Abstract
Profilin-1 (Pfn1) is a ubiquitously expressed actin-monomer binding protein that has been linked to many cellular activities ranging from control of actin polymerization to gene transcription. Traditionally, Pfn1 has been considered to be an essential control element for actin polymerization and cell migration. Seemingly contrasting this view, a few recent studies have shown evidence of an inhibitory action of Pfn1 on motility of certain types of carcinoma cells. In this review, we summarize biochemistry and functional aspects of Pfn1 in normal cells and bring in newly emerged action of Pfn1 in cancer cells that may explain its context-specific role in cell migration.
Keywords: :
Introduction
Profilins (Pfns) constitute a class of evolutionarily conserved small actin-binding proteins which have been linked to almost every aspect of cellular functions including proliferation, survival, motility, endocytosis and membrane trafficking, mRNA splicing and gene transcription. In mammals, four Pfn genes (Pfn1, -2, -3 and -4) have been discovered to date. Pfn1 is the founding member of the family and ubiquitously expressed in all cell types except skeletal muscle.Citation1-Citation3 Gene knockout of Pfn1 in mice leads to embryonic lethality at a very early stage (2-cell stage) of development thus indicating its absolutely essential role in embryonic cell survival and/or proliferation.Citation4 In vertebrates, two alternatively spliced variants of Pfn2 (Pfn2a and Pfn2b) have been identified. Even though Pfn2a (the major form) transcript is detected in a wide variety of tissues, the actual protein expression occurs predominantly in brain. The rare isoform Pfn2b is not expressed during embryogenesis but its mRNA is detectable in a limited number of adult tissues including skin, kidney and liver.Citation5,Citation6 Two additional splice variants which are capable of coding truncated forms of Pfn2 have also been documented in the literature; whether these variants are actually expressed is not known.Citation6 An exception to this expression pattern of Pfn2 is seen in birds where this protein is ubiquitously expressed.Citation7 Unlike Pfn1, genetic ablation of Pfn2 in mice does not compromise the overall embryonic development but leads to neurological phenotypes further confirming importance of Pfn2 function predominantly in neuronal system in vertebrates.Citation8 Finally, expressions of Pfn3 and Pfn4 appear to be restricted to testis with the latter being implicated during spermatogenesis.Citation9,Citation10
In this review, we will first summarize biochemical and functional aspects of Pfn1, the most widely studied Pfn isoform in the literature. For brevity and focus, we will limit our discussion primarily to its role in the regulation of actin dynamics and cell motility, introducing recently emerged counter-intuitive findings on Pfn1’s effect on certain cancer cells and how that could translate to new models of cell motility regulation by Pfn1.
Pfn1 Biochemistry
Despite a fairly low sequence homology, a remarkable structural similarity exists between the different variants of Pfn suggesting conserved ligand interactions of these members. In general, Pfns bind to three major classes of ligands: actin, proteins containing poly-l-proline (PLP) stretches and phosphoinositide (PPI)-based lipids (the only exception to this is Pfn4, which does not bind the first two types of ligands). Biochemical features of each of these interactions are summarized below.
Dual effects of Pfn1 on actin polymerization in vitro and in vivo
Pfn1 (mol wt: 12–15 kD) was originally identified as an actin-sequestering protein that forms a 1:1 complex with monomeric actin (G-actin) and prevents actin from polymerization.Citation11 Consistent with this initially proposed actin-sequestering function of Pfn, it was shown that simultaneous gene disruption of Pfn1 and Pfn2 elevates F-actin content in Dictyostelium amoebae, and conversely, increasing Pfn1 level in yeasts and NRK (normal rat kidney) fibroblasts results in loss of F-actin.Citation12-Citation14 However, several important biochemical findings related to Pfn1’s effect on actin polymerization suggested that Pfn1’s action on actin dynamics is more complex than it was originally conceived. Markey et al. demonstrated that the nucleation (lag) phase of actin polymerization in the presence of Pfn1 is shortened by spectrin-actin-band 4.1 complex (a nucleating complex that allows actin polymerization from the barbed end), but is lengthened by vilin (a nucleator that allows actin polymerization from the pointed end).Citation15 This observation for the first time suggested a possible asymmetric action of Pfn1 on kinetic regulation of barbed vs. pointed ends of actin filaments. Subsequently, a number of studies from different laboratories revealed that while Pfn1 strongly inhibits spontaneous nucleation and elongation at the slow-growing pointed ends of actin filaments, Pfn1:actin complex can add to the barbed ends and elongate actin filaments.Citation16-Citation18 Since artificially cross-linked Pfn1:actin complex interferes with filament elongation, it has been postulated that during filament elongation, Pfn1:actin complex initially binds to the barbed ends of actin filaments and subsequently, Pfn1 dissociates leaving the actin molecule behind.Citation19,Citation20 Two other biochemical properties of Pfn1 presumably further contribute to its actin polymerizing ability. First, Pfn1 can stimulate nucleotide exchange (ADP to ATP) on G-actinCitation21 and this potentially accelerates regeneration of polymerization-competent actin monomers (i.e., ATP-G-actin) from disassembling filaments. Second, Pfn1, when bound to actin, inhibits the ATPase activity of actin and this could also effectively increase the concentration of ATP-actin thus facilitating actin filament elongation.Citation22 Given that intracellular concentration of Pfn1 in most cells is also not sufficient to account for G-actin content via sequestration activity, it was therefore proposed that the primary in vivo function of Pfn1 might be to actually promote actin polymerization. Several cell-based experimental observations are in alignment with this alternatively proposed function of Pfn1. For example, when Pfn1 expression is silenced, there is an overall reduction in the F-actin content in several cell types (example: human vascular endothelial cells, lung epithelial cells, breast cancer cells) and conversely, F-actin level increases in response to overexpression of Pfn1 as demonstrated in CHO (chicken hamster ovary) cells.Citation23-Citation27
In summary, Pfn1 can have a dual effect on actin polymerization depending on its concentration relative to that of G-actin and free barbed ends of actin filaments. If the barbed ends of actin filaments are blocked by capping proteins, Pfn1 can simply function as an actin-sequestering protein preventing actin from polymerizing. However, when the barbed ends are free, Pfn1 can promote actin assembly provided sufficient pool of ATP-actin is available for polymerization. Cell-specific differences in the abundance of other G-actin-sequestering proteins (such as thymosin β4) and expression/activity of barbed-end capping proteins may explain the basis for differential effects of loss of Pfn1 on the overall actin polymerization between different cell types.
