Abstract
The Eph receptor tyrosine kinase family includes many members, which are often expressed together in various combinations and can promiscuously interact with multiple ephrin ligands, generating intricate networks of intracellular signals that control physiological and pathological processes. Knowing the entire repertoire of Eph receptors and ephrins expressed in a biological sample is important when studying their biological roles. Moreover, given the correlation between Eph receptor/ephrin expression and cancer pathogenesis, their expression patterns could serve important diagnostic and prognostic purposes. However, profiling Eph receptor and ephrin expression has been challenging. Here we describe a novel and straightforward approach to catalog the Eph receptors present in cultured cells and tissues. By measuring the binding of ephrin Fc fusion proteins to Eph receptors in ELISA and pull-down assays, we determined that a mixture of four ephrins is suitable for isolating both EphA and EphB receptors in a single pull-down. We then used mass spectrometry to identify the Eph receptors present in the pull-downs and estimate their relative levels. This approach was validated in cultured human cancer cell lines, human tumor xenograft tissue grown in mice, and mouse brain tissue. The new mass spectrometry approach we have developed represents a useful tool for the identification of the spectrum of Eph receptors present in a biological sample and could also be extended to profiling ephrin expression.
Keywords: :
Introduction
The Eph family of receptor tyrosine kinases regulates many physiological functions, such as nervous system development and neural plasticity, angiogenesis, insulin secretion in the pancreas, bone maintenance, intestinal homeostasis and immune function.Citation1 In addition, the Eph receptors have been implicated in numerous pathologies, including cancer progression, tumor angiogenesis, neurological disorders and inhibition of nerve regeneration after injury.Citation1,Citation2
The Eph receptors interact with membrane-bound ligands, the ephrins, at sites of cell-cell contact.Citation3 This causes clustering of Eph receptor-ephrin complexes, triggering “forward” signaling pathways that depend on Eph receptor kinase activity and “reverse” signaling pathways that propagate in the ephrin-expressing cells. The Eph family comprises 14 receptors in mammals, which are subdivided into two classes. The nine EphA receptors preferentially bind to five glycosylphosphatidylinositol-linked ephrin-A ligands, while the five EphB receptors preferentially bind to the three transmembrane ephrin-B ligands. Interactions within each class are promiscuous and, in addition, some interclass interactions can occur.Citation4
Multiple Eph receptors are often co-expressed in the same cells, where they can be activated by the same ephrins and likely function in concert.Citation5-Citation7 The difficulties in identifying the full repertoire of Eph receptors expressed in cells or tissues of interest has hindered progress in characterizing the signaling mechanisms and activities of individual Eph receptors in normal physiology and disease.Citation1,Citation2 Eph receptor and ephrin expression is also disregulated during cancer progression, and in many cases knowing the expression levels of specific Eph receptors and ephrins could be useful for diagnostic and prognostic purposes.Citation2,Citation8-Citation10 Profiling Eph receptor and ephrin expression in tumors may also be helpful in the future to design targeted and personalized therapies as well as to assess their effectiveness.
Comprehensive characterization of Eph receptor and ephrin expression has been challenging. Many different techniques can be used to analyze Eph receptor and ephrin mRNA expression in cells and tissues, including quantitative PCR (qPCR), northern blotting and in situ hybridization.Citation7,Citation11-Citation18 However, these approaches are time consuming, use a different probe for each family member and only assess mRNA levels, which do not always correlate with protein levels. Immunoblot analysis and immunohistochemistry can be used to detect Eph receptor and ephrin protein expression, but they are limited by the availability and quality of validated antibodies specifically recognizing each member of the Eph receptor or ephrin families.Citation2 In addition, detection with antibodies does not provide reliable information on the relative expression of different family members. Given the promiscuity of the interactions between Eph receptors and ephrins belonging to the same class, ephrins (or Eph receptors) fused to the Fc portion of human IgG1 or to alkaline phosphatase (AP) can be used as probes to reveal the combined expression of Eph receptors (or ephrins) of the A or B class.Citation19,Citation20 However, this approach cannot identify the specific Eph receptors or ephrins expressed.
Mass spectrometry is routinely used for the identification of specific proteins from complex mixtures. We took advantage of the ephrin binding promiscuities to develop an approach where binding to ephrin Fc fusion proteins immobilized on beads is used to isolate the repertoire of Eph receptors expressed in a biological sample. Analysis by mass spectrometry then identifies the individual Eph receptors and estimates their relative levels. This targeted mass spectrometry approach represents a straightforward method to catalog the Eph receptors expressed in cultured cells and tissues. By performing pull-downs with Eph receptor Fc fusion proteins, the approach could likely be extended to identify the repertoire of expressed ephrins.
Results and Discussion
Binding of ephrin Fc proteins to Eph receptors
Ephrins fused to the Fc portion of human IgG1 are commercially available and can be immobilized on protein A or protein G sepharose beads to pull down Eph receptors from cell or tissue lysates.Citation21 Although binding interactions between Eph receptors and ephrins are well known to be promiscuous,Citation4,Citation22 there is some selectivity in the interactions between not only Eph receptors and ephrins of different classes, but also of the same class. Therefore, to define the combination of ephrins that would be most suitable for the comprehensive isolation of the spectrum of Eph receptors expressed in a biological sample, we measured the binding of biotinylated ephrin Fc fusion proteins to Eph receptor Fc fusion proteins immobilized on ELISA plates. We examined all mammalian ephrins and Eph receptors, except for the poorly characterized EphA10 receptor. It should be noted that Fc fusion proteins are dimeric, which increases their binding avidity.Citation23,Citation24 Nevertheless, the apparent KD values obtained with this method can be used to compare the binding strengths of different Eph receptor-ephrin combinations.
We found that ephrin-A4 Fc and ephrin-A5 Fc bind with high apparent affinity to most EphA receptors with the exception of EphA1, to which ephrin-A5 binds poorly in agreement with previous reports ().Citation19,Citation25 However, ephrin-A1 Fc binds better than ephrin-A4 Fc and ephrin-A5 Fc to EphA2. Ephrin-A2 Fc and ephrin-A3 Fc interact more weakly with most EphA receptors. However, ephrin-A3 can also bind to heparan sulfate proteoglycans, which enhances the ability of this ephrin to interact with EphA receptors in cells.Citation26
Figure 1. Binding interactions of ephrin Fc fusion proteins with Eph receptors. Apparent dissociation constant (KD) values were obtained from curves measuring the binding of biotinylated ephrin Fc proteins to Eph receptor Fc proteins immobilized on ELISA plates. h, human; m, mouse; r, rat; nd, not determined.
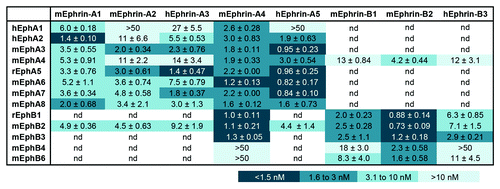
Because ephrin-A5 has also been reported to bind to EphB2,Citation21 we examined the ability of the other ephrin-A Fc proteins to bind to EphB2. This not only confirmed the interaction of ephrin-A5 with EphB2, but also revealed an interaction of all other ephrin-A Fc proteins with this receptor. Moreover, we unexpectedly found that ephrin-A4 Fc binds to EphB2 as well as the ephrin-B Fc proteins. This is in contrast with previous studies where different approaches were used, which did not reveal a high binding affinity of ephrin-A4 for EphB2.Citation21 Further experiments will be needed to resolve this discrepancy. In addition, we found that ephrin-A4 Fc also binds with high apparent affinity to EphB1 and EphB3, whereas its binding to EphB4 and EphB6 is undetectable (). This suggests that ephrin-A4 is the most promiscuous of the ephrins and efficiently binds both EphA and EphB receptors. It will be interesting to characterize the structural basis and physiological significance for the binding of ephrin-A4 to EphB receptors, and whether ephrin-A4 may function as an agonist or an antagonist for these receptors.
