Abstract
Stromal chemokine gradients within the breast tissue microenvironment play a critical role in breast cancer cell invasion, a prerequisite to metastasis. To elucidate which chemokines and mechanisms are involved in mammary cell migration we determined whether mesenchymal D1 stem cells secreted specific chemokines that differentially promoted the invasion of mammary tumor cells in vitro. Results indicate that mesenchymal D1 cells produced concentrations of CCL5 and CCL9 4- to 5-fold higher than the concentrations secreted by 4T1 tumor cells (P < 0.01). Moreover, 4T1 tumor cell invasion toward D1 mesenchymal stem cell conditioned media (D1CM), CCL5 alone, CCL9 alone or a combination CCL5 and CCL9 was observed. The invasion of 4T1 cells toward D1 mesenchymal stem CM was dose-dependently suppressed by pre-incubation with the CCR1/CCR5 antagonist met-CCL5 (P < 0.01). Furthermore, the invasion of 4T1 cells toward these chemokines was prevented by incubation with the broad-spectrum MMP inhibitor GM6001. Additionally, the addition of specific MMP9/MMP13 and MMP14 inhibitors prevented the MMP activities of supernatants collected from 4T1 cells incubated with D1CM, CCL5 or CCL9. Taken together these data highlight the role of CCL5 and CCL9 produced by mesenchymal stem cells in mammary tumor cell invasion.
Introduction
In addition to tumor cells, solid tumors are composed of multiple stroma cell types that modulate the density and composition of a complex extracellular matrix (ECM) and of a large array of soluble and bound molecules creating a unique microenvironment that influence tumor progression.Citation1-Citation3 Within the breast tumor microenvironment mesenchymal-derived cells, including adipose cells, the most abundant of the stromal cells of the breast, and fibroblasts secrete a variety of chemokines, growth factors, and cytokines that modulate cancer progression and metastasis.Citation4-Citation6 In particular, the chemokine network can greatly affect the progression and metastasis of breast cancer through the modulation of tumor cell death, proliferation, ECM attachment, proteolysis of the basement membrane, locomotion, and colony formation, but also cell trafficking and adhesion, the local inflammatory response and the redirection to the tumor site of immune effector and stem cells.Citation7-Citation12
Among chemokines that modulate breast cancer progression, CCL5 (RANTES), which binds to mostly to CCR5 but also to CCR1 has been extensively studied.Citation13-Citation16 Increased expressions of CCL5 have been correlated with breast cancer progression, particularly with triple negative breast cancer.Citation17-Citation19 Indeed, CCL5 overexpressed in tumors promote cancer cell proliferation, migration, invasion, and survival in a dose-dependent manner mostly through functional CCR5 receptors.Citation20-Citation24 In particular, CCL5 is also produced by tissue resident stem cells under the influence of cancer cells.Citation15,Citation25-Citation27 The secretion of CCL9 (MIP-1γ) is increased in colorectal carcinoma cells along with the heightened expression of its the cognate receptor CCR1 onto myeloid cells.Citation28,Citation29 These interactions between tumor cells and mesenchymal stem cells highlight a feedback mechanism that promotes tumor cell motility, invasion, growth, survival, and the overall metastatic potency.Citation14,Citation16 Furthermore, the concomitant expression of multiple chemokines in the human breast cancer microenvironment may contribute independently to breast malignancy.Citation30
Chemokines, CCL5 and CCL9 especially, modulates changes in the adhesion of the tumor cells in part through stimulating the release of matrix metalloproteinases (MMPs) leading to proteolytic degradation of the extracellular matrix.Citation31,Citation32 CCL-5 promoted tumor metastasis through stimulation of MMP-2 and MMP-9 secretions in both leukocytes and tumor cells,Citation33-Citation35 and in 4T1 tumor mass in vivo.Citation36 Mostly, stroma-derived MMP-9 and MMP-13 and also tumor-derived MMP-1, MMP-2, and MMP-14 have been shown to contribute to invasive tumor growth and metastasis.Citation37-Citation39
Given the complexity of the interactions between stroma and tumor cells within the breast tumor microenvironment, in vitro analyses of cell–cell interactions are essential models to provide a detailed understanding of the role of chemokines. Despite their limitations, in vitro models using cell lines have been utilized extensively and are invaluable. Recently, various 3D models of breast tissues have been described and used to investigate the stroma tumor cell interactions and anticancer drug efficacy.Citation40-Citation42 We, and others, have also investigated parameters from the local oxygen concentrations, the density of the ECM to the formation of chemokine heterodimers that can modulate cancer progression in in vitro models.Citation43-Citation45 Furthermore, normal murine mammary gland (NMuMG) cells formed acini only in the presence of murine D1 mesenchymal stem cells whereas 4T1 murine tumor cells did not.Citation46
To determine whether chemokines present within the breast tumor microenvironment, especially CCL5 and CCL9, promoted the invasion of mammary tumor cells, we first determined the effects of molecules secreted by epithelial (NMuMG), tumor (4T1) cells, and mesenchymal D1 stem cells on NMuMG and 4T1 cell invasions using 2D and 3D culture conditions. Further, we identified the predominant role of CCL5 and CCL9 in the invasion of 4T1 tumor cells through CCR1 and CCR5 receptor activation and the secretion of MMPs.
Results
Conditioned media from mammary epithelial NMuMG cells and D1 mesenchymal stem cells differentially affects the invasion of NMuMG and 4T1 cells
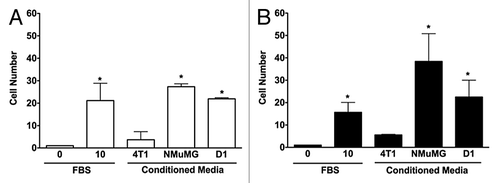
The 3D invasions of NMuMG and 4T1 cells were significantly different in response to various CMs tested in 3D cultures. As shown in , cells migrated within a 0.8% agarose 3D matrix toward the different CMs tested. Both NMuMG and 4T1 cells migrated toward mammary epithelial NMuMG cell and mesenchymal stem D1 cell CMs. However, CMs derived from mammary tumor 4T1 cells did not have any effect on the invasion of either NMuMG or 4T1 cells. In contrast, using wound healing assays (data not shown) no changes were noticed in the migration speed of 4T1 or NMuMG cells compared with the negative control (0% FBS) in monolayer repair over time measured following a 6 h incubation.
Figure 1. 3D invasions of NMuMG and 4T1 cells are increased by conditioned media from both mammary epithelial cells and D1 mesenchymal stem cells. Migration of NMuMG (A) and 4T1 cells (B) following a 5-d incubation with CMs derived from 4T1, NMuMG, and the mesenchymal stem D1 cells were quantified using 3D migration assays (see Materials and Methods section for details). Serum-free and 10% FBS–DMEM media were used as negative and positive controls, respectively. Representative microphotographs of cells migrating out of the wells for each condition were captured following the 5-d incubation. In experiments repeated at least 3 times, the cells moving toward different control and CMs were counted. Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05 compared with negative control (0% FBS).