PLP interaction
A distinguishing biochemical feature of Pfn is its ability to bind PLP sequences and this enables use of PLP-affinity chromatography for purification of Pfn from cellular extracts. PLP binding involves both N and C termini of Pfn1, and does not interfere with its actin-binding activity. Most bona fide cellular ligands of Pfn contain multiple PLP motifs, each comprising of at most 3–5 prolines flanked by a glycine or an alanine residue.Citation1,Citation28 On a chronological scale, VASP (vasodilator stimulated phosphoprotein) was the first PLP-domain bearing protein to be identified as a Pfn ligand.Citation29 Since then, a large number of PLP ligands of Pfn1 have been identified which vary with respect to their sub-cellular localization and/or function. In the cytoplasmic compartment, Pfn1 interacts with important regulators of actin dynamics and organization [N-WASP (neural Wiskott-Aldrich syndrome protein), WAVE (WASP-associated verprolin homology protein), Diaphanous, Mena (mammalian enabled)/VASP/Evl (enabled/VASP like), palladin, RIAM (Rap1 GTP-interacting adaptor molecule)/lamellipodin],Citation30-Citation36 cell-cell adhesion (AF-6),Citation37 and membrane trafficking [VCP (valocin containing protein), clathrin, huntingtin].Citation38,Citation39 Nuclear ligands of Pfn1 identified thus far include p42-POP (a myb-family transcription factor),Citation40 SMN (survival motor neuron protein—involved in small ribonucleoprotein processing and transport)Citation41 and exportin-6 (involved in nuclear export of actin).Citation42 Functional diversity of these PLP ligands places Pfn1 at the crossroads of many physiological activities in cells ranging from actin cytoskeletal organization to gene transcription.
PPI interaction
Pfn1’s binding to phosphatidylinositol-mono-phosphate [PI(4)P] and phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] reconstituted into lipid vesicles in vitro initially suggested that Pfn1 may have an ability to interact with membrane PPIs in cells.Citation43 The actual membrane localization of Pfn1 was confirmed by ultrastructural immunolocalization in human leukocytes and platelets which showed that a small sub-cellular pool of Pfn1 dynamically associates with the inner leaflet of the plasma membrane.Citation44 In yeasts, it was subsequently shown that inositol depletion results in translocation of Pfn1 from the plasma membrane to the cytosol thus suggesting that membrane localization of Pfn1 is mainly conferred by its PPI-interaction.Citation45 Besides PI(4)P and PI(4,5)P2, Pfn1 can also bind to lipid products of PI3-kinase or D3-PPIs {PPIs containing phosphates at D3-position of the inositol ring; example: phosphatidylinositol-3,4-bisphosphate [PI(3,4)P2], and phosphatidylinositol-3,4,5-triphosphate [PI(3,4,5)P3 or PIP3]}, and in fact, with much higher affinity than that of PI(4,5)P2, at least, in vitro.Citation46,Citation47 Since none of these binding experiments have been performed in actual cells, which of these PPI species is (are) responsible for membrane association of Pfn1 in vivo has not been conclusively determined yet. Given that PI(4,5)P2 is present in cells at a much higher abundance than D3-PPIs, we can speculate that in vivo membrane association of Pfn1 is predominantly through its PI(4,5)P2 binding, and other types of PPI interaction may occur transiently under conditions which promote D3-PPI synthesis, such as, in response to growth-factor (EGF, PDGF and IGF) stimulation.
PPIs not only bind to Pfn1, but can also bind to Pfn1:actin complex causing rapid dissociation of this complex.Citation43 Supporting this observation, a mutagenesis study further revealed that a point mutation on human Pfn1 that disrupts its PPI-binding also dramatically impairs its actin interaction.Citation48 Based on these findings, it was concluded that that PPI-binding region of Pfn1 overlaps with its actin-binding site. A similar competitive interaction between PI(4,5)P2 and PLP-ligands for Pfn1 was discovered through mutagenesis experiments and it was proposed that an additional region neighboring the PLP binding site in the C-terminal helix of Pfn1 may also be involved in PPI binding.Citation49,Citation50 Existence of this additional PPI-binding region on Pfn1 explains the basis for a ternary complex formation between covalently cross-linked Pfn1:actin and PI(4,5)P2.Citation51
Context-Specific Effect of Pfn1 in Cell Motility
Pfn1’s role in cell migration has been examined in both lower and higher eukaryotic model systems using various loss-of-function strategies. Gene deletion of Pfn1 and Pfn2 resulting in impaired motility of Dictyostelium amoebae produced the first direct evidence of Pfn’s requirement in cell migration. In this unicellular eukaryotic organism, Pfn1 knockout (K/O) alone failed to produce any phenotype likely because of compensatory action of Pfn2.Citation12 Effect of Pfn depletion in multi-cellular organism was first analyzed in drosophila where deletion of chickadee (encodes Pfn1) led to late-stage embryonic lethality; viable chickadee alleles exhibited aberrant actin assembly and impaired cell migration.Citation52 Similarly, gastrulation defects were observed in Xenopus and zebrafish when Pfn1 expression was downregulated through antisense morpholinos.Citation53,Citation54 Relevant to mammalian cell systems, it has been shown that loss of expression or disruption of ligand interactions of Pfn1 causes impaired migration/invasion and capillary morphogenesis of human vascular endothelial cells, and defects in neurite outgrowth.Citation24,Citation26,Citation55,Citation56 Very recently, cre/lox-based gene deletion approach has been utilized to study the effect of tissue-specific loss of Pfn1 in vivo. For example, Pfn1 deletion in chondrocyte leads to impaired migration, and results in chondrodysplasia and stunted bone growth in mouse.Citation57 Pfn1 ablation in brain inhibits radial migration of cerebellar granular neurons and causes cerebellar hypoplasia in mouse.Citation58 All of these studies point to a general pro-migratory function of Pfn1.