Of the three ephrin-Bs, ephrin-B2 Fc binds best to all EphB receptors as well as EphA4 (), which can bind both A and B class ephrins.Citation4,Citation19,Citation27 Ephrin-B3 Fc binds most weakly to all EphB receptors but, like ephrin-A3, this ephrin-B can bind heparan sulfate proteoglycans.Citation28
Identification of Eph receptors expressed in cancer cell lines
We next performed pull-down experiments with ephrin Fc proteins and analyzed them by immunoblotting and 1 dimensional LC/MS/MS mass spectrometry. We initially used PC3 prostate cancer cells, which have been reported to express mainly EphA2 among the EphA receptors.Citation6,Citation29,Citation30 By immunoblotting we found that EphA2 is pulled down by ephrin-A1 Fc and also, although less efficiently, by ephrin-A4 Fc and ephrin-A5 Fc (), consistent with the results of the ELISA assays (). We also readily detected EphB4 in pull-downs with ephrin-B2 Fc, in agreement with the reported substantial expression of this receptor in PC3 cells.Citation6,Citation30,Citation31
Figure 2. Identification of Eph receptors expressed in PC3 cells. (A) EphA2 and EphB4 were pulled down from PC3 prostate cancer cells by using the indicated ephrin Fc proteins and detected by immunoblot analysis. (B) Schematic representation of the EphA2 peptide coverage obtained from the 1 dimensional LC/MS/MS analysis of EphA2 pulled down with ephrin-A1 Fc in experiment #1 in (C). The extracellular, transmembrane (tm) and cytoplasmic regions of EphA2 are indicated. The peptides are represented as black lines with thickness proportional to the number of peptides with the same sequence identified in the sample (spectral counts). (C) The Eph receptors expressed in PC3 cells were pulled down with the indicated ephrin Fc proteins and identified by 1 dimensional LC/MS/MS analysis. The histograms show the spectral counts obtained for each Eph receptor, with the dark bottom portion of the bars representing the spectral counts that were assigned only to the indicated Eph receptor and the light top portion of the bars representing the spectral counts that could also correspond to other Eph receptors identified in the same experiment. The percentage of sequence coverage by all the peptides identified for each receptor is indicated above the bars. Three independent experiments are shown for ephrin-A1; the ephrin-A1 #3, ephrin-A5 and ephrin-B2 pull-downs were analyzed in parallel in the same experiment.
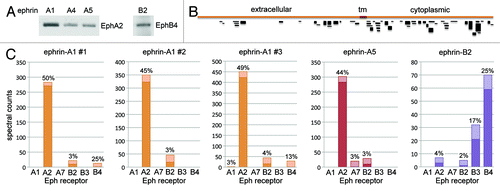
For mass spectrometry analysis, we separated the proteins associated with ephrin-A1 Fc, ephrin-A5 Fc or ephrin-B2 Fc by SDS-PAGE and used the 80–180 kDa region of the gel for trypsin digestion and LC/MS/MS mass spectrometry analysis. This enabled removal of the ephrin Fc proteins used for pull-down, which would interfere with the identification of the less abundant associated Eph receptors. Comparison of the spectra obtained with theoretical spectra from the International Protein Index human sequence database enabled identification of the tryptic peptides from the Eph receptors associated with each ephrin. In a first experiment with ephrin-A1 Fc, we obtained 283 spectra covering 50% of the EphA2 sequence (, ephrin-A1 #1). The majority of the peptides identified are from the EphA2 intracellular region (), perhaps because of the higher density of arginine and lysine residues in this portion of EphA2. Glycosylation in the extracellular region may also affect the efficiency of trypsin digestion and/or the identification of the peptides in the mass spectrometer. No other EphA receptor was identified, although 12 of the spectra are not unique for EphA2 (top, lighter portion of the bar in , ephrin-A1 #1). Some ambiguity in peptide assignments would be expected, given the conservation in the sequences of different Eph receptors. We also obtained 10 unique EphB2 spectra, in agreement with the ability of ephrin-A1 Fc to bind EphB2 (, ephrin-A1 #1) and possibly also due to an association of EphB2 with EphA2.Citation32 These results were highly reproducible in two subsequent independent experiments (, ephrin-A1 #2 and #3). The third ephrin-A1 Fc pull-down achieved a slightly higher sensitivity, with 423 unique EphA2 spectra and 16 unique EphB2 spectra, and also yielded 2 unique EphA1 spectra and 1 unique EphB4 spectrum. The ephrin-A5 Fc pull-down also produced similar results, identifying 283 unique EphA2 spectra, 10 unique EphB2 spectra and one unique EphA7 spectrum ().
A different pattern of Eph receptors was identified with ephrin-B2 Fc, with a predominance of EphB4 and EphB3 spectra, one unique EphB2 spectrum and three unique EphA2 spectra. Surprisingly, ephrin-B2 Fc pulls down EphB2 from PC3 lysates slightly less efficiently compared with ephrin-A1 Fc and ephrin-A5 Fc, which however show weaker EphB2 binding in ELISA assays (). Whether this may be due to an association of EphB2 with the large amounts of EphA2 present in the ephrin-A pull-downs remains to be determined. Importantly, parallel pull-downs using equimolar amounts of human Fc did not yield any Eph receptor-derived peptides in any of the experiments (not shown). Because spectral counts give information about the relative abundance of the Eph receptors in a sample,Citation33 these data confirm that EphA2 is by far the most abundant EphA receptor expressed in PC3 cells. They also indicate that EphB3 and EphB4 are the most abundant B class receptors in these cells.
Immunoblot analyses of pull-downs from the H460 human lung cancer cell line revealed that ephrin-A1 Fc preferentially pulls down EphA2 while other ephrin-A Fc proteins pull down several EphA receptors, including EphA2, EphA3, EphA4 and EphA5, with preference for different receptors (). This is in good agreement with the results of the ELISA binding assays (). A mixture of all five ephrins pulled down more even amounts of the different EphA receptors. These results, combined with the ELISA binding assays, suggest that a mixture of ephrin-A1 Fc, ephrin-A4 Fc and ephrin-A5 Fc would be a good combination of ephrins to pull down all the EphA receptors. Indeed, a mixture of these ephrins yielded a pattern of EphA receptors similar to the mixture of all the ephrin-As (). Interestingly, substantial levels of tyrosine phosphorylation were detected in the pull-downs with ephrin-A3 Fc, ephrin-A4 Fc, ephrin-A5 Fc and the two ephrin-A Fc mixtures (). This suggests that some of the EphA receptors expressed in the H460 cells are activated by endogenous ephrin ligands. If this is the case, then the ephrin-A Fc proteins incubated with cell lysates displace endogenous ephrins bound to the Eph receptors to enable their pull down.
Figure 3. Identification of Eph receptors expressed in the H460 and A549 lung cancer cell lines. (A) The Eph receptors expressed in H460 lung cancer cells were pulled down by using the indicated ephrin Fc fusion proteins, or mixtures of fusion proteins, and detected by immunoblot analysis with the indicated antibodies. (B and C) H460 or A549 cell lysates were subjected to pull-downs with a mixture of ephrin-A1 Fc, ephrin-A4 Fc, ephrin-A5 Fc and ephrin-B2 Fc and the associated Eph receptors were identified by 1-dimensional LC/MS/MS analysis. The histograms show the spectral counts obtained for each Eph receptor, with the dark bottom portion of the bars representing the spectral counts that were assigned only to the indicated Eph receptor. The percentage of sequence coverage for each receptor is indicated above the bars.