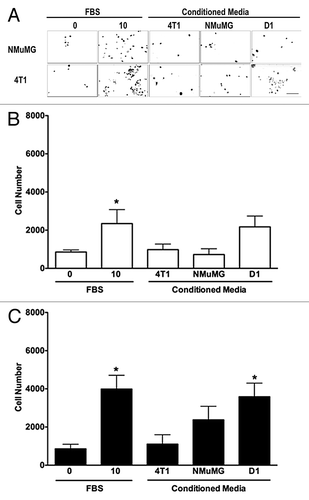
Conditioned media collected from D1 mesenchymal stem cells increases the invasion of 4T1 cells but not of NMuMG cells
To further define the effects of D1CM on cell migrations, the invasion of both NMuMG and 4T1 cells toward various CMs were determined using transwell migration assays. The D1 mesenchymal stem cell CM significantly increased the invasion of 4T1 cells (P < 0.05, ). No significant change in invasion toward any of the D1CM tested was observed with NMuMG cells (P > 0.05, ).
Figure 2. D1 mesenchymal stem cell conditioned media promotes the invasion of 4T1 cells but not NMuMG cells. Invasion of NMuMG and 4T1 breast cells placed in the upper chamber of Matrigel®-coated transwells toward a bottom chamber filled with conditioned media (1:1 dilution) collected from 4T1, NMuMG, or mesenchymal stem D1 cells was evaluated following a 24-h incubation. Representative microphotographs of NMuMG and 4T1 cells labeled with the vital nuclear dye Hoechst that migrated through Matrigel®-coated transwell membranes toward lower compartments filled with each treatment are presented (A). All microphotographs were taken in the same conditions of fluorescence and magnification (bar scale = 100 μm, lower right), converted to gray scale and inverted. Serum-free (0% FBS) and 10% FBS media were used as negative and positive control, respectively. The invasion was evaluated by counting the number of NMuMG (B) and 4T1 (C) cells on at least 5 representative pictures for each condition. The number of cells was then normalized to the surface area of the transwell membrane. Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05 compared with cells migrating toward media (0% FBS) alone.
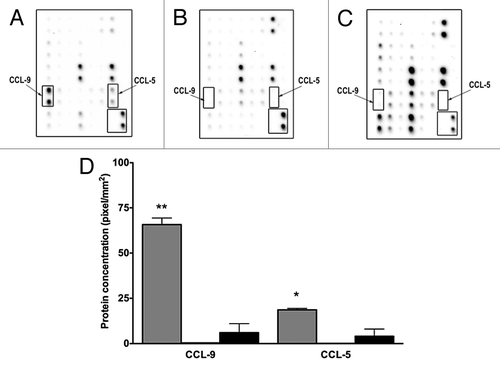
Chemokines CCL-5 and CCL-9 concentrations were higher in conditioned media derived from D1 cells
To determine the specific molecules differentially present in mesenchymal stem D1 cell CM compared with MNuMG and 4T1 CMs, the expression of chemokines and cytokines was evaluated using antibody arrays. Expressions of both CCL-5 and CCL-9 chemokines were significantly increased in mesenchymal stem D1 cell CM compared with the CMs derived from 4T1 cells (P < 0.05, ). Additionally, CXCL-16, MIP-1α and soluble TNF α receptor 2 were also decreased in CM derived from D1 cells (not shown).
Figure 3. D1 mesenchymal stem cell conditioned media contains higher CCL-5 and CCL-9 concentrations than conditioned media collected from NMuMG mammary epithelial and 4T1 tumor cells. Higher concentrations of CCL5 and CCL9 were detected in CM collected from mesenchymal stem cells (D1) (A), than in CM obtained from mammary epithelial cells (NMuMG) (B) and mammary tumor cells (4T1) (C) using cytokine protein arrays. This increase was semi-quantified (D) in CMs collected from D1 (gray bars), and in 4T1 (black bars) cells. Both CCL5 AND CCL9 expressions were very low in NMuMG conditioned media (below the detection limit, not shown). Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05, **P < 0.01 compared with 4T1 conditioned media.
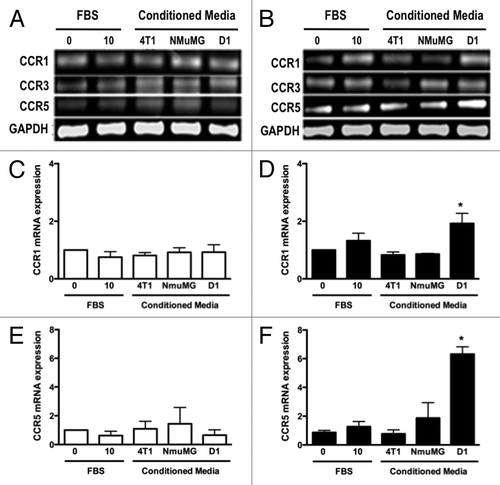
D1 mesenchymal stem cell conditioned media promoted the CCR1 and CCR5 mRNA in 4T1 cells but not NMuMG cells
We next determined whether high concentrations of CCL5 and CCL9 present in D1 mesenchymal stem cell CM influenced the mRNA and protein expression of CCL5 and CCL9 receptors by mammary epithelial NMuMG and mammary tumor 4T1 cells. The mRNA expression of CCR 1 and 5 receptors for CCL5, and CCR1, 3 and 5 receptors for CCL9, respectively, in 4T1 and NMuMG cells were evaluated. As shown , CCR3 mRNA expression was not altered regardless of the CMs or the cells tested. In contrast, while the expression of CCR1 and CCR5 receptors in the NMuMG cells remained unchanged following D1CM treatment, the expression of CCR1 and CCR5 was significantly increased in the 4T1 cells treated with D1CM compared with the 4T1 cells treated with either NMuMG CM or 4T1 CM and control conditions (P < 0.05, ).
Figure 4. D1 mesenchymal stem cell conditioned media stimulated the mRNA expressions of CCR1 and CCR5 in 4T1 cells but not in NMuMG cells. The mRNA expression for CCR1, CCR3 and CCR5 was evaluated and quantified by RT-PCR in RNA collected from NMuMG (A, C, and E) and 4T1 (B, D, and F) cells following a 3 h incubation with controls and CMs from 4T1, NMuMG and D1 mesenchymal stem cells, respectively. The primer sequences are listed in . Serum-free (0% FBS) and 10% FBS media served as negative and positive control, respectively. Using experiments repeated at least three times, CCR1 (C and D) and CCR5 (E and F) mRNA expressions in NMuMG (C and E) and 4T1 (E and F) cells normalized to loading control (GAPDH) mRNA expression were quantified. The expression of CCR3 mRNA was unaffected regardless of the treatments (P > 0.05, not shown). Results expressed as fold increase compared with 0% FBS control were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05 compared with negative control (0% FBS).
D1 mesenchymal stem cell conditioned media increased the CCR5 cell surface expression in 4T1 cells
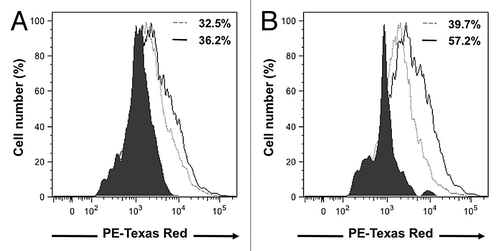
The cell surface expressions of CCR1 and CCR5 receptor on 4T1 cells in serum-free media or following incubations with D1CM were determined by flow-cytometry (). Compared with control conditions, the percentage of 4T1 cells positive for CCR1 receptors following incubation with D1CM appeared to remain unchanged (). In contrast, incubation with D1CM led to an increase in the percentage of 4T1 cells expressing CCR5 on their cell surface compared with 4T1 cells incubated in control conditions ().