Seemingly contrary to the essential role of Pfn1 in cell migration in the physiological contexts, a number of studies have reported that various invasive adenocarcinomas (breast, hepatic and bladder) exhibit Pfn1 downregulation when compared with their normal counterparts.Citation59-Citation62 We surprisingly found that breast cancer cells and even normal human mammary epithelial cells (HMEC) display hypermotile phenotype when Pfn1 expression is suppressed, and conversely, overexpression of Pfn1 suppresses breast cancer cell motility.Citation25,Citation63 Similar apparent inverse correlation between Pfn1 expression and tumor cell motility has also been documented for hepatocarcinoma cells.Citation61 These unexpected findings in carcinoma cells have implied that how loss of Pfn1 alters cellular phenotype is highly context-dependent.
Actin-Dependent Function of Pfn1 in Cell Motility
Cell motility is a complex integration of several discrete biomechanical events including lamellipodial or pseudopodial protrusion (powered by actin polymerization), stabilization of protrusion by formation and maturation of integrin-based adhesions, forward propulsion of cell body (driven by actomyosin contractility) and finally, release of cell rear (mediated by de-adhesion). Because of its critical role in actin polymerization, Pfn1’s function has been most widely studied in the context of regulation of membrane protrusion. Actin-driven intracellular motility of bacterial pathogens has been an attractive model system for studying biochemical and biophysical aspects of lamellipodial protrusion of motile cells. In these biomimetic assays, propulsion speed of bacterial pathogens (analogous to velocity of membrane protrusion in motile cells) is dramatically reduced in the absence or upon functional loss of Pfn1.Citation64-Citation66 Consistent with these observations, it has been further shown that Pfn1 depletion can cause defects in membrane extension and slower velocity of protrusion, and these phenotypes can only be rescued by re-expression of fully functional Pfn1 but not mutants that lack actin or PLP binding. These data demonstrated that Pfn1 is an essential driver of membrane protrusion during cell migration, and further show the importance of actin and PLP interactions in this aspect of Pfn1’s function.Citation24,Citation26
Membrane protrusion requires de novo actin nucleation followed by filament elongation and/or elongation of pre-existing filaments, catalyzed by at least three major classes of actin-binding proteins including N-WASP/WAVE, Ena/VASP and formins. A common feature of these different classes of proteins is the presence of PLP domains which allows them to interact with Pfn1. In vitro kinetic experiments have shown that low concentration of Pfn1 (~1–5 μM) can stimulate actin polymerization by N-WASP and Ena/VASP.Citation36,Citation67 Functional co-operativity of Pfn1 with these proteins also appears to be true in vivo. For example, N-WASP-induced membrane protrusion and bacterial pathogen motility requires functional actin and PLP interactions of Pfn1.Citation65,Citation68 Overexpression of a dictyostelium homolog of WASP with PLP deletion dramatically impairs the protrusive ability and chemotaxis of the organism.Citation69 Similarly, Ena/VASP-induced motility of bacterial pathogens is attenuated when its PLP domain is deleted.Citation70 Consistent with these data, colocalization and FRET studies have shown evidence of prominent VASP–Pfn1 interaction at the sites of dynamic actin polymerization near the leading edge in migrating cells.Citation71,Citation72 Together, these findings support a widely accepted model that interaction of Pfn1:actin complex to the promoters of actin nucleation and elongation at the leading edge facilitates sub-membranous actin polymerization through barbed-end elongation and promotes membrane protrusion during cell migration. An alternative mechanism of how Pfn1 regulates membrane protrusion has been proposed in the literature based on the effect of Pfn1 in millimolar concentration range on actin polymerization. Even though the overall cellular concentration of Pfn1 ranges from 10–50 μM in most cells, it has been suggested that presence of multiple Pfn1-binding sites on various actin-regulatory proteins (16–20 on VASP, 12 on N-WASP) and membrane PI(4,5)P2 (up to 5) can potentially generate a local milieu of Pfn1 in the millimolar concentration range near the leading edge in cells. Pfn1 at this concentration range increases the off-rate of actin monomers at the barbed ends of actin filaments and actually causes F-actin depolymerization via competing with barbed-end capping proteins. Paradoxically, this faster rate of deplomerization can promote net polymerization by accelerating ATP-replacement of actin monomers.Citation73-Citation76 Even though there is a mechanistic difference between the two models, both build upon the fundamental idea of Pfn1 utilizing its actin and PLP interactions to promote net actin polymerization at the leading edge and in turn, driving membrane protrusion during cell migration.
Actin-Independent Function of Pfn1 in Cell Motility
Cumulative findings of two recent studies from our group have uncovered a novel actin-independent mechanism by which Pfn1 can negatively regulate breast cancer cell motility.Citation63,Citation77 Specifically, those studies showed that the Pfn1 depletion in breast cancer cells leads to a hypermotile phenotype through enhancing membrane targeting of lamellipodin (Lpd—a PPI-binding protein which recruits Ena/VASP to the leading edgeCitation33,Citation78) and in turn, augmenting Ena/VASP localization at the leading edge. Interestingly, all of these phenotypes of Pfn1-deficient cells were rescued by re-expression of Pfn1 or even its mutants that were deficient in actin or PLP binding, but not a mutant defective in PPI interaction. This suggested that Pfn1 suppresses breast cancer cell motility through its PPI interaction via negatively regulating recruitment of Lpd-Ena/VASP complex to the leading edge. Lpd plays an important role in membrane protrusion through downstream action of Ena/VASP,Citation33,Citation78 and among the different Ena/VASP proteins, at least, Mena has been shown to be a critical player in breast cancer invasion and metastasis.Citation79-Citation83 Thus, controlling Lpd-Ena/VASP recruitment to the leading edge clearly seems to be an attractive mechanism to regulate breast cancer cell motility which Pfn1-deficient condition takes advantage of.