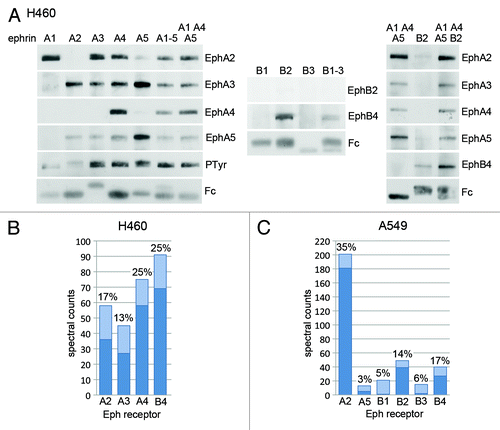
Pull-downs performed using ephrin-B2 Fc, but not ephrin-B1 Fc or ephrin-B3 Fc, identified EphB4 in H460 cells (), consistent with the ELISA results and the known preference of EphB4 for ephrin-B2.Citation34 Ephrin-B2 Fc also pulled down EphB4 more effectively than the mixture of all three ephrin-B Fc proteins, where the amount of ephrin-B2 Fc is less. In contrast, EphB2 was not detected either in the immunoblots or by mass spectrometry. A combination of 1/6 ephrin-A1 Fc, 1/6 ephrin-A4 Fc, 1/6 ephrin-A5 Fc and 1/2 ephrin-B2 Fc yielded the EphA receptors as well as EphB4 in a single pull-down (). Mass spectrometry analysis of a pull-down obtained from H460 cells with the mixture of the four ephrin Fc proteins identified approximately similar levels of EphA2, EphA3, EphA4 and EphB4 (). Although EphA5 was detected by immunoblotting, it was not identified by the proteomics data analysis software (which indicates that less than two unique peptides corresponding to this receptor were identified by the search algorithm). However, 40 of 214 total spectral counts represent conserved peptides that could be derived from EphA5 (besides one or more of the other Eph receptors identified). Thus, more comprehensive sequence coverage revealing unique peptides may be required to identify EphA5. The discrepancy could also reflect a higher sensitivity of the EphA5 antibody used as compared with mass spectrometry. Mass spectrometry analysis of the A549 human lung cancer cell line revealed high levels of EphA2 and much lower levels of EphB2 and EphB4, with only 1–5 unique peptides identified for EphA5, EphB1 and EphB3 (). Although EphB3 has been detected by immunoblotting and qPCR in both H460 and A549 cell lines (),Citation35 mass spectrometry analysis did not identify this Eph receptor in H460 cells (although 28 of the 214 spectral counts could be derived from EphB3), while in A549 cells it only identified 2 unique EphB3 peptides and 13 peptides that could be derived from EphB3 but also other Eph receptors. This suggests low levels of EphB3 protein expression in the two lung cancer cell lines under our experimental conditions.
Identification of Eph receptors expressed in PC3M prostate cancer xenograft tissue
To determine whether the combination of pull-down and mass spectrometry analysis can also be used to identify the repertoire of Eph receptors expressed in tissue samples, we used PC3M-luc-C6 xenografts grown in nude mice. Pull-downs with the mixture of four ephrin Fc proteins were performed using xenograft tissue lysed in Triton X-100-containing buffer (which is the same buffer that was used to solubilize the cultured cells) or in modified RIPA buffer (which may more effectively solubilize Eph receptors from tissue samples because it contains not only Triton X-100 but also deoxycholate and SDS as detergents). Immunoblot analysis of the pull-downs shows that similar levels of EphA2 and EphB4 were isolated from tissue solubilized with the two lysis buffers (). Mass spectrometry analysis using xenograft tissue lysed in modified RIPA buffer revealed the presence of a number of Eph receptors, including higher levels of EphA2, EphB2 and EphB4 and lower levels of EphA3, EphA5 and EphA7 (). These results are somewhat different from those obtained with cultured PC3 cells, from which PC3M-luc-C6 cells are derived.Citation36 Mass spectrometry analysis did not identify EphA3 and EphA5 in cultured PC3 cells, and EphB2 appears to be present at much lower levels than EphB4 in the PC3 cells () compared with PC3M-luc-C6 tumor xenograft tissue ().
Figure 4. Identification of Eph receptors expressed in PC3M-luc-C6 tumor xenografts. (A) PC3M xenograft tissue lysed in Triton X-100-containing buffer or modified RIPA buffer was used for pull-downs with a mixture of ephrin-A1 Fc, ephrin-A4 Fc, ephrin-A5 Fc and ephrin-B2 Fc or Fc as a control (indicated by C). Immunoblot analysis reveals similar levels of EphA2 and EphB4 pulled down from the two tumor samples. The two top arrows in the panel showing proteins stained with amido black indicate the ephrin Fc fusion proteins, while the bottom arrow indicates the Fc protein. (B) PC3M-luc-C6 xenografts lysed in RIPA buffer were subjected to pull-down with a mixture of ephrin-A1 Fc, ephrin-A4 Fc, ephrin-A5 Fc (10 μg each) and ephrin-B2 Fc (30 μg each) and the associated Eph receptors were identified by 1 dimensional LC/MS/MS analysis. The histogram shows the spectral counts obtained for each Eph receptor, with the dark bottom portion of the bars representing the spectral counts that were assigned only to the indicated Eph receptor. The percentage of sequence coverage for each receptor is indicated above the bars. (C) Comparison of Eph receptors expression in cultured PC3 and PC3M-luc-C6 cells and in PC3M-luc-C6 tumor xenografts. The PC3M-luc-C6 cells and tumor xenograft lysates were analyzed in duplicate. All samples were lysed in RIPA buffer. (D and E) PC3M-luc-C6 xenografts lysed in RIPA buffer were subjected to pull-down with a mixture of ephrin-A1 Fc, ephrin-A4 Fc, ephrin-A5 Fc (3 μg each) and the associated Eph receptors were identified by 1 dimensional LC/MS/MS analysis as in (B). The histogram in (D) shows the spectral counts obtained for each Eph receptor, as described in (B). The histogram in E shows the spectral counts for EphA2 (for which 13 unique human spectral counts and four unique mouse spectral counts were obtained), EphA3 (for which only spectral counts that could be derived from either the human or mouse receptor were obtained) and EphA5 (for which one unique human spectral count and two unique mouse spectral counts were obtained). The bottom portion of the bars represents the spectral counts that correspond only to the human sequence (green) or mouse sequence (orange) and the top portion of the bars represents the spectral counts that could correspond to either the mouse or the human sequence. The percentage of sequence coverage for each receptor is indicated above the bars. h, human; m, mouse.