Figure 5. D1 mesenchymal stem cell conditioned media increased cell surface expression of CCR1 and CCR5 receptors in 4T1 cells. Representative flow-cytometry histograms that depict the CCR1 (A) and CCR5 (B) receptors expressed on the cell surface of 4T1 cell following a 24 h incubation in the presence of control media (dotted gray line) or D1CM (solid dark line). Staining with the secondary antibody alone (Texas red conjugated anti-goat antibody) is depicted as solid black area. These results are representative of at least three separate experiments. Results indicate that incubation with D1CM led to a limited and a larger cell surface expression of CCR1 and CCR5, respectively.
The antagonist to CCR5 and CCR1 Met-CCL5 inhibited D1CM-driven 4T1 cell invasion
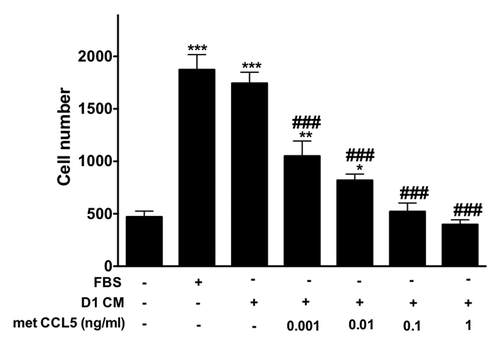
To confirm the effects of CCL5 and CCL9 on the migration of 4T1 cells, we measured the invasion of 4T1 cells in response to CM obtained from D1 cells in the presence of different concentrations of Met-CCL5 an antagonist to both chemokine receptors CCR1 and CCR5 (). Treatment with Met-CCL5 reduced the invasion of 4T1 cells in response to D1CM in a dose-dependent manner (P < 0.05, ). Met-CCL5 above 0.01 ng/ml caused significant reductions in 4T1 cell invasion ().
Figure 6. The inhibitor of CCR1 and CCR5 Met-CCL5 inhibits 4T1 cells invasion toward D1 mesenchymal stem cell conditioned media. The number of 4T1 cells migrating through uncoated and Matrigel®-coated transwell chamber was used to evaluate the effects of increasing concentrations of met-CCL5, an inhibitor of both CCR1 and CCL5 on the invasion promoted by mesenchymal stem D1 cell CM (for details see materials and Methods section). Serum-free (0% FBS) and 10% FBS media serve as negative and positive control, respectively. The numbers of migrating 4T1 cells normalized to the surface area of the transwell membrane are presented. Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001 compared with 0% FBS control; #P < 0.05, ##P < 0.01, ###P < 0.001, compared with migration of 4T1 cells toward D1CM alone.
4T1 cell invasion was promoted by CCL5 and CCL9 alone or combined and inhibited by MMP inhibitors
4T1 cell invasion toward CCL5 or CCL9 alone or in combination increased (P < 0.01 compared with media alone, ) but remained lower than the invasion observed with D1CM. Interestingly, the 4T1 invasion toward the combination CCL5 and CCL9 was not different from the 4T1 invasion toward CCL5 or CCL9 alone ().
Figure 7. The increased 4T1 cell invasion promoted by CCL5 and CCL9 alone was prevented through pre-treatment with the MMP inhibitor GM6001. Briefly, 4T1 tumors cells were pre-incubated without (black bars) or with GM6001 (1 μM, hachured bars) for 30 min prior to invasion assays. Invasion assays were conducted using transwell chambers (for details see Materials and Methods section). The bottom chamber was filled with either media alone, CCL5 (12.5μM), CCL9 (12.5μM) the combination of both or DICM. The numbers of migrating 4T1 cells normalized to the surface area of the transwell membrane are presented. Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001 compared with migration of 4T1 cells toward 0% FBS alone.
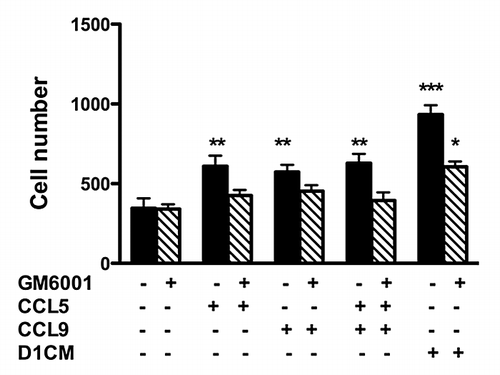
Next we investigated the role of MMPs in 4T1 cells invasions. First, in 4T1 cells incubated with D1CM, the addition of the chemokine receptor antagonist Met-CCL5 lead decreased MMP-9, MMP-13, and MMP-14 expression as detected by western blots analyses (not shown). To further assess the role of MMPs, 4T1 cell invasion in the presence of the wide-spectrum MMP inhibitor GM6001 were conducted. As shown (hachured bars), the pre-incubation of 4T1 cells with GM6001 (1 μM) led to the inhibition of the invasion toward CCL5, CCL9, or the combination CCL5–CCL9. Only, 4T1 invasion toward D1CM was observed (). However, GM6001 treatment significantly inhibited 4T1 cell invasion even toward D1CM ().
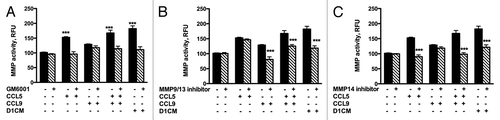
Changes in the expressions of specific MMPs were further assessed using functional assays (). No detectable amount of any of the MMP’s tested was measured in D1CMs (not shown). The concentrated supernatants collected from 4T1 cells incubated with D1CM, CCL5, CCL9 or the combination CCL5 and CCL9 were tested for their abilities to cleave the fluorescent MMP substrate. Supernatants of 4T1 cells incubated with D1CM, CCL5, CCL9, or the combination CCL5 and CCL9 had significantly higher MMP activities (darken bars) compared with media alone (P < 0.001, ). The addition of GM6001 (10 nM) to the enzymatic reaction decreased MMP activities to levels similar to those observed following incubation with control media (, ns). The addition of the MMP9/MMP13 inhibitor was associated with decreases in MMP activities of 4T1 cell supernatants collected following treatments with CCL9, the combination CCL5/CCL9 and D1CM but not with CCL5 alone (P < 0.001, ). The addition of the MMP14 inhibitor led to decrease in MMP activities of 4T1 supernatants collected following treatments with CCL5, the combination CCL5–CCL9 and D1CMs but not with CCL9 alone (P < 0.001, ).
Figure 8. D1 mesenchymal stem cell CM, CCL5, and CCL9 promoted the activities of MMPs including MMP9/13 and MMP14. MMP activities of supernatants collected from 4T1 cells incubated 24 h with either media alone, CCL5, CCL9, or D1CM were freeze-dried and concentrated to 1/10 of their initial volume were assayed according to the manufacturer’s recommendations (see Materials and Methods for details). To determine whether the activities of MMP9/13 or MMP14 were altered by either CCL5, CCL9, the combination CCL5–CCL9 or D1CM, specific inhibitors of MMP9/13 (10 nM) and MMP14 (50 nM), along with the general MMP inhibitor GM6001 (10 nM) were added to the reaction. Data are expressed in relative fluorescence unit (RFU). Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. ***P < 0.001 compared with MMP activities of 4T1 cells incubated with 0% FBS alone.