So, how does Pfn1:PPI interaction control membrane recruitment of Lpd? Lpd contains a pleckstrin-homology (PH) domain that displays selective affinity for PI(3,4)P2.Citation33 PI(3,4)P2 is also generated at the sites of Lpd recruitment in cells.Citation84 Thus, PI(3,4)P2 appears to be a key PPI for membrane docking of Lpd. Since Pfn1 can bind to PI(3,4)P2, one possibility is that Pfn1 can compete with Lpd for PI(3,4)P2 binding and therefore, in the absence of Pfn1, Lpd:PI(3,4)P2 interaction is increased. While this model is conceptually simple, the actual demonstration of this mechanism in cells is not trivial and has not been examined yet. However, correlated with Pfn1’s inhibitory effect on Lpd recruitment to the membrane, we demonstrated that membrane accumulation of PI(3,4)P2 at the leading edge in breast cancer cells is downregulated by Pfn1:PPI interaction.Citation77 At present, the underlying mechanism of how Pfn1 influences membrane accumulation of PI(3,4)P2 remains unknown. Based on previous evidence of Pfn1’s ability to inhibit PI(4,5)P2 hydrolysis in vitro,Citation85,Citation86 and reduced PtdIns(3,4,5)P3 generating ability in breast cancer cells upon Pfn1 overexpression,Citation87 one can speculate that Pfn1:PI(4,5)P2 interaction somehow protects PI(4,5)P2 from its natural turnover including PI3K-mediated conversion to PIP3 and subsequent generation of downstream PPIs such as PI(3,4)P2. Overall, these findings present an alternative actin-independent modality of Pfn1’s control of cell motility in which Pfn1:PPI interaction negatively regulates PI(3,4)P2 [possibly by putting a brake on PI(4,5)P2 turnover] and blocks PI(3,4)P2-dependent recruitment of other motility-regulatory protein complexes (such as Lpd/Ena-VASP complex) to the plasma membrane.
Interestingly, Pfn1’s inhibitory action on Lpd recruitment to the membrane does not appear to be restricted to only those cell types where loss of Pfn1 creates a hypermotile phenotype (e.g., breast cancer and normal human mammary epithelial cells). In fact, Pfn1 depletion causes similar membrane enrichment of Lpd-Ena/VASP in human vascular endothelial cells even though the overall motility is actually impaired suggesting that Pfn1’s control of Lpd localization is a conserved mechanism.Citation63,Citation77 Although increased Ena/VASP activity at the leading edge generally correlates with faster membrane protrusion, its net effect on cell motility has been shown to be context-dependent. For example, in the case of fibroblasts, Ena/VASP-promoted lamellipodial protrusions are unproductive for forward movement because of their increased tendency of withdrawal and this translates to a slower overall migration speed. At the molecular level, this is thought to be due to an antagonistic action of Ena/VASP on the capping protein.Citation88,Citation89 Essentially, higher Ena/VASP activity at the front leads to generation of longer actin filaments which also have propensity to buckle causing withdrawal of protrusions. By contrast, in rapidly moving fish scale keratocytes, Ena/VASP localization to the leading edge positively correlates with the protrusion speed as well as the overall speed of migration.Citation90 Analyses of protrusion dynamics in breast cancer cells showed that Pfn1 depletion causes a reduction in the actual protrusion speed but this is offset by a marked increase in the persistence of protrusion causing a net result of increased forward movement. However, inhibiting Ena/VASP function virtually obliterates the protrusive ability of Pfn1-deficient cells.Citation63 Therefore, it appears that Pfn1-deficient breast cancer cells utilizes Ena/VASP enrichment at the leading edge as a strategy to overcome protrusion defects which would have occurred otherwise. So, why should Ena/VASP-driven protrusion in Pfn1-depleted breast cancer cells display increased persistence while in fibroblasts, it does not? When a cell generates a lamellipodial protrusion, whether the protrusion will sustain or undergo withdrawal depends on how efficiently it is stabilized by cell-substrate adhesion. Therefore, adhesion dynamics may play a key role in determining how Ena/VASP recruitment to the leading edge translates to the overall cell motility. Perhaps in Pfn1-depleted breast cancer cells, the intrinsic adhesion dynamics sets a stage for Ena/VASP-driven activity at the leading edge favorable for overall cell motility.
Is There a Unifying Theory to Explain the Context-Specific Role of Pfn1 in Cell Motility?
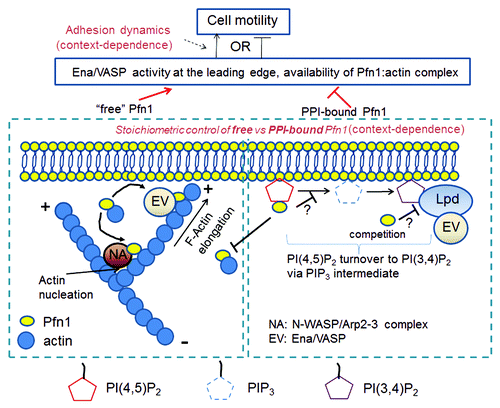
We can construct a simple mechanistic model describing two potential mechanisms by which Pfn1 can regulate cell motility. On one hand, Pfn1, when not bound to PPIs, facilitates sub-membranous actin polymerization at the leading edge catalyzed by various PLP-domain bearing actin regulators (e.g., N-WASP and Ena/VASP) and this action of Pfn1 has a positive effect on the actual velocity of membrane protrusion. On the other hand, Pfn1, when bound to PPIs, not only prevents its interaction with actin, but also inhibits Ena/VASP recruitment to the leading edge through limiting membrane availability of PI(3,4)P2 for Lpd, and this has a negative impact on the actual velocity of protrusion. Therefore, the net action of Pfn1 on the overall cell motility should depend on the relative stoichiometry of PPI-bound vs. -unbound pool of Pfn1 in the immediate vicinity of the plasma membrane, and how effectively protrusion is coupled to adhesion to generate productive motility (). Cell-specific differences in adhesion dynamics and intracellular signaling that regulate stoichiometric control of PPI-bound vs. -unbound pool of Pfn1 could very well explain the context-specific role of Pfn1 in cell migration.