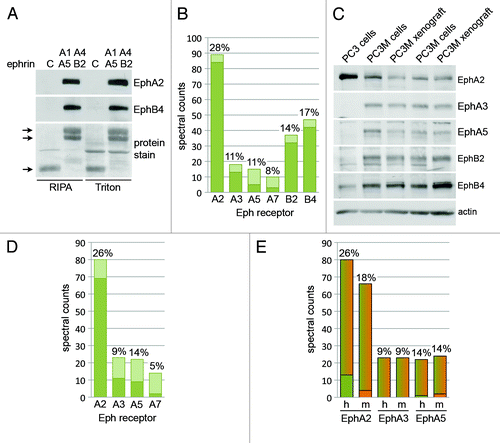
By immunoblotting we confirmed that cultured PC3M-luc-C6 cells express higher levels of EphA3, EphA5 and EphB2 than PC3 cells (). EphB4 levels are also higher in PC3M-luc-C6 than PC3 cells, as previously reported for PC3M cells,Citation31 while EphA2 levels are lower. On the other hand, expression of the five Eph receptors is similar in PC3M-luc-C6 cultured cells and tumor xenografts, suggesting that 2-dimensional growth in tissue culture does not substantially alter the pattern of Eph receptor expression in these cells compared with 3-dimensional growth in vivo. A second mass spectrometry experiment performed using a different tumor sample and a mixture of ephrin-A1 Fc, ephrin-A4 Fc and ephrin-A5 Fc to pull down the EphA receptors closely reproduced these results (). The spectral counts in this second experiment were also comparable to those in the first experiment even though approximately one-third as much ephrin Fc proteins were used for the pull-down, indicating that lower amounts of the ephrins can be used without compromising the results.
A search of the mouse protein database using the mass spectrometry data from the experiment shown in did not identify any peptides unique to mouse Eph receptor sequences, although it identified a number of peptides that are common to the mouse and human Eph receptor sequences and some that are unique to the human sequences (not shown). This is likely due to the low percentage of stromal cells present in the tumor sample and to the high conservation in amino acid sequences between human and mouse Eph receptor orthologs (for example, the amino acid identity between mouse and human receptors is 92% for EphA2, 99% for EphB2 and 93% for EphB4). Although the majority of peptides identified in the second experiment could also be derived from the human or mouse Eph receptor sequences, some peptides unique to the mouse sequences were identified for EphA2 and EphA5 (), suggesting that these receptors are present in the stromal compartment of the tumors. Furthermore, comparing the number of mouse and human unique peptides suggests that a substantial amount of the shared spectral counts could be derived from the mouse Eph receptors. In contrast, because all the EphA3 peptides identified could be derived from either the human or the mouse sequence, their origin cannot be unequivocally determined. Increasing the sensitivity of the mass spectrometry approach to obtain better sequence coverage or using sorted stromal cells could help overcome this limitation.
Identification of Eph receptors that bind ephrin-A3 in the mouse hippocampus
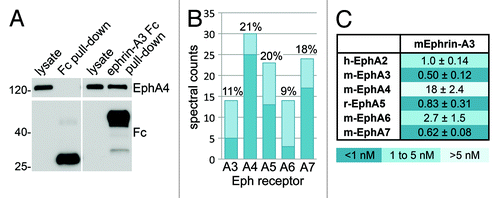
The interaction between ephrin-A3 and EphA4 in the adult mouse hippocampus has an important role in the regulation of dendritic spine morphology, glutamate uptake by astrocytes and synaptic plasticity.Citation12,Citation37,Citation38 However, other EphA receptors have also been reported to be expressed in this brain regionCitation39-Citation42 and could represent additional ephrin-A3 signaling partners. Immunoblot analysis confirmed the presence of EphA4 in pull-downs performed with mouse ephrin-A3 Fc but not Fc (). The same samples were analyzed by mass spectrometry, which led to the identification of EphA4 as the most abundant receptor, followed by EphA5 and EphA7 and lower amounts of EphA3 and EphA6 (). Because the ELISA binding assays using the commercial human ephrin-A3 Fc—which lacks the most C-terminal region—show weakest binding to EphA4, we also generated a mouse ephrin-A3 AP fusion protein containing the entire sequence except for the signal for glycosylphosphatidylinositol linkage. This is the same region contained in the mouse ephrin-A3 Fc construct. The ephrin-A3 AP was used in ELISA assays to determine the apparent binding affinity of this ligand for the EphA receptors known to be expressed in the hippocampus and, as a comparison, for EphA2 (). The results show that ephrin-A3 AP binds best to EphA3, EphA5 and EphA7 followed by EphA2 and EphA6. Overall, the relative binding strength of mouse ephrin-A3 AP for the different EphA receptors is consistent with that of biotinylated human ephrin-A3 Fc (). The discrepancy in the apparent KD values measured with ephrin-A3 AP and Fc proteins, both of which are dimeric, may be due to the inaccuracy in determining the concentration of the AP fusion protein based on AP activity. In any case, the data show that despite the physiological relevance of the EphA4-ephrin-A3 interaction and the predominance of EphA4 in the pull-downs, ephrin-A3 binds most weakly to EphA4 than other Eph receptors. Likely the high expression of EphA4 in the hippocampusCitation12,Citation43 compensates for the lower binding affinity. These findings also suggest that besides EphA4, other EphA receptors could be activated by ephrin-A3 as well as likely contribute to stimulating ephrin-A3 reverse signaling in the adult mouse hippocampus.
Figure 5. Identification of Eph receptors that bind ephrin-A3 in mouse hippocampus. (A) Ephrin-A3 Fc or Fc as a control were used for pull-downs from adult mouse hippocampus, which were probed by immunoblotting with anti-EphA4 and anti-Fc antibodies. (B) A mouse hippocampal lysate was subjected to pull-down with ephrin-A3 Fc and the associated Eph receptors were identified by 1 dimensional LC/MS/MS analysis. The histogram shows the spectral counts obtained for each Eph receptor, with the dark bottom portion of the bars representing the spectral counts that were assigned only to the indicated Eph receptor. The percentage of sequence coverage for each receptor is indicated above the bars. (C) Apparent dissociation constant (KD) values obtained from curves measuring ephrin-A3 AP binding to Eph receptor Fc proteins immobilized on ELISA plates; the value for EphA4 is approximate since the ephrin-A3 AP concentration was insufficient to reach maximal binding.
Conclusions and Perspectives
We have developed a straightforward mass spectrometry-based method that can be used for the identification of the Eph receptors expressed in culture cells and tissue samples. By using a mixture of ephrin Fc proteins to pull down Eph receptors, this approach can simultaneously profile the entire repertoire of Eph receptors expressed in a sample, yielding information that could be useful for research and diagnostic purposes. Analysis of the simpler mixture of pulled down proteins rather than the whole proteome increases sensitivity and facilitates bioinformatic analysis. In addition, the spectrum of Eph receptors that interact with a specific ephrin can be identified in a cell line or tissue of interest. The approach is most suitable to analyze samples that express substantial levels of Eph receptors and are available in adequate quantities, while it may not be suitable for primary cells or tissues available in very limited quantities. For detection of Eph receptors in such samples more sensitive methods, such as antibody-based methods, remain preferable provided that validated antibodies are available. However, antibody detection will not provide information on the relative abundance of the receptors. Further refinements of the approach, such as developing ways to analyze the pulled down Eph receptors without performing gel separation, may lead to increased sensitivity and therefore reduce the amount of sample needed.
This new approach could also be modified to identify Eph receptor post-translational modifications, such as tyrosine and serine/threonine phosphorylation sites, proteins that interact with the pulled down Eph receptors, and ephrin binding partners other than Eph receptors. Another useful application would be the identification of ephrin ligands through pull-downs performed with Eph receptor Fc proteins. However, this may prove more challenging than the identification of Eph receptors, due to the smaller size of the ephrins and thus the smaller number of peptides generated by protease digestion. In addition, the peptides identified for the Eph receptors are preferentially derived from the cytoplasmic region, which is absent or much smaller in the ephrin ligands. Perhaps the use of other proteases besides trypsin or removal of sugar groups before protease digestion would allow more efficient identification of peptides from the extracellular regions. Because the extracellular sequences are more divergent, this may also enable better distinction of human and mouse sequences, which would be useful in discriminating Eph receptors and ephrins expressed in tumor cells vs. the stromal compartment of tumor xenografts.