Discussion
Within the breast tumor microenvironment, stromal cells including mesenchymal stem cells contribute to the development of breast cancer.Citation5,Citation14,Citation47,Citation48 For example, D1 mesenchymal stem cells in culture with NMuMG cells promoted the generation of 3D acinus and duct-like structures whereas NMuMG mammary epithelial cells alone did not.Citation46 Here we investigated the effects of stroma mesenchymal D1 stem cells’ secretions especially the chemokines CCL5 and CCL9 on 4T1 invasion. Our results indicate that both CCL5 and CCL9 chemokines were produced at high concentrations by D1 mesenchymal stem cells and promoted tumor cell invasion likely through CCR1 and CCR5 receptors present on mammary 4T1 tumor cells. The invasion of 4T1 cells was dependent on MMP activities including MMP9 and/or MMP13 and MMP14 stimulated by CCL5 and CCL9 through the activation of CCR1 and/or CCR5 receptors.
In vivo, the role of stroma cells in the promotion of breast tumor progression has been demonstrated in immune-compromised mice.Citation14-Citation16 Indeed, mammary carcinoma co-implanted with mesenchymal stem cells in immune compromised mice led to significant increase in metastatic lesions.Citation14,Citation16 Stroma cell including mesenchymal stem cells and tumor cell interactions through cell-to-cell contacts or soluble mediators control most aspects of tumor formation and progression.Citation8,Citation14 Our observations also indicate that D1 mesenchymal stem cells express high concentrations of chemokines that promoted the invasion of the 4T1 mammary tumor cells. Furthermore, D1 cell secretions tended to differently stimulate NMuMG epithelial mammary cells and 4T1 tumor cells, highlighting the role of mesenchymal stem cell secretion in malignant cells dissemination.Citation14,Citation49,Citation50 These observations confirmed previously published in vivo dataCitation13,Citation14 and thus support the use in vitro models systems to study the invasion steps of the breast cancer progression using mammary tumor cells and mesenchymal cell secretion and/or conditioned media.Citation45,Citation46
Our data confirm that D1 mesenchymal stem cells secrete and/or shed many different molecules including the CCL5 and CCL9 chemokines at concentrations much higher than mammary epithelial and tumor cells and that CCL5 and CCL9 in particular promoted 4T1 mammary cell invasion. Those chemokines, CCL5 especially, have been shown to play key roles in breast cancer cell migration as major regulators of cell trafficking and adhesion including of tumor cells, immune cells, and stem cells.Citation51,Citation52 The CCL5 production by D1 mesenchymal stem cells shown here appears to mimic the CCL5 production by tissue resident stem cells under the influence of cancer cells leading to the promotion of breast cancer cell proliferation, dissemination and invasion described earlier.Citation14,Citation15,Citation25-Citation27 In immune competent mice, 4T1 cells expressing high levels of CCL5 led to significantly higher lung and liver metastasis.Citation36 In vivo CCL5 secreted by breast tumor cells promoted monocyte migration to the tumor site and stimulated the expression of pro-inflammatory cytokines thereby facilitating metastasis formation,Citation53 highlighting the multiple targets and mechanisms of CCL5 within the tumor microenvironment. CCL9 has been shown to promote colon cancer progression,Citation28 and our data suggest that CCL9 may also have a role in the invasion of breast cancer cells.
Furthermore, since the combination CCL5–CCL9 was not associated with significant increase in 4T1 cells migration when compared with each chemokine alone may indicate that both chemokine share similar signaling pathways.Citation5 Moreover, CCL5 or CCL9 alone or in combination only partially generated the invasion observed in the presence of D1CM, suggesting that other molecules including chemokines and mechanisms are likely involved in the promotion of 4T1 cell invasion. Whether physical interactions through the formation of heterodimers may also modulate the breast tumor cell responses to CCL5 and CCL9 combinations remains to be defined.Citation44 The secretion and shedding of multiple molecules in addition to CCL5 and CCL9 may explain the higher 4T1 cell invasion observed in the presence of D1CM compared with CCL5 and CCL9 alone or in combination. In addition to CCL5 and CCL9 chemokines, D1 mesenchymal stem cells like other stromal cells also produce other molecules including interleukin 6, that also promote migration and invasion of breast cancer cells.Citation4,Citation6 And some of those molecules including the chemokines such as CCL5 and CCL2 have been shown to contribute, independently, to breast malignancy.Citation30
Here treatments with Met-CCL5 an antagonist to primarily CCR5, CCR1 lead to a dose-dependent inhibition of 4T1 cell invasion supporting the involvement of those receptor in the signaling promoting cell invasion including MMP expression as described previously.Citation13,Citation14 The antagonist Met-CCL5 has been shown to inhibits multiple cellular processes in part through slower internalization and altered trafficking.Citation13,Citation54 CCL5 inhibitors have been shown to block metastasis of breast cancer cells,Citation55 and Met-CCL5 inhibited breast tumor growth,Citation13 highlighting the therapeutic potential of CC chemokine receptor antagonists.
The chemokines present in the D1CM including CCL5 and CCL9 promoted the expression and activities of MMPs essential for the invasion of 4T1 cells. MMPs through the remodeling of the extracellular matrix and of cell–ECM and cell–cell contacts, facilitate the detachment of tumor cells from the surrounding tissue within both the primary or the metastatic site.Citation31,Citation56-Citation58 Here, inhibitions of the MMP activities led to reduced invasion. The use of specific inhibitors to MMP9/MMP13 or MMP14 indicated that these MMPs, all of which have been associated with tumor invasion and metastasis,Citation37,Citation38 were involved in 4T1 invasion. Possibly associated with the MMP detection methods used,Citation59,Citation60 the differences observed here between 4T1 cell invasion following a large-spectrum MMP inhibitor and the in vitro activities of MMPs secreted and/or shed by 4T1 cells, and in the specific in vitro activities themselves, notably following incubations with CCL5 and CCL9, remains to be fully investigated. Nonetheless, the mechanisms of 4T1 invasion observed here through chemokines, including CCL5 and CCL9 produced by mesenchymal cells that upon binding to CC receptors, led to increased MMP activities including of MMP9/MMP13 and MMP14. These observations parallel increases in CCL9 that promoted the recruitment of stroma cells expressing CCR1 and secreting MMPs in colon cancer,Citation28 and the CCL5 stimulated expression of MMPs by multiple stroma cells including immune cells.Citation35,Citation61
Taken together the data presented strongly suggest that 4T1 mammary tumor cell invasion in vitro is promoted by D1 mesenchymal stem cell secretions, especially the chemokines CCL5 and CLC9. They also indicate that this invasion is dose-dependent of the activation of CC chemokine receptors and result in increased MMP activities, especially of MMP9 or MMP13 and MMP14. Furthermore, they highlight the utility of in vitro models in the dissection of the complex interactions between cell types within the tumor microenvironment. These observations further emphasize the key role of stroma cell secretions in breast tumor cell invasion. Indeed, tumor cell chemotactic responses may influence the migratory traits of sub-populations within the tumor and potentially contribute to their in vivo behavior, growth, and survival as suggested earlier.Citation62 Furthermore, these chemokines are also produced in large amounts by other stroma cells including adipocytes and given the role and complexity of the interactions chemokine–chemokine and chemokine–chemokine receptor in the breast tumor microenvironment, their actions on the tumor cell behavior need to be carefully assessed.