Figure 1. A schematic model integrating actin-dependent and -independent modes of Pfn1’s regulation of cell motility. In this model, actin-dependent function refers to the action of Pfn1 pool in the vicinity of the membrane which is not physically interacting with PPIs and this mainly involves positive regulation of N-WASP-Arp2/3- and Ena/VASP-mediated actin polymerization. The actin-independent function refers to the action of PPI-bound pool of Pfn1, and this involves negative regulation of Ena/VASP recruitment to the leading edge through limiting membrane availability of PI(3,4)P2 for Lpd, either by affecting the metabolic turnover of PI(4,5)P2 and thus downregulating PI(3,4)P2 generation and/or direct binding competition.
Conclusion and Outlook
Even after many years of extensive research devoted to Pfn1, it continues to be an enigmatic molecule to cell biologists. Last three decades of research heavily focused on Pfn1’s control of actin polymerization. Recent studies demonstrating differential effect of Pfn1 on motility of normal vs. cancer cells now force us to look beyond the traditional cytoskeletal function of Pfn1, particularly stimulating thoughts on an alternative role of Pfn1 as a modulator of PPI signaling at the membrane-cytosol interface and how that might impact cell motility.
Given the functional diversity of PLP ligands of Pfn1, it is also unlikely that Pfn1 controls cell migration solely through regulating lamellipodial dynamics. For example, it is known that Pfn1 regulates nucleo-cytoplasmic shuttling of actin via its interaction with exportin (a nuclear exporter of actin).Citation42 Activation of serum-response factor (SRF), a transcription factor that regulates many of serum-inducible genes, is inhibited by G-actin. Consistent with actin polymerization activity of Pfn1, a previous study had reported that SRF activation in cells can be upregulated dramatically by overexpression of wild-type but not an actin-binding deficient mutant of Pfn1.Citation91 Pfn1 has also been shown to interact directly with transcription factor in the nucleus and modulate gene expression.Citation40 Therefore, nuclear activities of Pfn1 may also play significant role in cell migration through influencing gene expression either in SRF-dependent or -independent mechanisms. A comprehensive understanding of these functions may provide further insights on the molecular basis for context-specific effect on Pfn1 in cell migration.
Acknowledgments
This is supported by grants from the National Cancer Institute from the National Institute of Health (2R01-CA108607-06) and Women’s Cancer Research Center, University of Pittsburgh to P.R.
References
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol 2004; 14:461 - 9; http://dx.doi.org/10.1016/j.tcb.2004.07.003; PMID: 15308213
- Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol 2007; 159:131 - 49; http://dx.doi.org/10.1007/112_2007_704; PMID: 17682948
- Karlsson R, Lindberg U. Profilin, an essential control element for actin polymerization. Actin monomer binding proteins (ed Pekka Lapplainen): Landes Biosciences and Springer 2007;Chapter 3:29-44.
- Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci U S A 2001; 98:3832 - 6; http://dx.doi.org/10.1073/pnas.051515498; PMID: 11274401
- Di Nardo A, Gareus R, Kwiatkowski D, Witke W. Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J Cell Sci 2000; 113:3795 - 803; PMID: 11034907
- Lambrechts A, Braun A, Jonckheere V, Aszodi A, Lanier LM, Robbens J, et al. Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol Cell Biol 2000; 20:8209 - 19; http://dx.doi.org/10.1128/MCB.20.21.8209-8219.2000; PMID: 11027290
- Murk K, Buchmeier S, Jockusch BM, Rothkegel M. In birds, profilin-2a is ubiquitously expressed and contributes to actin-based motility. J Cell Sci 2009; 122:957 - 64; http://dx.doi.org/10.1242/jcs.041715; PMID: 19258389
- Pilo Boyl P, Di Nardo A, Mulle C, Sassoè-Pognetto M, Panzanelli P, Mele A, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J 2007; 26:2991 - 3002; http://dx.doi.org/10.1038/sj.emboj.7601737; PMID: 17541406
- Braun A, Aszódi A, Hellebrand H, Berna A, Fässler R, Brandau O. Genomic organization of profilin-III and evidence for a transcript expressed exclusively in testis. Gene 2002; 283:219 - 25; http://dx.doi.org/10.1016/S0378-1119(01)00855-1; PMID: 11867228
- Obermann H, Raabe I, Balvers M, Brunswig B, Schulze W, Kirchhoff C. Novel testis-expressed profilin IV associated with acrosome biogenesis and spermatid elongation. Mol Hum Reprod 2005; 11:53 - 64; http://dx.doi.org/10.1093/molehr/gah132; PMID: 15591451
- Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol 1977; 115:465 - 83; http://dx.doi.org/10.1016/0022-2836(77)90166-8; PMID: 563468
- Haugwitz M, Noegel AA, Karakesisoglou J, Schleicher M. Dictyostelium amoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell 1994; 79:303 - 14; http://dx.doi.org/10.1016/0092-8674(94)90199-6; PMID: 7954798
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol 1994; 125:1289 - 301; http://dx.doi.org/10.1083/jcb.125.6.1289; PMID: 8207058
- Cao LG, Babcock GG, Rubenstein PA, Wang YL. Effects of profilin and profilactin on actin structure and function in living cells. J Cell Biol 1992; 117:1023 - 9; http://dx.doi.org/10.1083/jcb.117.5.1023; PMID: 1577865
