the gel in chilled 0.1 M KCl solutions and the desired band was excised. The excised gel was pulverized, mixed with Freund’s complete adjuvant, emulsified, and injected into the New Zealand white rabbit. Overall three booster doses were administered at two week intervals. Blood was drawn from the immunized rabbit two weeks after the third booster; serum was separated and stored in at –20 °C.
Western blot analysis
Western blotting was performed by transfer of protein separated in SDS-PAGE on PVDF membrane (Sigma) according to standard protocol. SDS was included in the transfer buffer to increase efficiency of high-molecular weight protein. Five percent bloto (Genotech) dissolved in TBST were used to block the membrane. Respective antiserum was used to probe the transferred protein on membrane. Primary antibody bound membrane was incubated with 1:10000 dilution of HRP-conjugated secondary antibody (Bio-Rad) and detected with enhanced chemiluminescence (ECL) method.
Fluorescence microscopy
Fluorescence microscopy (IFM) of H. pylori cells was performed as reported earlier, with some minor modifications.Citation24 One loop full of freshly grown H. pylori cells (wild type/mutants) was scraped from plate and re-suspended in BHI media, spotted on poly-L-lysin coated coverslip fixed with p-formaldehyde, and kept in humid chamber for 30 min. In the next step, the coverslip containing fixed bacteria was blocked with 5% BSA in PBS (pH 7.4) for 2 h. BSA blocked coverslip was incubated with respective antibodies (1:500) diluted in PBS containing BSA for 4 h, washed three times with PBS, incubated with cy3 conjugated anti-rabbit antiserum (1:500) in PBS containing 5% BSA for 1.5 h and finally washed three times with PBS. The coverslip was mounted and visualized under the fluorescence microscope. Cells were viewed by Carl Zeiss florescence microscope. Images were captured and analyzed with Axio-vision-4.8 software.
MBP pull down assay
MBP-tagged CagX was expressed in E. coli strain DH10β, whereas polyhistidine tagged CagX was expressed in Bl21 (DE3).Citation25 Cell pellets containing MBP-taged proteins were re-suspended in suspension buffer (50 mM Tris.HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 10% glycerol) and cells were lysed with lysozyme by sonication. The lysed cell suspension was centrifuged at 10 K for 20 min at 4 °C. The resulting supernatants were collected and used for pull-down experiment. Cell lysates containing MBP-tagged cagX and MBP alone were mixed with pre-equilibrated Amylose resin and incubated for 1 h in vertical rocker at 4 °C. Resin bound proteins were washed twice with binding buffer followed by blocking with 0.1% BSA in binding buffer. In the subsequent step, measured amount of sarcosine solubilized recombinant His-CagX was added to the respective Amylose resin bound MBP-tagged proteins/MBP, and incubated for 1 h at 4 ᴼC. Protein-bound resins were washed 3–4 times with binding buffer, and bound proteins were eluted by 20 mM maltose containing buffer and analyzed by western blotting. Anti-His and anti-MBP (NEB, USA) antibodies were used for detecting His-CagX (prey) and MBP/ MBP-tagged bait.
Blue native gradient page/SDS PAGE (2D-PAGE)
Blue Native PAGE was performed according to Schägger and von Jagow (1991).Citation26 Briefly, 2% DDM solubilized H. pylori cell extract in 20 mM Bis-Tris, 750 mM 6-aminocaproic acid (ACA), 5 mM EDTA, and 0.1 mM PMSF was mixed with 10 X BN-PAGE sample buffer (100 mM Bis-Tris.HCl pH 7.0, 750 mM 6-aminocaproic acid, 0.05% Coomassie brilliant blue G-250, and 1 mM PMSF). Samples were applied on 4–14% gradient polyacrilamide gel. Anode buffer contained 50 mM Bis-TRIS-HCl pH 7.0; cathode buffer contained 50 mM Tricine, 15 mM Bis-Tris.HCl pH7.0 and 0.02% CBB-250. Apoferitin (450 and 900 kDa), Urease (270 and 540 kDa) and BSA (66 and 132 kDa) (Sigma) were used as molecular mass standards. Proteins separated by blue native gel electrophoresis were either transferred to PVDF membrane for western blotting or gel strips, corresponding to wells, and were excised and used for separation through SDS-PAGE in the second dimension. Protein bands were probed with respective antibody, washed, treated with HRP conjugated secondary antibody, and finally detected with ECL method.