Materials and Methods
Cell cultures and media
Normal mouse mammary gland cells (NMuMG), metastatic mouse mammary cells (4T1), and mesenchymal murine stem (D1) cells were obtained from ATCC. Cells were cultured at 37 °C and 5% CO2 in DMEM media supplemented with 10% fetal bovine serum (FBS), gentamycin, and amphotericin B. Media for NMuMG cells was supplemented with 10 μg/ml of insulin (Sigma). Other media supplies were obtained from Mediatech.
Conditioned media collection
NMuMG, 4T1 and D1 cells were cultured at 37 °C and 5% CO2 in DMEM media supplemented with 10% FBS, gentamycin, and amphotericin B. Upon confluence, cells were cultured for 24 h in media without FBS and then incubated with RPMI media without phenol red. After a 48 h incubation, conditioned media (CMs) were collected and centrifuged to eliminate debris. Following a filtration (0.2 μm filter, BD Biosciences) step, CMs were stored at −20 °C until use. For migration and invasion assays, the CM concentration used was a 1:1 (1 part media, 1 part CM) dilution to prevent the possible masking effects of lower nutrient (e.g., amino acids and glucose) contents in CMs. For western blots, CMs were freeze-dried and reconstituted in a small volume of sterile water concentrating the proteins secreted from each cell supernatant by 40-fold.
Wound healing assays
NMuMG and 4T1 cells were plated in 6-well plates and incubated in DMEM media supplemented with 10% FBS for 24 h. The plates were then incubated in serum free media for 24 h. Cell monolayers were scratched using sterile 20 μl pipette tips, and cell cultures were washed with PBS. CMs (1:1 dilution) were added and the wound size recorded (micro-photographs) over time at 0, 3, 6, and 24 h. For each time point and treatment, migratory distances (μm) were measured at 8 different locations per condition following image acquisition using a BioChemi Camera (2/3” Cooled Monochrome CCD/High-Res/12bit) and the VisionWorks software (UVP imaging system, UVP).
3D migration assays
To study invasion in 3D conditions, NMuMG and 4T1cells (8 × 103 cells/well) were seeded in the center well made in a sterile 0.8% agarose gel composed of multiple wells prepared as described previously.Citation63 Prior to seeding, cells were briefly incubated with the vital nuclear dye Hoechst (1:2000 dilution, Invitrogen) to enable cell visualization using fluorescent microscopes (excitation 350 nm, emission 450 nm). Cells were cultured with DMEM media without FBS and supplemented with gentamycin and amphotericin B. CMs (1:1 dilution) were placed in the wells surrounding the center well, which contained either 4T1 or NMuMG cells. Serum-free (0% FBS) and 10% FBS-DMEM media were used as negative and positive controls, respectively. At least five random representative microphotographs of the cells migrating out of the center well toward each of the conditions were recorded after 5 d of culture. The numbers of cells moving toward each chemotactic condition were numerated and the 3D chemotactic potential of each conditioned medium defined.
Invasion assays
Following an incubation with the vital nuclear dye Hoechst (1:2000 dilution), NMuMG, or 4T1 cells (5 × 104 cells per well) were seeded in serum free media in the upper compartment of 24 well plate transwell migration chambers (BD Biosciences). Upper compartments were coated with growth factor reduced Matrigel® (50 μl, BD-Biosciences) to evaluate the invasion associated with different conditions. The lower chamber was filled with 300 μL of either CCL5 (12.5 μM; R&D Systems) alone, CCL9 (12.5 μM; R&D Systems) alone, the combination CCL5 (12.5 μM) and CCL9 (12.5 μM), or D1CM (1:1 dilution) in serum free (0%) media. Serum-free (0% FBS) and media supplemented with 10% FBS served as negative and positive control, respectively. After a 24 h incubation at 37 °C, 5% CO2, cells that did not migrate to the lower chamber were removed from the upper surface of the transwell membrane. Cells at the lower surface of the membrane were counted in at least 5 different random fields for each condition.
Inhibition of CCR1 and CCR5 receptors
4T1 cells were pretreated with increasing concentrations (0–100 ng/ml) of the antagonist to CCR1 and CCR5 Met-CCL5 (R&D Systems) for 30 min. The modified CCR5 ligand Met-CCL5 has been shown to antagonize receptor activation and function in response to its natural ligands in part through a slower internalization and a trafficking independent of recycling endosomes.Citation13,Citation54 Although Met-CCL5 binding to CCR5 has been documented in multiple cells, binding of Met-CCL5 to CCR1 chemokine receptor has also been shown to inhibit multiple eosinophil functions including Ca2+ intracellular trafficking, actin polymerization, reactive oxygen species release.Citation64 Following an incubation with the live nuclear dye Hoechst 4T1 cells were seeded in the upper chamber of the transwell in serum-free media (0% FBS). Invasion studies were conducted in a 24-well plate in which the transwell membrane upper compartments were coated with growth factor reduced Matrigel® (50 μl) to evaluate the invasion of 4T1 cells treated with Met-CCL5 associated with D1CM. The lower chamber was filled with 300 μl of 1:1 dilution of D1CM or control media, i.e., media with 0% FBS and media with 10% FBS for negative and positive controls, respectively. After a 24 h incubation at 37 °C, 5% CO2, cells that did not migrate to the lower chamber were removed from the upper chamber. The fluorescent nuclear vital dye allowed the count of the cells present on the lower surface of the membrane in at least five different random high-power microscope fields for each treatment condition.
Cytokine antibody arrays
The levels of cytokines and chemokines in conditioned media derived from various cell types were determined using antibody-based arrays (RayBiotech, Inc.). Following a blocking step, cytokine array membranes were incubated with CMs (1:1 dilution). Experiments were performed following the manufacturer’s recommendations. Briefly, the different CMs were incubated with the cytokine arrays for 2 h at room temperature and the presence of cytokines or chemokines was detected (sensitivity: pg/ml) using chemiluminescence. The intensity of the signals obtained were quantified and expressed as pixels/mm2 using the Quantity One software (Bio-Rad). After normalization to the number of cells present when CMs were collected, the intensity for each cytokine and chemokine tested were calculated using the formula (density of the sample − density of negative control)/(density of the positive control − density of the negative control) × 100.
CCL5 receptor and CCL9 receptor RNA expressions
NMuMG and 4T1 cells were grown to confluency and treated with 1:1 dilutions of the CMs tested for 3 h following a 24 h serum starvation. Total RNA was isolated using a one-step lysis reagent (Invitrogen Life Technologies). After quantification, mRNA expression levels of CCR1, CCR3, and CCR5 receptors for CCL9, and CCL5, respectively, were detected using specific primers (see ) and the One Step RT-PCR kit per manufacturer’s instructions (Promega). The mRNA expressions of these chemokine receptors were normalized to the expression of the housekeeping gene glyceraldehyde phosphate 3 dehydrogenase (GAPDH). Amplicons were identified following electrophoresis on 2% agarose gels containing ethidium bromide. Each band was semi-quantified based on the ethidium bromide signal using Quantity One (Biorad).