- Markey F, Larsson H, Weber K, Lindberg U. Nucleation of actin polymerization from profilactin. Opposite effects of different nuclei. Biochim Biophys Acta 1982; 704:43 - 51; http://dx.doi.org/10.1016/0167-4838(82)90130-3; PMID: 7201325
- Pring M, Weber A, Bubb MR. Profilin-actin complexes directly elongate actin filaments at the barbed end. Biochemistry 1992; 31:1827 - 36; http://dx.doi.org/10.1021/bi00121a035; PMID: 1737036
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry 1984; 23:6631 - 41; http://dx.doi.org/10.1021/bi00321a054; PMID: 6543322
- Tilney LG, Bonder EM, Coluccio LM, Mooseker MS. Actin from Thyone sperm assembles on only one end of an actin filament: a behavior regulated by profilin. J Cell Biol 1983; 97:112 - 24; http://dx.doi.org/10.1083/jcb.97.1.112; PMID: 6863386
- Hájková L, Nyman T, Lindberg U, Karlsson R. Effects of cross-linked profilin:beta/gamma-actin on the dynamics of the microfilament system in cultured cells. Exp Cell Res 2000; 256:112 - 21; http://dx.doi.org/10.1006/excr.1999.4786; PMID: 10739658
- Nyman T, Page R, Schutt CE, Karlsson R, Lindberg U. A cross-linked profilin-actin heterodimer interferes with elongation at the fast-growing end of F-actin. J Biol Chem 2002; 277:15828 - 33; http://dx.doi.org/10.1074/jbc.M112195200; PMID: 11844798
- Goldschmidt-Clermont PJ, Furman MI, Wachsstock D, Safer D, Nachmias VT, Pollard TD. The control of actin nucleotide exchange by thymosin beta 4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol Biol Cell 1992; 3:1015 - 24; PMID: 1330091
- Tobacman LS, Korn ED. The regulation of actin polymerization and the inhibition of monomeric actin ATPase activity by Acanthamoeba profilin. J Biol Chem 1982; 257:4166 - 70; PMID: 6461652
- Bitko V, Oldenburg A, Garmon NE, Barik S. Profilin is required for viral morphogenesis, syncytium formation, and cell-specific stress fiber induction by respiratory syncytial virus. BMC Microbiol 2003; 3:9 - 20; http://dx.doi.org/10.1186/1471-2180-3-9; PMID: 12740026
- Ding Z, Gau D, Deasy B, Wells A, Roy P. Both actin and polyproline interactions of profilin-1 are required for migration, invasion and capillary morphogenesis of vascular endothelial cells. Exp Cell Res 2009; 315:2963 - 73; http://dx.doi.org/10.1016/j.yexcr.2009.07.004; PMID: 19607826
- Zou L, Jaramillo M, Whaley D, Wells A, Panchapakesa V, Das T, et al. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br J Cancer 2007; 97:1361 - 71; http://dx.doi.org/10.1038/sj.bjc.6604038; PMID: 17940506
- Ding Z, Lambrechts A, Parepally M, Roy P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J Cell Sci 2006; 119:4127 - 37; http://dx.doi.org/10.1242/jcs.03178; PMID: 16968742
- Finkel T, Theriot JA, Dise KR, Tomaselli GF, Goldschmidt-Clermont PJ. Dynamic actin structures stabilized by profilin. Proc Natl Acad Sci U S A 1994; 91:1510 - 4; http://dx.doi.org/10.1073/pnas.91.4.1510; PMID: 8108438
- Holt MR, Critchley DR, Brindle NPJ. The focal adhesion phosphoprotein, VASP. Int J Biochem Cell Biol 1998; 30:307 - 11; http://dx.doi.org/10.1016/S1357-2725(97)00101-5; PMID: 9611773
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, et al. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J 1995; 14:1583 - 9; PMID: 7737110
- Boukhelifa M, Moza M, Johansson T, Rachlin A, Parast M, Huttelmaier S, et al. The proline-rich protein palladin is a binding partner for profilin. FEBS J 2006; 273:26 - 33; http://dx.doi.org/10.1111/j.1742-4658.2005.05036.x; PMID: 16367745
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J 1998; 17:6932 - 41; http://dx.doi.org/10.1093/emboj/17.23.6932; PMID: 9843499
- Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, et al. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell 2004; 7:585 - 95; http://dx.doi.org/10.1016/j.devcel.2004.07.021; PMID: 15469846
- Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell 2004; 7:571 - 83; http://dx.doi.org/10.1016/j.devcel.2004.07.024; PMID: 15469845
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, et al. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J 1997; 16:3044 - 56; http://dx.doi.org/10.1093/emboj/16.11.3044; PMID: 9214622
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 1996; 87:227 - 39; http://dx.doi.org/10.1016/S0092-8674(00)81341-0; PMID: 8861907
- Suetsugu S, Miki H, Takenawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO J 1998; 17:6516 - 26; http://dx.doi.org/10.1093/emboj/17.22.6516; PMID: 9822597
- Boettner B, Govek EE, Cross J, Van Aelst L. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc Natl Acad Sci U S A 2000; 97:9064 - 9; http://dx.doi.org/10.1073/pnas.97.16.9064; PMID: 10922060
- Shao J, Welch WJ, Diprospero NA, Diamond MI. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol Cell Biol 2008; 28:5196 - 208; http://dx.doi.org/10.1128/MCB.00079-08; PMID: 18573880
- Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, et al. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J 1998; 17:967 - 76; http://dx.doi.org/10.1093/emboj/17.4.967; PMID: 9463375
- Lederer M, Jockusch BM, Rothkegel M. Profilin regulates the activity of p42POP, a novel Myb-related transcription factor. J Cell Sci 2005; 118:331 - 41; http://dx.doi.org/10.1242/jcs.01618; PMID: 15615774
- Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch JW, Buchmeier S, Jockusch BM, et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J Biol Chem 1999; 274:37908 - 14; http://dx.doi.org/10.1074/jbc.274.53.37908; PMID: 10608857