A possible limitation of the pull-down approach is that receptors that bind best to multiple ephrins may be preferentially enriched. We have not identified EphA8, EphA10 and EphB6 in the limited set of samples examined and we identified only low levels of other Eph receptors. This may reflect lower expression levels compared with other Eph receptors in the samples. Alternatively, it may be the consequence of a weaker ability of some of these receptors to bind the ephrins used for the pull-downs. As a consequence, relative expression levels deduced by pull-down/mass spectrometry analysis may not be completely accurate, but would nevertheless provide an indication of Eph receptor abundance. In addition, the method allows comparison of the abundance of the same Eph receptor in different samples. In summary, the advantage of our approach, albeit not perfect, is that it can provide a comprehensive comparison of the relative Eph receptor protein levels in a straightforward manner and represents a new tool, complementary to others available, to characterize Eph receptor expression.
Materials and Methods
Ephrin-A3 Fc and AP constructs
To generate the ephrin-A3 Fc expression vector, the cDNA sequence encoding amino acids 32 to 213 of mouse ephrin-A3 (GeneBank accession number NM_010108) was cloned between the NheI and BamHI sites of a pcDNA3 vector containing the CD5 signal peptide sequence upstream of the NheI site and the Fc portion of human IgG1 downstream of the BamHI site, as done previously to generate Fc fusion proteins of other ephrins.Citation44,Citation45 Amino acids 32–213 represent the entire extracellular domain of mouse ephrin-A3 after cleavage of the signal peptide, except for the sequence for glycosylphophatidylinositol anchor attachment. To generate the pAPtag-2-ephrin-A3 vector, the sequence encoding the CD5 signal peptide and amino acids 32–213 of ephrin-A3 were subcloned into the pAPtag-2 vector (GenHunter).Citation46
ELISA assays
To obtain curves for the binding of ephrins to Eph receptors, Eph receptor Fc fusion proteins (R&D Systems) were immobilized on high binding capacity 96-well polystyrene plates (Corning Glass, #3690) by overnight incubation at approximately 4 μg/ml (see next paragraph) in phosphate buffered saline (PBS) at 4°C. The wells were blocked for 1 h with 5 mg/ml bovine serum albumin (Sigma-Aldrich, #7888) in PBS at room temperature. After washing, the wells were incubated for 2 h with different concentrations of biotinylated ephrin Fc proteins (R&D Systems) in TBST (150 mM NaCl, 50 mM TRIS-HCl, pH 7.5 with 0.01% Tween 20) at room temperature, washed, and then incubated for 30 min with 0.5 μg/ml streptavidin conjugated to horseradish peroxidase (HRP) (Thermo Fisher Scientific, #21127) in TBST. Bound HRP was quantified by adding 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS, Sigma-Aldrich, #A1888) in citric acid as a substrate and reading the absorbance at 405 nm. Absorbance in wells where no ephrins were added was subtracted as background.
The relative amounts of the Eph receptor Fc fusion proteins and biotinylated ephrin Fc fusion proteins to be used in the binding assays described above were verified by coating the proteins on high binding capacity wells as described above. After blocking with bovine serum albumin, the wells were incubated with 1 μg/ml anti-human Fc antibody coupled to alkaline phosphatase (Promega, #S382B) in TBST. One milligram per milliliter p-nitrophenyl phosphate (PNPP, Thermo Fisher Scientific, #34045) in SEAP buffer (105 mM diethanolamine, 0.5 mM MgCl2, pH 9.8) was then added as the substrate, and the absorbance at 405 nm was measured.
Ephrin-A3 AP binding to EphA receptors was measured using cell culture medium from HEK293 cells transiently transfected with the pAPtag-2-ephrin-A3 vector. Medium was collected after 3–4 d and concentrated using an Amicon Ultra centrifugal unit (Millipore, #UFC903024). Ephrin-A3 AP concentration in the medium was calculated from the AP activity.Citation47 EphA receptor Fc fusion proteins were immobilized on protein A-coated 96-well plates (Thermo Fisher Scientific, #15132) at 1 μg/ml and for 1 h at room temperature. Concentrated culture supernatant containing ephrin-A3 AP was diluted in TBST and incubated with the EphA receptors at room temperature for 1 h. After washing away unbound ephrin-A3 AP, 1 mg/ml PNPP in SEAP buffer was added as the substrate and the absorbance at 405 nm was measured. All binding curves were analyzed using nonlinear regression and the program Prism (GraphPad Software Inc.).
Ephrin Fc pull-down assays from cells and tumor tissue
PC3 prostate cancer cells and H460 and A549 lung cancer cells were obtained from ATCC and PC3-3M-luc-C6 Bioware® prostate cancer cells were obtained from Caliper. The cells were grown in RPMI 1640 medium (Mediatech, #SH20027) with 10% FBS (Tissue Culture Biologicals, #101) and pen/strep (Omega Scientific, #AA-40). After reaching confluency, the cells were collected in Triton lysis buffer (PBS containing 1% Triton X-100 and 10 μg/ml aprotinin, 5 μg/ml pepstatin, 10 μg/ml leupeptin and 0.75 mM phenylmethylsulfonylfluoride as protease inhibitors). In the experiment shown in , phosphatase inhibitors (10 μM NaF and 1 μM sodium pervanadate) were also included. PC3-3M-luc-C6 tumors grown subcutaneously in nude mice were collected in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health when they reached a volume of ~1 mm3. The tumors were homogenized using a Polytron PT1600 E homogenizer (Kinematica) in four volumes of Triton lysis buffer or modified RIPA buffer (1% Triton X-100, 1% Na deoxycholate, 0.1% SDS, 20 mM Tris, 150 mM NaCl, 1 mM EDTA with protease inhibitors). Tumor lysates were centrifuged at 16,000 g for 15 min and the supernatant was used for pull-down experiments. Protein concentrations were measured using the BCA protein assay kit (Thermo Fisher Scientific, #23227).
Twenty micrograms of protein lysates were used for detection of Eph receptors in whole cell or tumor lysates by immunoblotting. For pull-downs to be analyzed by immunoblotting, we used 0.4–0.5 mg protein lysates and 3.75 μg ephrin-A Fc or ephrin-B Fc. In pull-downs with combinations of multiple ephrin-A or ephrin-B Fc proteins, we used mixtures containing equal amounts of each ephrin Fc and a total of 3.75 μg. In pull-downs with both ephrin-A Fc proteins and ephrin-B2 Fc, we used a total of 7.5 μg ephrin Fc proteins, including 1.25 μg ephrin-A1 Fc, 1.25 μg ephrin-A4 Fc, 1.25 μg ephrin-A5 Fc, and 3.75 μg ephrin-B2. For pull-downs to be analyzed by mass spectrometry, we used 3–4 mg protein lysates and 30 μg Fc proteins (for ephrin-A Fc or ephrin-B Fc separate pull-downs) or 60 μg Fc proteins (for combined ephrin-A Fc and ephrin-B Fc pull-downs). Since only one third of each sample was analyzed in the mass spectrometer, approximately 3–4-fold more protein lysate is needed for mass spectrometry analysis as compared with what would be needed to profile all 14 Eph receptors by immunoblotting. Cell and tumor lysates were incubated for 2–18 h with ephrin Fc fusion proteins or equimolar amounts of control Fc immobilized on 10–20 μl GammaBind Sepharose beads (GE Healthcare Life Sciences, #17-0886). The GammaBind sepharose was then washed 4 times with lysis buffer and the bound proteins were eluted by incubation in Laemmli sample buffer (60 mM Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue) for 6 min at 100°C.