Table 1. Primer sequences used in PCR amplifications and resulting amplicon sizes
Western blots for CCR1, CCR5, and MMP protein expressions
Following culture and treatments, protein extracts from NMuMG and 4T1 cells were obtained by homogenization in a lysis buffer (Tissue Protein Extraction Reagent, Thermo-Scientific) supplemented with protease inhibitors (Santa Cruz Biotechnology). For MMPs, both cell extracts and cell supernatants were collected. Cell supernatants were freeze-dried and resuspended in 40-fold lower equal volumes of DI water. Protein concentrations were evaluated using protein assays (Pierce Protein Research Products). Following denaturation, protein samples (40 μg) were ran onto 8% and 10% polyacrylamide gels for cell extracts and cell supernatants, respectively, and transferred onto nitrocellulose membranes. After a blocking step, membranes were incubated overnight with specific antibodies to CCR1, CCR5, MMP9, MMP13, or MMP14 used at a dilution of 1/500 (Santa Cruz Biotechnology). Following washing steps and an hour incubation with horseradish peroxidase conjugated anti-goat, anti-mouse or anti-rabbit secondary antibodies (Jackson Laboratory), the intensity and bands recognized by the specific primary antibodies were detected using chemiluminescence (Biorad) and recorded using the UVP biochemiluminescence system. All blots were stripped (Chemicon) and reprobed for β-actin (1:5000) (Sigma). For each condition, the intensity of each protein band was determined using QuantityOne (Biorad) and normalized to the β-actin intensity.
Flow-cytometry analyses of CCR1 and CCR5 receptors
Following a 24 h incubation with media alone or D1CM (1:1 with media), 4T1 cells detached and fixed in 1% PFA for 30 min at room temperature. Next 4T1 cells resuspended in PBS supplemented with 1% BSA were incubated with antibodies specific for CCR1 and CCR5 (goat-raised antibodies, Santa Cruz Biotechnology), for 45 min at 4 °C. After washing with PBS, 4T1 cells were incubated an anti-goat PE-Texas red-conjugated secondary antibody (Invitrogen). Control stain included the secondary antibody alone. Following additional washes, the presence of cell surface CCR1 and of CCR5 was monitored by flow-cytometry (Fortessa cytometer, Becton Dickinson). Analyses were conducted using the CellQuest software and graphical representations were obtained using FlowJo software (FlowJo). Data are presented as percentage of positive cells for either CCR1 or CCR5. The control corresponding to the background stain associated with the secondary antibody is displayed on each histogram.
Inhibition of MMP activities
The MMP activity of 4T1 cells during invasion assays was determined. In invasion assays conducted as described above, 4T1 cells were incubated with or without the potent, cell-permeable, broad-spectrum hydroxamic acid inhibitor of matrix metalloproteinases (MMPs) MMP inhibitor GM6001 (1 μM, EMD Millipore) for 30 min prior to the assays. Following a 24 h incubation, the number of invading cells was determined as described above.
Additionally, 4T1 cells seeded in tissue culture dishes (100 mm, Corning Inc.) seeded to confluence were first cultured in serum-free (0% FBS) media for 24 h prior to treatments. 4T1 cells were then incubated with either serum-free (0% FBS) media, CCL5 (12.5 μM) alone, CCL9 (12.5 μM) alone, the combination CCL5 (12.5 μM) and CCL9 (12.5 μM), or D1CM for 24 h and the supernatants collected, filtered (0.2 μm filters; Fischer Scientific) and stored at −20 °C. Following freeze-drying, the supernatant powders were resuspended in 1/10 of the initial volume concentrating each supernatant 10-folds. These concentrated supernatants were assayed for MMP activities using fluorometric MMP activity assays according to the manufacturer’s recommendations (Abcam). Briefly, each sample was first activated through a 2 h incubation with 4-Aminophenylmercuric Acetate (APMA, 1 mM) at 37 °C and then incubated with the substrate a fluorescence resonance energy transfer intact peptide. The fluorescence of this substrate is quenched as long as the peptide is intact. The more MMPs cleave the peptide, the more quenching is removed and the more fluorescence is detected (Excitation 540 nm; Emission 590 nm). Fluorescence was recorded on a Synergy HT fluorometer (Bio-Tek). The enzymatic reactions were performed without inhibitors or in the presence of the broad-spectrum MMP inhibitor GM6001 (10 nM; EMD Millipore), the MMP9/MMP13 inhibitor (10 nM; EMD Millipore, 444252), or the MMP14 inhibitor (50 nM; EMD Millipore, NSC405020). Data are presented as relative fluorescence units (RFU).
Statistical analyses
All experiments and treatments were conducted at least three times. Data are presented as mean ± SEM. Statistics for all assays were analyzed using one-way ANOVA and the post-hoc Student Newman–Keuls test or the Student t test (Sigma). Significance was set at P < 0.05 a priori.
Acknowledgments
This work was supported by grants from the Department of Defense (Era of Hope program BC044778) and the National Science Foundation (EFRI program CBE0736007). The authors would also like to acknowledge the help provided by the Institute for Biological Interfaces of Engineering (IBIOE, Clemson University).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell 2010; 18:884 - 901; http://dx.doi.org/10.1016/j.devcel.2010.05.012; PMID: 20627072
- Pérez-Moreno M. When neighbourhood matters: tumour microenvironment. Clin Transl Oncol 2009; 11:70 - 4; http://dx.doi.org/10.1007/s12094-009-0316-z; PMID: 19211370
- Mareel M, Oliveira MJ, Madani I. Cancer invasion and metastasis: interacting ecosystems. Virchows Arch 2009; 454:599 - 622; http://dx.doi.org/10.1007/s00428-009-0784-0; PMID: 19471961
- Walter M, Liang S, Ghosh S, Hornsby PJ, Li R. Interleukin 6 secreted from adipose stromal cells promotes migration and invasion of breast cancer cells. Oncogene 2009; 28:2745 - 55; http://dx.doi.org/10.1038/onc.2009.130; PMID: 19483720
- Mishra P, Banerjee D, Ben-Baruch A. Chemokines at the crossroads of tumor-fibroblast interactions that promote malignancy. J Leukoc Biol 2011; 89:31 - 9; http://dx.doi.org/10.1189/jlb.0310182; PMID: 20628066
- Jotzu C, Alt E, Welte G, Li J, Hennessy BT, Devarajan E, et al. Adipose tissue-derived stem cells differentiate into carcinoma-associated fibroblast-like cells under the influence of tumor-derived factors. Anal Cell Pathol (Amst) 2010; 33:61 - 79; PMID: 20978328