- Stüven T, Hartmann E, Görlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J 2003; 22:5928 - 40; http://dx.doi.org/10.1093/emboj/cdg565; PMID: 14592989
- Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature 1985; 314:472 - 4; http://dx.doi.org/10.1038/314472a0; PMID: 2984579
- Hartwig JH, Chambers KA, Hopcia KL, Kwiatkowski DJ. Association of profilin with filament-free regions of human leukocyte and platelet membranes and reversible membrane binding during platelet activation. J Cell Biol 1989; 109:1571 - 9; http://dx.doi.org/10.1083/jcb.109.4.1571; PMID: 2793934
- Ostrander DB, Gorman JA, Carman GM. Regulation of profilin localization in Saccharomyces cerevisiae by phosphoinositide metabolism. J Biol Chem 1995; 270:27045 - 50; http://dx.doi.org/10.1074/jbc.270.45.27045; PMID: 7592954
- Lu PJ, Shieh WR, Rhee SG, Yin HL, Chen CS. Lipid products of phosphoinositide 3-kinase bind human profilin with high affinity. Biochemistry 1996; 35:14027 - 34; http://dx.doi.org/10.1021/bi961878z; PMID: 8909300
- Moens PD, Bagatolli LA. Profilin binding to sub-micellar concentrations of phosphatidylinositol (4,5) bisphosphate and phosphatidylinositol (3,4,5) trisphosphate. Biochim Biophys Acta 2007; 1768:439 - 49; http://dx.doi.org/10.1016/j.bbamem.2006.12.012; PMID: 17275780
- Sohn RH, Chen J, Koblan KS, Bray PF, Goldschmidt-Clermont PJ. Localization of a binding site for phosphatidylinositol 4,5-bisphosphate on human profilin. J Biol Chem 1995; 270:21114 - 20; http://dx.doi.org/10.1074/jbc.270.36.21114; PMID: 7673143
- Lambrechts A, Jonckheere V, Dewitte D, Vandekerckhove J, Ampe C. Mutational analysis of human profilin I reveals a second PI(4,5)-P2 binding site neighbouring the poly(L-proline) binding site. BMC Biochem 2002; 3:12; http://dx.doi.org/10.1186/1471-2091-3-12; PMID: 12052260
- Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J 1997; 16:484 - 94; http://dx.doi.org/10.1093/emboj/16.3.484; PMID: 9034331
- Skare P, Karlsson R. Evidence for two interaction regions for phosphatidylinositol(4,5)-bisphosphate on mammalian profilin I. FEBS Lett 2002; 522:119 - 24; http://dx.doi.org/10.1016/S0014-5793(02)02913-7; PMID: 12095630
- Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development 1994; 120:717 - 28; PMID: 7600952
- Sato A, Khadka DK, Liu W, Bharti R, Runnels LW, Dawid IB, et al. Profilin is an effector for Daam1 in non-canonical Wnt signaling and is required for vertebrate gastrulation. Development 2006; 133:4219 - 31; http://dx.doi.org/10.1242/dev.02590; PMID: 17021034
- Lai SL, Chan TH, Lin MJ, Huang WP, Lou SW, Lee SJ. Diaphanous-related formin 2 and profilin I are required for gastrulation cell movements. PLoS One 2008; 3:e3439; http://dx.doi.org/10.1371/journal.pone.0003439; PMID: 18941507
- Lambrechts A, Jonckheere V, Peleman C, Polet D, De Vos W, Vandekerckhove J, et al. Profilin-I-ligand interactions influence various aspects of neuronal differentiation. J Cell Sci 2006; 119:1570 - 8; http://dx.doi.org/10.1242/jcs.02884; PMID: 16569658
- Michaelsen K, Murk K, Zagrebelsky M, Dreznjak A, Jockusch BM, Rothkegel M, et al. Fine-tuning of neuronal architecture requires two profilin isoforms. Proc Natl Acad Sci U S A 2010; 107:15780 - 5; http://dx.doi.org/10.1073/pnas.1004406107; PMID: 20798032
- Böttcher RT, Wiesner S, Braun A, Wimmer R, Berna A, Elad N, et al. Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J 2009; 28:1157 - 69; http://dx.doi.org/10.1038/emboj.2009.58; PMID: 19262563
- Kullmann JA, Neumeyer A, Gurniak CB, Friauf E, Witke W, Rust MB. Profilin1 is required for glial cell adhesion and radial migration of cerebellar granule neurons. EMBO Rep 2012; 13:75 - 82; http://dx.doi.org/10.1038/embor.2011.211; PMID: 22081137
- Grønborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics 2006; 5:157 - 71; http://dx.doi.org/10.1074/mcp.M500178-MCP200; PMID: 16215274
- Janke J, Schlüter K, Jandrig B, Theile M, Kölble K, Arnold W, et al. Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J Exp Med 2000; 191:1675 - 86; http://dx.doi.org/10.1084/jem.191.10.1675; PMID: 10811861
- Wu N, Zhang W, Yang Y, Liang YL, Wang LY, Jin JW, et al. Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics 2006; 6:6095 - 106; http://dx.doi.org/10.1002/pmic.200500321; PMID: 17051635
- Zoidakis J, Makridakis M, Zerefos PG, Bitsika V, Esteban S, Frantzi M, et al. Profilin 1 is a potential biomarker for bladder cancer aggressiveness. Mol Cell Proteomics 2012; 11:M111 - , 009449; http://dx.doi.org/10.1074/mcp.M111.009449; PMID: 22159600
- Bae YH, Ding Z, Zou L, Wells A, Gertler F, Roy P. Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J Cell Physiol 2009; 219:354 - 64; http://dx.doi.org/10.1002/jcp.21677; PMID: 19115233
- Theriot JA, Rosenblatt J, Portnoy DA, Goldschmidt-Clermont PJ, Mitchison TJ. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell 1994; 76:505 - 17; http://dx.doi.org/10.1016/0092-8674(94)90114-7; PMID: 8313471
- Mimuro H, Suzuki T, Suetsugu S, Miki H, Takenawa T, Sasakawa C. Profilin is required for sustaining efficient intra- and intercellular spreading of Shigella flexneri. J Biol Chem 2000; 275:28893 - 901; http://dx.doi.org/10.1074/jbc.M003882200; PMID: 10867004
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 1999; 401:613 - 6; http://dx.doi.org/10.1038/44183; PMID: 10524632
- Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol 2010; 191:571 - 84; http://dx.doi.org/10.1083/jcb.201003014; PMID: 21041447