The eluted proteins were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane for immunoblotting or silver stained using the SilverQuest staining kit (Invitrogen/Life Technologies, #LC6070) for mass spectrometry analysis. Pull-downs and cell lysates were probed by immunoblotting with an anti-phosphotyrosine antibody (Millipore #05321), rabbit anti-EphA2 and mouse anti EphA4 antibodies (Invitrogen/Life Technologies, #347400 and 371600), rabbit anti-EphA3 and anti-EphA5 antibodies (Santa Cruz Biotechnology, #sc919 and sc1014), an anti-human Fc antibody (Jackson Immunoresearch #309-006-008), and an anti-β-actin antibody (Sigma-Aldrich, #A3853). Rabbit antibodies to EphB2 and EphB4 were made using as antigens glutathione S-transferase (GST) fusion proteins of the carboxyl-terminal tails (including the SAM domain) of chicken EphB2 and human EphB3.Citation48,Citation49 The immune sera were purified on affinity columns containing the antigen used for immunization and absorbed on columns containing a GST fusion protein of the equivalent region of a related Eph receptor.
Ephrin-A3 Fc pull-down assay from mouse hippocampus
For hippocampal extracts preparation, adult mouse hippocampi (from 2 mo old C57BL/6 mice) were collected in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and washed quickly with ice cold PBS. Extracts were prepared by homogenization of half hippocampus in 1.5 ml Triton lysis buffer, using a Polytron PT1600 E homogenizer. Homogenates were sonicated and centrifuged at 14,000 g for 10 min at 4°C; 10 μg of the supernatant were used for immunoblotting and 0.5 mg for each pull-down. Since only one-fourth of each sample was analyzed in the mass spectrometer, approximately the same amount of tissue lysate is needed for mass spectrometry analysis as compared with what would be needed to profile all 14 Eph receptors by immunoblotting.
To obtain ephrin-A3 protein fused to Fc, HEK293AD cells (Cell Biolabs Inc. #AD-100) were grown in DMEM (Mediatech, #10-017-CV) with 10% FBS, pen/strep and 4 mM L-glutamine (Omega Scientific#GS-60). Cells were transfected with pcDNA3 encoding ephrin-A3 Fc, using Lipofectamine 2000 (Invitrogen, #11668) according to the manufacturer instructions. Stable clones were selected with 1.5 mg/ml G-418 (Roche Applied Science, #04727878001) and ephrin-A3 Fc levels in their culture medium were measured in ELISA assays. The clone producing the highest amount of ephrin-A3 Fc was grown to 70% confluency in 15 cm plates and then grown for 3 d in OptiMEM reduced serum medium (Invitrogen/Life Science Technologies, #31985). The medium containing ephrin-A3 Fc was then collected, buffered with Hepes pH 7.6 to a final concentration of 10 mM, centrifuged at 1,800 g for 5 min, and incubated with protein A beads (GenScript, #L00210) for 1 h at 4°C. Approximately 100 μg ephrin-A3 Fc or Fc bound to protein A beads were incubated with hippocampal extracts for 2 h at 4°C, washed with lysis buffer and incubated for 5 min at 95°C with Laemmli sample buffer. The samples were probed by immunoblotting with mouse anti-EphA4 antibody (Life Technologies/Invitrogen, #37-1600) and rabbit anti-human Fc antibody (Jackson Immuno Research, #309-006-008) or separated by SDS-PAGE and silver stained as described above for mass spectrometry analysis.
Protein identification from silver stained gel bands by LC/MS/MS
SDS-PAGE silver-stained gel lanes corresponding to molecular weights between ~80–180 kDa were cut into 1 mm × 1 mm pieces and de-stained. Following vacuum drying, proteins were reduced and alkylated by 5 mM DTT and 15 mM iodoacetamide treatment prior to digestion with 25 ng/μl trypsin (Promega, #V5111) in 50 mM ammonium bicarbonate for 1 h at 4°C followed by 16 h at 37°C. The resulting tryptic peptides were extracted from the gel by adding 300 μl 5% formic acid in water, sonicating 10 min in a water bath, and then washing 4 times with 50% acetonitrile in 5% formic acid in water and once with 100% acetonitrile. The extracted peptides were pooled together, vacuum dried and re-dissolved in 18 μl of 0.1% trifluoroacetic acid (TFA), before concentrating and desalting them using a Millipore C18 Zip Tip (Millipore, #ZTC18S096). The eluted peptides were then vacuum dried and re-dissolved in 20–24 μl LC/MS loading buffer (2% acetonitrile in 0.1% formic acid in water).
Five to eight microliters (corresponding to 25–30%) of tryptic digested samples were analyzed using an automated Nano LC-LTQ MS/MS mass spectrometer (Thermo Fisher Scientific) with an ADVANCE ESI source (Bruke-Michrom). The samples were loaded on the mass spectrometer using an Eksigent Nano 2D LC system, which includes a C18 trap column (Agilent, #ZORBAX 300SB-C18) in combination with a capillary reverse-phase column (15 cm in length, 100 μm id) packed with 5 μm Magic C18 AQ resin (Bruker-Michrom, #CP3/61271/00). A linear gradient of elution from buffer A (2% acetonitrile in H2O plus 0.1% formic acid) to 15% buffer A plus 85% buffer B (80% acetonitrile plus 0.1% formic acid) over 120 min was used. For samples derived from mouse hippocampus, 12 μl (corresponding to 25%) of tryptic digested sample were loaded and analyzed using an automated microLC-LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) with an ADVANCE ESI source. The peptides were separated using a Michrom LC system, which includes a C18 trap column (Bruker-Michrom) in combination with a capillary reverse-phase column (15 cm in length, 200 μm id) packed with 5 μm Magic C18 resin. A linear gradient of elution from 90% buffer A (0.1% formic acid) to 80% buffer B (acetonitrile plus 0.1% formic acid) over 120 min was used. The LC/MS run was operated in the data dependent mode. For each MS spectrum, MS/MS fragmentation data for the four strongest ions above an ion relative intensity of 5 × 104 were collected with dynamic exclusion enabled and collision energy set at 35%.
The MS/MS spectra were analyzed by Sorcerer Enterprise v.3.5 release (Sage-N Research Inc.) with the SEQUEST algorithm as the search program for peptide/protein identification. SEQUEST was set up to search the target-decoy IPI.Human.v3.73 or IPI.mouse.v3.73 databases (www.ebi.ac.uk/IPI/IPIhelp.html) with the allowance of up to two missed cleavages, Semi Tryptic search and precursor mass tolerance of 1.5 atomic mass units. Differential searches included 16 Da for methonine oxidation and 57 Da for cysteines alkylation. The search results were viewed, sorted, filtered, and statically analyzed by using the comprehensive proteomics data analysis software Peptide/Protein Prophet v.4.4.1 (Institute for System Biology). To minimize false positive identifications by MS/MS, the minimum trans-proteomic pipeline (TPP) probability scores for proteins and peptides were set at 0.95 and 0.9, respectively, to assure low error with reasonably good sensitivity. In addition, the threshold of cross correlation (Xcorr) scores was set to 1.5, 2.0 and 2.5 for singly, doubly and triply charged fully digested peptides, respectively.