- Pupa SM, Ménard S, Forti S, Tagliabue E. New insights into the role of extracellular matrix during tumor onset and progression. J Cell Physiol 2002; 192:259 - 67; http://dx.doi.org/10.1002/jcp.10142; PMID: 12124771
- Calorini L, Bianchini F. Environmental control of invasiveness and metastatic dissemination of tumor cells: the role of tumor cell-host cell interactions. Cell Commun Signal 2010; 8:24; PMID: 20822533
- Slettenaar VI, Wilson JL. The chemokine network: a target in cancer biology?. Adv Drug Deliv Rev 2006; 58:962 - 74; http://dx.doi.org/10.1016/j.addr.2006.03.012; PMID: 16996642
- Dubinett SM, Lee JM, Sharma S, Mulé JJ. Chemokines: can effector cells be redirected to the site of the tumor?. Cancer J 2010; 16:325 - 35; http://dx.doi.org/10.1097/PPO.0b013e3181eb33bc; PMID: 20693843
- Hembruff SL, Cheng N. Chemokine signaling in cancer: Implications on the tumor microenvironment and therapeutic targeting. Cancer Ther 2009; 7:A 254 - 67; PMID: 20651940
- Beider K, Abraham M, Peled A. Chemokines and chemokine receptors in stem cell circulation. Front Biosci 2008; 13:6820 - 33; http://dx.doi.org/10.2741/3190; PMID: 18508696
- Robinson SC, Scott KA, Wilson JL, Thompson RG, Proudfoot AE, Balkwill FR. A chemokine receptor antagonist inhibits experimental breast tumor growth. Cancer Res 2003; 63:8360 - 5; PMID: 14678997
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007; 449:557 - 63; http://dx.doi.org/10.1038/nature06188; PMID: 17914389
- Pinilla S, Alt E, Abdul Khalek FJ, Jotzu C, Muehlberg F, Beckmann C, et al. Tissue resident stem cells produce CCL5 under the influence of cancer cells and thereby promote breast cancer cell invasion. Cancer Lett 2009; 284:80 - 5; http://dx.doi.org/10.1016/j.canlet.2009.04.013; PMID: 19427114
- Albarenque SM, Zwacka RM, Mohr A. Both human and mouse mesenchymal stem cells promote breast cancer metastasis. Stem Cell Res 2011; 7:163 - 71; http://dx.doi.org/10.1016/j.scr.2011.05.002; PMID: 21763624
- Lv D, Zhang Y, Kim HJ, Zhang L, Ma X. CCL5 as a potential immunotherapeutic target in triple-negative breast cancer. Cell Mol Immunol 2013; In press http://dx.doi.org/10.1038/cmi.2012.69; PMID: 23376885
- Zhang Y, Yao F, Yao X, Yi C, Tan C, Wei L, et al. Role of CCL5 in invasion, proliferation and proportion of CD44+/CD24- phenotype of MCF-7 cells and correlation of CCL5 and CCR5 expression with breast cancer progression. Oncol Rep 2009; 21:1113 - 21; PMID: 19288016
- Luboshits G, Shina S, Kaplan O, Engelberg S, Nass D, Lifshitz-Mercer B, et al. Elevated expression of the CC chemokine regulated on activation, normal T cell expressed and secreted (RANTES) in advanced breast carcinoma. Cancer Res 1999; 59:4681 - 7; PMID: 10493525
- Zhang J, Patel L, Pienta KJ. Targeting chemokine (C-C motif) ligand 2 (CCL2) as an example of translation of cancer molecular biology to the clinic. Prog Mol Biol Transl Sci 2010; 95:31 - 53; http://dx.doi.org/10.1016/B978-0-12-385071-3.00003-4; PMID: 21075328
- Zhang J, Patel L, Pienta KJ. CC chemokine ligand 2 (CCL2) promotes prostate cancer tumorigenesis and metastasis. Cytokine Growth Factor Rev 2010; 21:41 - 8; http://dx.doi.org/10.1016/j.cytogfr.2009.11.009; PMID: 20005149
- Hembruff SL, Jokar I, Yang L, Cheng N. Loss of transforming growth factor-beta signaling in mammary fibroblasts enhances CCL2 secretion to promote mammary tumor progression through macrophage-dependent and -independent mechanisms. Neoplasia 2010; 12:425 - 33; PMID: 20454514
- Lapteva N, Huang XF. CCL5 as an adjuvant for cancer immunotherapy. Expert Opin Biol Ther 2010; 10:725 - 33; http://dx.doi.org/10.1517/14712591003657128; PMID: 20233026
- Soria G, Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett 2008; 267:271 - 85; http://dx.doi.org/10.1016/j.canlet.2008.03.018; PMID: 18439751
- Murooka TT, Rahbar R, Fish EN. CCL5 promotes proliferation of MCF-7 cells through mTOR-dependent mRNA translation. Biochem Biophys Res Commun 2009; 387:381 - 6; http://dx.doi.org/10.1016/j.bbrc.2009.07.035; PMID: 19607806
- Kroeze KL, Jurgens WJ, Doulabi BZ, van Milligen FJ, Scheper RJ, Gibbs S. Chemokine-mediated migration of skin-derived stem cells: predominant role for CCL5/RANTES. J Invest Dermatol 2009; 129:1569 - 81; http://dx.doi.org/10.1038/jid.2008.405; PMID: 19122644
- Forst B, Hansen MT, Klingelhöfer J, Møller HD, Nielsen GH, Grum-Schwensen B, et al. Metastasis-inducing S100A4 and RANTES cooperate in promoting tumor progression in mice. PLoS One 2010; 5:e10374; http://dx.doi.org/10.1371/journal.pone.0010374; PMID: 20442771
- Kitamura T, Taketo MM. Keeping out the bad guys: gateway to cellular target therapy. Cancer Res 2007; 67:10099 - 102; http://dx.doi.org/10.1158/0008-5472.CAN-07-2100; PMID: 17974948
- Kitamura T, Fujishita T, Loetscher P, Revesz L, Hashida H, Kizaka-Kondoh S, et al. Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses colon cancer liver metastasis by blocking accumulation of immature myeloid cells in a mouse model. Proc Natl Acad Sci U S A 2010; 107:13063 - 8; http://dx.doi.org/10.1073/pnas.1002372107; PMID: 20616008
- Soria G, Yaal-Hahoshen N, Azenshtein E, Shina S, Leider-Trejo L, Ryvo L, et al. Concomitant expression of the chemokines RANTES and MCP-1 in human breast cancer: a basis for tumor-promoting interactions. Cytokine 2008; 44:191 - 200; http://dx.doi.org/10.1016/j.cyto.2008.08.002; PMID: 18790652
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2002; 2:161 - 74; http://dx.doi.org/10.1038/nrc745; PMID: 11990853
- Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr Opin Cell Biol 2004; 16:558 - 64; http://dx.doi.org/10.1016/j.ceb.2004.07.010; PMID: 15363807
- Herman JG, Stadelman HL, Roselli CE. Curcumin blocks CCL2-induced adhesion, motility and invasion, in part, through down-regulation of CCL2 expression and proteolytic activity. Int J Oncol 2009; 34:1319 - 27; PMID: 19360344
- Adler EP, Lemken CA, Katchen NS, Kurt RA. A dual role for tumor-derived chemokine RANTES (CCL5). Immunol Lett 2003; 90:187 - 94; http://dx.doi.org/10.1016/j.imlet.2003.09.013; PMID: 14687724
- Murooka TT, Rahbar R, Platanias LC, Fish EN. CCL5-mediated T-cell chemotaxis involves the initiation of mRNA translation through mTOR/4E-BP1. Blood 2008; 111:4892 - 901; http://dx.doi.org/10.1182/blood-2007-11-125039; PMID: 18337562
- Stormes KA, Lemken CA, Lepre JV, Marinucci MN, Kurt RA. Inhibition of metastasis by inhibition of tumor-derived CCL5. Breast Cancer Res Treat 2005; 89:209 - 12; http://dx.doi.org/10.1007/s10549-004-5328-3; PMID: 15692764