- Suetsugu S, Miki H, Takenawa T. Distinct roles of profilin in cell morphological changes: microspikes, membrane ruffles, stress fibers, and cytokinesis. FEBS Lett 1999; 457:470 - 4; http://dx.doi.org/10.1016/S0014-5793(99)01086-8; PMID: 10471831
- Myers SA, Han JW, Lee Y, Firtel RA, Chung CY. A Dictyostelium homologue of WASP is required for polarized F-actin assembly during chemotaxis. Mol Biol Cell 2005; 16:2191 - 206; http://dx.doi.org/10.1091/mbc.E04-09-0844; PMID: 15728724
- Geese M, Loureiro JJ, Bear JE, Wehland J, Gertler FB, Sechi AS. Contribution of Ena/VASP proteins to intracellular motility of listeria requires phosphorylation and proline-rich core but not F-actin binding or multimerization. Mol Biol Cell 2002; 13:2383 - 96; http://dx.doi.org/10.1091/mbc.E02-01-0058; PMID: 12134077
- Gau D, Ding Z, Baty C, Roy P. Fluorescence Resonance Energy Transfer (FRET)-based Detection of Profilin-VASP Interaction. Cell Mol Bioeng 2011; 4:1 - 8; http://dx.doi.org/10.1007/s12195-010-0133-z; PMID: 21566724
- Li Y, Grenklo S, Higgins T, Karlsson R. The profilin:actin complex localizes to sites of dynamic actin polymerization at the leading edge of migrating cells and pathogen-induced actin tails. Eur J Cell Biol 2008; 87:893 - 904; http://dx.doi.org/10.1016/j.ejcb.2008.06.003; PMID: 18707793
- Bubb MR, Yarmola EG, Gibson BG, Southwick FS. Depolymerization of actin filaments by profilin. Effects of profilin on capping protein function. J Biol Chem 2003; 278:24629 - 35; http://dx.doi.org/10.1074/jbc.M302796200; PMID: 12730212
- Yarmola EG, Bubb MR. How depolymerization can promote polymerization: the case of actin and profilin. Bioessays 2009; 31:1150 - 60; http://dx.doi.org/10.1002/bies.200900049; PMID: 19795407
- Yarmola EG, Dranishnikov DA, Bubb MR. Effect of profilin on actin critical concentration: a theoretical analysis. Biophys J 2008; 95:5544 - 73; http://dx.doi.org/10.1529/biophysj.108.134569; PMID: 18835900
- Yarmola EG, Bubb MR. Profilin: emerging concepts and lingering misconceptions. Trends Biochem Sci 2006; 31:197 - 205; http://dx.doi.org/10.1016/j.tibs.2006.02.006; PMID: 16542844
- Bae YH, Ding Z, Das T, Wells A, Gertler F, Roy P. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc Natl Acad Sci U S A 2010; 107:21547 - 52; http://dx.doi.org/10.1073/pnas.1002309107; PMID: 21115820
- Michael M, Vehlow A, Navarro C, Krause M. c-Abl, Lamellipodin, and Ena/VASP proteins cooperate in dorsal ruffling of fibroblasts and axonal morphogenesis. Curr Biol 2010; 20:783 - 91; http://dx.doi.org/10.1016/j.cub.2010.03.048; PMID: 20417104
- Roussos ET, Balsamo M, Alford SK, Wyckoff JB, Gligorijevic B, Wang Y, et al. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. J Cell Sci 2011; 124:2120 - 31; http://dx.doi.org/10.1242/jcs.086231; PMID: 21670198
- Roussos ET, Goswami S, Balsamo M, Wang Y, Stobezki R, Adler E, et al. Mena invasive (Mena(INV)) and Mena11a isoforms play distinct roles in breast cancer cell cohesion and association with TMEM. Clin Exp Metastasis 2011; 28:515 - 27; http://dx.doi.org/10.1007/s10585-011-9388-6; PMID: 21484349
- Roussos ET, Wang Y, Wyckoff JB, Sellers RS, Wang W, Li J, et al. Mena deficiency delays tumor progression and decreases metastasis in polyoma middle-T transgenic mouse mammary tumors. Breast Cancer Res 2010; 12:R101; http://dx.doi.org/10.1186/bcr2784; PMID: 21108830
- Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim HD, Giampieri S, et al. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell 2008; 15:813 - 28; http://dx.doi.org/10.1016/j.devcel.2008.09.003; PMID: 19081071
- Goswami S, Philippar U, Sun D, Patsialou A, Avraham J, Wang W, et al. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin Exp Metastasis 2009; 26:153 - 9; http://dx.doi.org/10.1007/s10585-008-9225-8; PMID: 18985426
- Smith K, Humphreys D, Hume PJ, Koronakis V. Enteropathogenic Escherichia coli recruits the cellular inositol phosphatase SHIP2 to regulate actin-pedestal formation. Cell Host Microbe 2010; 7:13 - 24; http://dx.doi.org/10.1016/j.chom.2009.12.004; PMID: 20114025
- Goldschmidt-Clermont PJ, Kim JW, Machesky LM, Rhee SG, Pollard TD. Regulation of phospholipase C-gamma 1 by profilin and tyrosine phosphorylation. Science 1991; 251:1231 - 3; http://dx.doi.org/10.1126/science.1848725; PMID: 1848725
- Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, Pollard TD. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science 1990; 247:1575 - 8; http://dx.doi.org/10.1126/science.2157283; PMID: 2157283
- Das T, Bae YH, Wells A, Roy P. Profilin-1 overexpression upregulates PTEN and suppresses AKT activation in breast cancer cells. J Cell Physiol 2009; 218:436 - 43; http://dx.doi.org/10.1002/jcp.21618; PMID: 18937284
- Bear JE, Loureiro JJ, Libova I, Fässler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell 2000; 101:717 - 28; http://dx.doi.org/10.1016/S0092-8674(00)80884-3; PMID: 10892743
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 2002; 109:509 - 21; http://dx.doi.org/10.1016/S0092-8674(02)00731-6; PMID: 12086607
- Lacayo CI, Pincus Z, VanDuijn MM, Wilson CA, Fletcher DA, Gertler FB, et al. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol 2007; 5:e233; http://dx.doi.org/10.1371/journal.pbio.0050233; PMID: 17760506
- Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 1999; 98:159 - 69; http://dx.doi.org/10.1016/S0092-8674(00)81011-9; PMID: 10428028