It should be noted that due to the high conservation in the sequences of different Eph receptors, some of the tryptic peptides identified could be derived from multiple receptors. The results from the search algorithm are filtered to select only the Eph receptors for which at least two of the identified peptides could be derived only from that receptor. Thus, some peptides could also be derived from Eph receptors that are not selected. These Eph receptors could nevertheless be present in the samples analyzed. For simplicity, we refer to the peptides that are not shared among the Eph receptors positively identified in an experiment as “unique.”
| Abbreviations: | ||
| AP | = | alkaline phosphatase |
| ELISA | = | enzyme-linked immunosorbent assay |
| HPLC | = | high-performance liquid chromatography |
| LC | = | liquid chromatography |
| MS | = | mass spectrometry |
| qPCR | = | quantitative polymerase chain reaction |
| RT-PCR | = | reverse-transcription PCR |
| SDS-PAGE | = | sodium dodecylsulfate-polyacrylamide gel electrophoresis |
Acknowledgments
The authors thank K. Motamedchaboki for performing the mass spectrometry analyses, D. Melendez and K. Murai for engineering the mouse ephrin-A3 Fc fusion construct, M. Koolpe for engineering the ephrin-A3 AP construct, J. Stebbins and M. Pellecchia for providing PC3M-luc-C6 cells and tumor xenograft tissue, W. Placzek and M. Pellecchia for providing H460 cells and R. Sano and J. Reed for providing A549 cells. This work was supported by National Institutes of Health grants P01CA138390 and P01 HD025938 (E.B.P.), funds from the Tobacco-Related Disease Research Grants Program Office of the University of California, grant number 18XT-0099 (E.B.P.), and by a postdoctoral fellowship from the Spanish Foundation for Science and Technology (FECYT; E.R.T.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell 2008; 133:38 - 52; http://dx.doi.org/10.1016/j.cell.2008.03.011; PMID: 18394988
- Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer 2010; 10:165 - 80; http://dx.doi.org/10.1038/nrc2806; PMID: 20179713
- Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol 2005; 6:462 - 75; http://dx.doi.org/10.1038/nrm1662; PMID: 15928710
- Pasquale EB. Eph-ephrin promiscuity is now crystal clear. Nat Neurosci 2004; 7:417 - 8; http://dx.doi.org/10.1038/nn0504-417; PMID: 15114347
- Flanagan JG. Neural map specification by gradients. Curr Opin Neurobiol 2006; 16:59 - 66; http://dx.doi.org/10.1016/j.conb.2006.01.010; PMID: 16417998
- Astin JW, Batson J, Kadir S, Charlet J, Persad RA, Gillatt D, et al. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat Cell Biol 2010; 12:1194 - 204; http://dx.doi.org/10.1038/ncb2122; PMID: 21076414
- Dorrell C, Schug J, Lin CF, Canaday PS, Fox AJ, Smirnova O, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia 2011; 54:2832 - 44; http://dx.doi.org/10.1007/s00125-011-2283-5; PMID: 21882062
- Batlle E, Bacani J, Begthel H, Jonkheer S, Gregorieff A, van de Born M, et al. EphB receptor activity suppresses colorectal cancer progression. Nature 2005; 435:1126 - 30; http://dx.doi.org/10.1038/nature03626; PMID: 15973414
- Müller-Tidow C, Diederichs S, Bulk E, Pohle T, Steffen B, Schwäble J, et al. Identification of metastasis-associated receptor tyrosine kinases in non-small cell lung cancer. Cancer Res 2005; 65:1778 - 82; http://dx.doi.org/10.1158/0008-5472.CAN-04-3388; PMID: 15753374
- Brantley-Sieders DM, Jiang A, Sarma K, Badu-Nkansah A, Walter DL, Shyr Y, et al. Eph/ephrin profiling in human breast cancer reveals significant associations between expression level and clinical outcome. PLoS One 2011; 6:e24426; http://dx.doi.org/10.1371/journal.pone.0024426; PMID: 21935409
- Vergara-Silva A, Schaefer KL, Berg LJ. Compartmentalized Eph receptor and ephrin expression in the thymus. Gene Expr Patterns 2002; 2:261 - 5; http://dx.doi.org/10.1016/S1567-133X(02)00054-6; PMID: 12617812
- Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. Nat Neurosci 2003; 6:153 - 60; http://dx.doi.org/10.1038/nn994; PMID: 12496762
- van Eyll JM, Passante L, Pierreux CE, Lemaigre FP, Vanderhaeghen P, Rousseau GG. Eph receptors and their ephrin ligands are expressed in developing mouse pancreas. Gene Expr Patterns 2006; 6:353 - 9; http://dx.doi.org/10.1016/j.modgep.2005.09.010; PMID: 16446123
- Hafner C, Schmitz G, Meyer S, Bataille F, Hau P, Langmann T, et al. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem 2004; 50:490 - 9; http://dx.doi.org/10.1373/clinchem.2003.026849; PMID: 14726470
- Alonso-C LM, Trinidad EM, de Garcillan B, Ballesteros M, Castellanos M, Cotillo I, et al. Expression profile of Eph receptors and ephrin ligands in healthy human B lymphocytes and chronic lymphocytic leukemia B-cells. Leuk Res 2009; 33:395 - 406; http://dx.doi.org/10.1016/j.leukres.2008.08.010; PMID: 18819711
- Sakamoto A, Sugamoto Y, Tokunaga Y, Yoshimuta T, Hayashi K, Konno T, et al. Expression profiling of the ephrin (EFN) and Eph receptor (EPH) family of genes in atherosclerosis-related human cells. J Int Med Res 2011; 39:522 - 7; PMID: 21672356
- Ishii M, Mueller I, Nakajima T, Pasquale EB, Ogawa K. EphB signaling inhibits gap junctional intercellular communication and synchronized contraction in cultured cardiomyocytes. Basic Res Cardiol 2011; 106:1057 - 68; http://dx.doi.org/10.1007/s00395-011-0219-3; PMID: 21892745
- Saeger BM, Suhm M, Neubüser A. Ephrin/ephrin receptor expression during early stages of mouse inner ear development. Dev Dyn 2011; 240:1578 - 85; http://dx.doi.org/10.1002/dvdy.22632; PMID: 21465626
- Gale NW, Holland SJ, Valenzuela DM, Flenniken A, Pan L, Ryan TE, et al. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron 1996; 17:9 - 19; http://dx.doi.org/10.1016/S0896-6273(00)80276-7; PMID: 8755474
- Flenniken AM, Gale NW, Yancopoulos GD, Wilkinson DG. Distinct and overlapping expression patterns of ligands for Eph-related receptor tyrosine kinases during mouse embryogenesis. Dev Biol 1996; 179:382 - 401; http://dx.doi.org/10.1006/dbio.1996.0269; PMID: 8903354
- Himanen JP, Chumley MJ, Lackmann M, Li C, Barton WA, Jeffrey PD, et al. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci 2004; 7:501 - 9; http://dx.doi.org/10.1038/nn1237; PMID: 15107857
- Eph Nomenclature Committee. Unified nomenclature for Eph family receptors and their ligands, the ephrins. Cell 1997; 90:403 - 4; http://dx.doi.org/10.1016/S0092-8674(00)80500-0; PMID: 9267020