- Vosseler S, Lederle W, Airola K, Obermueller E, Fusenig NE, Mueller MM. Distinct progression-associated expression of tumor and stromal MMPs in HaCaT skin SCCs correlates with onset of invasion. Int J Cancer 2009; 125:2296 - 306; http://dx.doi.org/10.1002/ijc.24589; PMID: 19610062
- Zhao T, Harada H, Teramura Y, Tanaka S, Itasaka S, Morinibu A, et al. A novel strategy to tag matrix metalloproteinases-positive cells for in vivo imaging of invasive and metastatic activity of tumor cells. J Control Release 2010; 144:109 - 14; http://dx.doi.org/10.1016/j.jconrel.2010.01.023; PMID: 20096316
- Joo CK, Seomun Y. Matrix metalloproteinase (MMP) and TGF beta 1-stimulated cell migration in skin and cornea wound healing. Cell Adh Migr 2008; 2:252 - 3; http://dx.doi.org/10.4161/cam.2.4.6772; PMID: 19262153
- Krause S, Maffini MV, Soto AM, Sonnenschein C. A novel 3D in vitro culture model to study stromal-epithelial interactions in the mammary gland. Tissue Eng Part C Methods 2008; 14:261 - 71; http://dx.doi.org/10.1089/ten.tec.2008.0030; PMID: 18694322
- Horning JL, Sahoo SK, Vijayaraghavalu S, Dimitrijevic S, Vasir JK, Jain TK, et al. 3-D tumor model for in vitro evaluation of anticancer drugs. Mol Pharm 2008; 5:849 - 62; http://dx.doi.org/10.1021/mp800047v; PMID: 18680382
- Kim JB, Stein R, O’Hare MJ. Three-dimensional in vitro tissue culture models of breast cancer-- a review. Breast Cancer Res Treat 2004; 85:281 - 91; http://dx.doi.org/10.1023/B:BREA.0000025418.88785.2b; PMID: 15111767
- Bland E, Dréau D, Burg KJ. Overcoming hypoxia to improve tissue-engineering approaches to regenerative medicine. J Tissue Eng Regen Med 2012; In press http://dx.doi.org/10.1002/term.540; PMID: 22761177
- Carlson J, Baxter SA, Dréau D, Nesmelova IV. The heterodimerization of platelet-derived chemokines. Biochim Biophys Acta 2013; 1834:158 - 68; http://dx.doi.org/10.1016/j.bbapap.2012.09.010; PMID: 23009808
- Lance A, Yang C-C, Swamydas M, Dean D, Deitch S, Burg KJ, et al. Increased extracellular matrix density decreases MCF10A breast cell acinus formation in 3D culture conditions. J Tissue Eng Regen Med 2013; In press http://dx.doi.org/10.1002/term.1675; PMID: 23404906
- Swamydas M, Eddy JM, Burg KJ, Dréau D. Matrix compositions and the development of breast acini and ducts in 3D cultures. In Vitro Cell Dev Biol Anim 2010; 46:673 - 84; http://dx.doi.org/10.1007/s11626-010-9323-1; PMID: 20585895
- Zhao M, Dumur CI, Holt SE, Beckman MJ, Elmore LW. Multipotent adipose stromal cells and breast cancer development: Think globally, act locally. Mol Carcinog 2010; 49:923 - 7; http://dx.doi.org/10.1002/mc.20675; PMID: 20842668
- Udagawa T, Wood M. Tumor-stromal cell interactions and opportunities for therapeutic intervention. Curr Opin Pharmacol 2010; 10:369 - 74; http://dx.doi.org/10.1016/j.coph.2010.06.010; PMID: 20638903
- Kuhn NZ, Tuan RS. Regulation of stemness and stem cell niche of mesenchymal stem cells: implications in tumorigenesis and metastasis. J Cell Physiol 2010; 222:268 - 77; http://dx.doi.org/10.1002/jcp.21940; PMID: 19847802
- Coghlin C, Murray GI. Current and emerging concepts in tumour metastasis. J Pathol 2010; 222:1 - 15; http://dx.doi.org/10.1002/path.2727; PMID: 20681009
- Sharma M. Chemokines and their receptors: orchestrating a fine balance between health and disease. Crit Rev Biotechnol 2010; 30:1 - 22; http://dx.doi.org/10.3109/07388550903187418; PMID: 19780653
- Liu J, Merritt JR. CC chemokine receptor small molecule antagonists in the treatment of rheumatoid arthritis and other diseases: a current view. Curr Top Med Chem 2010; 10:1250 - 67; http://dx.doi.org/10.2174/156802610791561192; PMID: 20536422
- Azenshtein E, Luboshits G, Shina S, Neumark E, Shahbazian D, Weil M, et al. The CC chemokine RANTES in breast carcinoma progression: regulation of expression and potential mechanisms of promalignant activity. Cancer Res 2002; 62:1093 - 102; PMID: 11861388
- Kiss DL, Longden J, Fechner GA, Avery VM. The functional antagonist Met-RANTES: a modified agonist that induces differential CCR5 trafficking. Cell Mol Biol Lett 2009; 14:537 - 47; http://dx.doi.org/10.2478/s11658-009-0017-1; PMID: 19448977
- Velasco-Velázquez M, Jiao X, De La Fuente M, Pestell TG, Ertel A, Lisanti MP, et al. CCR5 antagonist blocks metastasis of basal breast cancer cells. Cancer Res 2012; 72:3839 - 50; http://dx.doi.org/10.1158/0008-5472.CAN-11-3917; PMID: 22637726
- Radisky ES, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J Mammary Gland Biol Neoplasia 2010; 15:201 - 12; http://dx.doi.org/10.1007/s10911-010-9177-x; PMID: 20440544
- Cai KQ, Yang WL, Capo-Chichi CD, Vanderveer L, Wu H, Godwin AK, et al. Prominent expression of metalloproteinases in early stages of ovarian tumorigenesis. Mol Carcinog 2007; 46:130 - 43; http://dx.doi.org/10.1002/mc.20273; PMID: 17131304
- Martin MD, Matrisian LM. The other side of MMPs: protective roles in tumor progression. Cancer Metastasis Rev 2007; 26:717 - 24; http://dx.doi.org/10.1007/s10555-007-9089-4; PMID: 17717634
- Fields GB. Using fluorogenic peptide substrates to assay matrix metalloproteinases. Methods Mol Biol 2010; 622:393 - 433; http://dx.doi.org/10.1007/978-1-60327-299-5_24; PMID: 20135296
- Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, et al. Proteolytic Activity Matrix Analysis (PrAMA) for simultaneous determination of multiple protease activities. Integr Biol (Camb) 2011; 3:422 - 38; http://dx.doi.org/10.1039/c0ib00083c; PMID: 21180771
- Robinson SC, Scott KA, Balkwill FR. Chemokine stimulation of monocyte matrix metalloproteinase-9 requires endogenous TNF-alpha. Eur J Immunol 2002; 32:404 - 12; http://dx.doi.org/10.1002/1521-4141(200202)32:2<404::AID-IMMU404>3.0.CO;2-X; PMID: 11813159
- Prest SJ, Rees RC, Murdoch C, Marshall JF, Cooper PA, Bibby M, et al. Chemokines induce the cellular migration of MCF-7 human breast carcinoma cells: subpopulations of tumour cells display positive and negative chemotaxis and differential in vivo growth potentials. Clin Exp Metastasis 1999; 17:389 - 96; http://dx.doi.org/10.1023/A:1006657109866; PMID: 10651305
- Mousseau Y, Leclers D, Faucher-Durand K, Cook-Moreau J, Lia-Baldini AS, Rigaud M, et al. Improved agarose gel assay for quantification of growth factor-induced cell motility. Biotechniques 2007; 43:509 - 16; http://dx.doi.org/10.2144/000112557; PMID: 18019343
- Elsner J, Petering H, Höchstetter R, Kimmig D, Wells TN, Kapp A, et al. The CC chemokine antagonist Met-RANTES inhibits eosinophil effector functions through the chemokine receptors CCR1 and CCR3. Eur J Immunol 1997; 27:2892 - 8; http://dx.doi.org/10.1002/eji.1830271122; PMID: 9394815