Subcellular fractionation
H. pylori cells (strain Hp 26695) were grown on BHI-serum plate, washed with PBS twice and re-suspended in 500 μl of 20 mM Tris-HCl, pH 8.0 for cell fractionation. Re-suspended cells were sonicated on ice; and centrifuged at 8000 × g for 10 min at 4 °C. This supernatant was centrifuged again for the separation of membrane and cytoplasmic protein fraction as like Kumar et al. (2010).Citation23 Each fraction was dissolved in SDS sample buffer, boiled, and subjected to SDS-PAGE, followed by western blotting using appropriate antibodies.
Generation of H. pylori null mutant strain
Natural transformation of H. pylori was performed as described by Haas et al. (1993).Citation27 Briefly, bacteria were harvested from brain–heart infusion (BHI) agar plates and suspended in BHI medium containing 10% FCS to an OD550 of 0.1. Two to three micrograms of desired plasmid DNA was added and followed by plating on selective serum plates. CagA, CagT, and CagX mutant strain was made by transforming H. pylori with plasmid pJP-95 (plasmid construct to make hp0532 null mutant), pWs-30 (plasmid construct to make hp0547 null mutant), and pJP-90 (plasmid construct for CagX mutation) respectively and screened for a mutant strain using antibody. These plasmids were the kind gift from Prof. Haas (Max-Planck-Institut für Biologie, Germany).
| Abbreviations: | ||
| C-terminal | = | carboxy terminal |
| CBB | = | coomassie brilliant blue |
| H. pylori | = | Helicobacter pylori |
| IPTG | = | isopropyl β-D-thiogalactoside |
| kDa | = | kilo-dalton |
| MALDI-TOF/MS | = | matrix assisted laser desorption ionization-time of flight/mass spectroscopy |
| mM | = | mili molar |
| OD | = | optical density |
| ORF | = | open reading frame |
| PAI | = | pathogenicity island |
| TFSS | = | type IV secretion system |
| PAGE | = | poly acrylamide gel electrophoresis |
| SDS | = | sodium dodecyl sulphate |
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
G.J.G. acknowledges the fellowships from the Council for Scientific and Industrial Research (CSIR) and University Grant Commission (UGC), New Delhi, India. Plasmid constructs were a kind gift from Prof Haas (Max-Planck-Institut für Biologie, Abteilung Infektionsbiologie, Tübingen, Germany). The authors are thankful to Jawaharlal Nehru University, New Delhi, India, M.S. University of Baroda, Vadodara, Gujarat, India, National Institute of Technology, Raipur (Chhattisgarh), India. G.J.G. is grateful to Prof. Rakesh Bhatnagar (School of Biotechnology, Jawaharlal Nehru University [JNU], New Delhi, India) for providing valuable advice, encouragement, and support.
References
- Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest 2004; 113:321 - 33; PMID: 14755326
- Goodwin CS, Worsley BW. Microbiology of Helicobacter pylori.. Gastroenterol Clin North Am 1993; 22:5 - 19; PMID: 8449570
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984; 1:1311 - 5; http://dx.doi.org/10.1016/S0140-6736(84)91816-6; PMID: 6145023
- Parsonnet J. The incidence of Helicobacter pylori infection. Aliment Pharmacol Ther 1995; 9:Suppl 2 45 - 51; PMID: 8547528
- Parsonnet J, Hansen S, Rodriguez L, Gelb AB, Warnke RA, Jellum E, Orentreich N, Vogelman JH, Friedman GD. Helicobacter pylori infection and gastric lymphoma. N Engl J Med 1994; 330:1267 - 71; http://dx.doi.org/10.1056/NEJM199405053301803; PMID: 8145781
- Banić M, Franceschi F, Babić Z, Gasbarrini A. Extragastric manifestations of Helicobacter pylori infection. Helicobacter 2012; 17:Suppl 1 49 - 55; http://dx.doi.org/10.1111/j.1523-5378.2012.00983.x; PMID: 22958156
- Thirumurthi S, Graham DY. Helicobacter pylori infection in India from a western perspective. Indian J Med Res 2012; 136:549 - 62; PMID: 23168695
- Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 2001; 345:784 - 9; http://dx.doi.org/10.1056/NEJMoa001999; PMID: 11556297
- Dhar SK, Soni RK, Das BK, Mukhopadhyay G. Molecular mechanism of action of major Helicobacter pylori virulence factors. Mol Cell Biochem 2003; 253:207 - 15; http://dx.doi.org/10.1023/A:1026051530512; PMID: 14619971
- Terry CE, McGinnis LM, Madigan KC, Cao P, Cover TL, Liechti GW, Peek RM Jr., Forsyth MH. Genomic Comparison of cag pathogenicity island (PAI)-positive and -negative Helicobacter pylori strains: identification of novel markers for cag PAI-positive strains. Infect Immun 2005; 73:3794 - 8; http://dx.doi.org/10.1128/IAI.73.6.3794-3798.2005; PMID: 15908415
- Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A 1996; 93:14648 - 53; http://dx.doi.org/10.1073/pnas.93.25.14648; PMID: 8962108
- Fischer W, Püls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol 2001; 42:1337 - 48; http://dx.doi.org/10.1046/j.1365-2958.2001.02714.x; PMID: 11886563
- Christie PJ. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol Microbiol 2001; 40:294 - 305; http://dx.doi.org/10.1046/j.1365-2958.2001.02302.x; PMID: 11309113
- Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol 2003; 49:219 - 34; http://dx.doi.org/10.1046/j.1365-2958.2003.03549.x; PMID: 12823823
- Tanaka J, Suzuki T, Mimuro H, Sasakawa C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol 2003; 5:395 - 404; http://dx.doi.org/10.1046/j.1462-5822.2003.00286.x; PMID: 12780777
- Busler VJ, Torres VJ, McClain MS, Tirado O, Friedman DB, Cover TL. Protein-protein interactions among Helicobacter pylori cag proteins. J Bacteriol 2006; 188:4787 - 800; http://dx.doi.org/10.1128/JB.00066-06; PMID: 16788188
- Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol 2008; 190:2161 - 71; http://dx.doi.org/10.1128/JB.01341-07; PMID: 18178731
- Dyson HJ, Lerner RA, Wright PE. The physical basis for induction of protein-reactive antipeptide antibodies. Annu Rev Biophys Biophys Chem 1988; 17:305 - 24; http://dx.doi.org/10.1146/annurev.bb.17.060188.001513; PMID: 2456075
- Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature 2009; 462:1011 - 5; http://dx.doi.org/10.1038/nature08588; PMID: 19946264
- Fronzes R, Schäfer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science 2009; 323:266 - 8; http://dx.doi.org/10.1126/science.1166101; PMID: 19131631
- Simor AE, Shames B, Drumm B, Sherman P, Low DE, Penner JL. Typing of Campylobacter pylori by bacterial DNA restriction endonuclease analysis and determination of plasmid profile. J Clin Microbiol 1990; 28:83 - 6; PMID: 2153701
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680 - 5; http://dx.doi.org/10.1038/227680a0; PMID: 5432063
- Kumar A, Sisodia B, Misra P, Sundar S, Shasany AK, Dube A. Proteome mapping of overexpressed membrane-enriched and cytosolic proteins in sodium antimony gluconate (SAG) resistant clinical isolate of Leishmania donovani.. Br J Clin Pharmacol 2010; 70:609 - 17; http://dx.doi.org/10.1111/j.1365-2125.2010.03716.x; PMID: 20840452
- Cendron L, Couturier M, Angelini A, Barison N, Stein M, Zanotti G. The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion. J Mol Biol 2009; 386:204 - 17; http://dx.doi.org/10.1016/j.jmb.2008.12.018; PMID: 19109970
- Tolia NH, Joshua-Tor L. Strategies for protein coexpression in Escherichia coli.. Nat Methods 2006; 3:55 - 64; http://dx.doi.org/10.1038/nmeth0106-55; PMID: 16369554
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 1991; 199:223 - 31; http://dx.doi.org/10.1016/0003-2697(91)90094-A; PMID: 1812789
- Haas R, Meyer TF, van Putten JP. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol Microbiol 1993; 8:753 - 60; http://dx.doi.org/10.1111/j.1365-2958.1993.tb01618.x; PMID: 8332066