- Lackmann M, Mann RJ, Kravets L, Smith FM, Bucci TA, Maxwell KF, et al. Ligand for EPH-related kinase (LERK) 7 is the preferred high affinity ligand for the HEK receptor. J Biol Chem 1997; 272:16521 - 30; http://dx.doi.org/10.1074/jbc.272.26.16521; PMID: 9195962
- Pabbisetty KB, Yue X, Li C, Himanen JP, Zhou R, Nikolov DB, et al. Kinetic analysis of the binding of monomeric and dimeric ephrins to Eph receptors: correlation to function in a growth cone collapse assay. Protein Sci 2007; 16:355 - 61; http://dx.doi.org/10.1110/ps.062608807; PMID: 17322526
- Coulthard MG, Lickliter JD, Subanesan N, Chen K, Webb GC, Lowry AJ, et al. Characterization of the Epha1 receptor tyrosine kinase: expression in epithelial tissues. Growth Factors 2001; 18:303 - 17; http://dx.doi.org/10.3109/08977190109029118; PMID: 11519828
- Irie F, Okuno M, Matsumoto K, Pasquale EB, Yamaguchi Y. Heparan sulfate regulates ephrin-A3/EphA receptor signaling. Proc Natl Acad Sci U S A 2008; 105:12307 - 12; http://dx.doi.org/10.1073/pnas.0801302105; PMID: 18715996
- Bergemann AD, Zhang L, Chiang MK, Brambilla R, Klein R, Flanagan JG. Ephrin-B3, a ligand for the receptor EphB3, expressed at the midline of the developing neural tube. Oncogene 1998; 16:471 - 80; http://dx.doi.org/10.1038/sj.onc.1201557; PMID: 9484836
- Holen HL, Zernichow L, Fjelland KE, Evenroed IM, Prydz K, Tveit H, et al. Ephrin-B3 binds to a sulfated cell-surface receptor. Biochem J 2011; 433:215 - 23; http://dx.doi.org/10.1042/BJ20100865; PMID: 20925654
- Miao H, Burnett E, Kinch M, Simon E, Wang B. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat Cell Biol 2000; 2:62 - 9; http://dx.doi.org/10.1038/35000008; PMID: 10655584
- Fox BP, Tabone CJ, Kandpal RP. Potential clinical relevance of Eph receptors and ephrin ligands expressed in prostate carcinoma cell lines. Biochem Biophys Res Commun 2006; 342:1263 - 72; http://dx.doi.org/10.1016/j.bbrc.2006.02.099; PMID: 16516143
- Xia G, Kumar SR, Masood R, Zhu S, Reddy R, Krasnoperov V, et al. EphB4 expression and biological significance in prostate cancer. Cancer Res 2005; 65:4623 - 32; http://dx.doi.org/10.1158/0008-5472.CAN-04-2667; PMID: 15930280
- Janes PW, Griesshaber B, Atapattu L, Nievergall E, Hii LL, Mensinga A, et al. Eph receptor function is modulated by heterooligomerization of A and B type Eph receptors. J Cell Biol 2011; 195:1033 - 45; http://dx.doi.org/10.1083/jcb.201104037; PMID: 22144690
- Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics 2008; 7:2373 - 85; http://dx.doi.org/10.1074/mcp.M800203-MCP200; PMID: 18644780
- Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci 1998; 21:309 - 45; http://dx.doi.org/10.1146/annurev.neuro.21.1.309; PMID: 9530499
- Ji XD, Li G, Feng YX, Zhao JS, Li JJ, Sun ZJ, et al. EphB3 is overexpressed in non-small-cell lung cancer and promotes tumor metastasis by enhancing cell survival and migration. Cancer Res 2011; 71:1156 - 66; http://dx.doi.org/10.1158/0008-5472.CAN-10-0717; PMID: 21266352
- Kozlowski JM, Fidler IJ, Campbell D, Xu ZL, Kaighn ME, Hart IR. Metastatic behavior of human tumor cell lines grown in the nude mouse. Cancer Res 1984; 44:3522 - 9; PMID: 6744277
- Carmona MA, Murai KK, Wang L, Roberts AJ, Pasquale EB. Glial ephrin-A3 regulates hippocampal dendritic spine morphology and glutamate transport. Proc Natl Acad Sci U S A 2009; 106:12524 - 9; http://dx.doi.org/10.1073/pnas.0903328106; PMID: 19592509
- Filosa A, Paixão S, Honsek SD, Carmona MA, Becker L, Feddersen B, et al. Neuron-glia communication via EphA4/ephrin-A3 modulates LTP through glial glutamate transport. Nat Neurosci 2009; 12:1285 - 92; http://dx.doi.org/10.1038/nn.2394; PMID: 19734893
- Buchert M, Schneider S, Meskenaite V, Adams MT, Canaani E, Baechi T, et al. The junction-associated protein AF-6 interacts and clusters with specific Eph receptor tyrosine kinases at specialized sites of cell-cell contact in the brain. J Cell Biol 1999; 144:361 - 71; http://dx.doi.org/10.1083/jcb.144.2.361; PMID: 9922461
- Olivieri G, Miescher GC. Immunohistochemical localization of EphA5 in the adult human central nervous system. J Histochem Cytochem 1999; 47:855 - 61; http://dx.doi.org/10.1177/002215549904700702; PMID: 10375373
- Ciossek T, Ullrich A, West E, Rogers JH. Segregation of the receptor EphA7 from its tyrosine kinase-negative isoform on neurons in adult mouse brain. Brain Res Mol Brain Res 1999; 74:231 - 6; http://dx.doi.org/10.1016/S0169-328X(99)00285-5; PMID: 10640696
- Cooper MA, Crockett DP, Nowakowski RS, Gale NW, Zhou R. Distribution of EphA5 receptor protein in the developing and adult mouse nervous system. J Comp Neurol 2009; 514:310 - 28; http://dx.doi.org/10.1002/cne.22030; PMID: 19326470
- Grunwald IC, Korte M, Wolfer D, Wilkinson GA, Unsicker K, Lipp HP, et al. Kinase-independent requirement of EphB2 receptors in hippocampal synaptic plasticity. Neuron 2001; 32:1027 - 40; http://dx.doi.org/10.1016/S0896-6273(01)00550-5; PMID: 11754835
- Shao H, Lou L, Pandey A, Pasquale EB, Dixit VM. cDNA cloning and characterization of a ligand for the Cek5 receptor protein-tyrosine kinase. J Biol Chem 1994; 269:26606 - 9; PMID: 7929389
- Menzel P, Valencia F, Godement P, Dodelet VC, Pasquale EB. Ephrin-A6, a new ligand for EphA receptors in the developing visual system. Dev Biol 2001; 230:74 - 88; http://dx.doi.org/10.1006/dbio.2000.0109; PMID: 11161563
- Cheng HJ, Nakamoto M, Bergemann AD, Flanagan JG. Complementary gradients in expression and binding of ELF-1 and Mek4 in development of the topographic retinotectal projection map. Cell 1995; 82:371 - 81; http://dx.doi.org/10.1016/0092-8674(95)90426-3; PMID: 7634327
- Flanagan JG, Cheng HJ, Feldheim DA, Hattori M, Lu Q, Vanderhaeghen P. Alkaline phosphatase fusions of ligands or receptors as in situ probes for staining of cells, tissues, and embryos. Methods Enzymol 2000; 327:19 - 35; http://dx.doi.org/10.1016/S0076-6879(00)27264-9; PMID: 11044971
- Holash JA, Pasquale EB. Polarized expression of the receptor protein tyrosine kinase Cek5 in the developing avian visual system. Dev Biol 1995; 172:683 - 93; http://dx.doi.org/10.1006/dbio.1995.8039; PMID: 8612982
- Noren NK, Lu M, Freeman AL, Koolpe M, Pasquale EB. Interplay between EphB4 on tumor cells and vascular ephrin-B2 regulates tumor growth. Proc Natl Acad Sci U S A 2004; 101:5583 - 8; http://dx.doi.org/10.1073/pnas.0401381101; PMID: 15067119