Abstract
The Rho-family of p21 small GTPases are directly linked to the regulation of actin-based motile machinery and play a key role in the control of cell migration. Aside from the original and most well-characterized canonical Rho GTPases RhoA, Rac1, and Cdc42, numerous isoforms of these key proteins have been identified and shown to have specific roles in regulating various cellular motility processes. The major difficulty in addressing these isoform-specific effects is that isoforms typically contain highly similar primary amino acid sequences and thus are able to interact with the same upstream regulators and the downstream effector targets. Here, we will introduce the major members of each GTPase subfamily and discuss recent advances in the design and application of fluorescent resonance energy transfer-based probes, which are at the forefront of the technologies available to directly probe the differential, spatiotemporal activation dynamics of these proteins in live single cells. Currently, it is possible to specifically detect the activation status of RhoA vs. RhoC isoforms, as well as Cdc42 vs. TC-10 isoforms in living cells. Clearly, additional efforts are still required to produce biosensor systems capable of detecting other isoforms of Rho GTPases including RhoB, Rac2/3, RhoG, etc. Through such efforts, we will uncover the isoform-specific roles of these near-identical proteins in living cells, clearly an important area of the Rho GTPase biology that is not yet fully appreciated.
Introduction
Cell motility is a complex process that is essential for many aspects of life. The correct functioning of the immune system, the development of the embryo and the process of wound healing all rely on cell migration.Citation1-6 Furthermore, aberrant cell motility is a hallmark of many pathological conditions, including cancer metastasis and atherosclerosis.Citation7,8 In order to move, cells initiate a program of events termed the “Cell Motility Cycle.” Once an external signal is detected, the cell extends an actin-rich protrusion in the direction of motion, which adheres to the substrate in order to pull the cell forward. The rear of the cell then contracts, drawing the rest of the cell forward. The activity of Rho GTPases is essential to coordinate the stages of this cycle by virtue of their central role in regulating actin polymerization.Citation9-13
Rho GTPases are a Ras-related family of proteins that play a role in many physiological processes such as cell migration, cell division and gene expression.Citation14-17 The Rho GTPases act as molecular switches that cycle between an inactive GDP-bound conformation and an active GTP-bound conformation.Citation18,19 When active, the GTPase can interact with a variety of downstream signaling molecules or “effectors” to propagate different signaling outcomes. Rho GTPases are also prenylated at their C-terminus and this allows for their insertion into cellular membranes, which is essential for their correct localization and function.Citation20 Regulation of the activity of Rho GTPases is controlled by three types of proteins – GEFs, GAPs and GDI.Citation21 GEFs (Guanine nuecleotide exchange factors) promote the exchange of GDP for GTP, thereby activating the Rho GTPase, while GAPs (GTPase activating proteins) enhance the intrinsic GTP hydrolysis activity of the GTPase resulting in its inactivation.Citation22,23 The third class of regulator, the RhoGDI, binds to the GDP-bound GTPase masking the prenylated C-terminus and sequestering the inactive Rho GTPase in the cystosol.Citation24
Due to the importance of both the localization and activity of Rho GTPases in their functioning, traditional biochemical techniques and imaging studies are not sufficient to gain detailed insight into their roles and regulation. In the last decade the introduction of the technique of FRET (Fluorescence Resonance Energy Transfer) has gained increasing popularity as it allows the simultaneous measurement of both the localization of the GTPase and its activation status. FRET is a non-radiative energy transfer process between two compatible fluorophores that require certain characteristics, including: 1) sufficient quantum efficiency of the donor fluorophore; 2) sufficiently long fluorescence lifetime of the donor fluorophore to allow for non-radiative transfer; 3) sufficient overlap in the emission spectra of the donor fluorophore and the excitation spectra of the acceptor fluorophore; and 4) correct coupling angles of the dipoles of the donor and the acceptor fluorophores.Citation25 While the theoretical aspects of FRET is beyond the scope of the current review, it is sufficient to say that the reversibility and the sensitivity of FRET is ideally suited to the live-cell imaging experiments in which the dynamics of the protein activation states can reversibly modulate in the order of seconds. This is due primarily to the fact that FRET is exquisitely sensitive to the relative distance and the dipole-orientations of the FRET donor and acceptor fluorophores.Citation26 Unlike approaches including bifluorescence complementation,Citation27,28 proximity ligation assays,Citation29,30 and simple monitoring of fluorescent protein-labeled binding domain accumulations, FRET-based measurements offer live-cell, rapid, dynamic, reversible and sensitive measurements, even when bulk-accumulations and bulk-changes of the signaling molecules do not likely take place, such as in the case of the majority of Rho GTPase signaling.Citation31,32 FRET studies involving Rho GTPases primarily utilize biosensors which are typically single or dual chain constructs encompassing the GTPase, an effector binding domain and two fluorescent moieties, for example CFP and YFP, that are capable of FRET transfer.Citation33 Activation of the GTPase results in an interaction with the effector binding domain and a consequent alteration in the relative orientations of the two fluorescent molecules. This leads to a measurable change in the efficiency of FRET in the biosensor and provides a spatial and temporal readout for the activation of the GTPase.
The Rho family of GTPases consists of 22 members of which the best characterized are RhoA, Rac1 and Cdc42. These proteins have all been classically associated with the regulation of actin cytoskeleton and cell motility and it is well established that each of these GTPases plays a specific role in regulating the signaling pathways that control actin reorganization. Classically, Cdc42 is thought to promote filopodia formation, while Rac1 regulates the protrusion of lamellipodia and RhoA is associated with cell contraction.Citation9-11 However, that is not the whole story, each of these major GTPases also has different isoforms, which besides sharing much of their sequence, also share the same effectors and upstream regulators—GEFs, GAPs and GDIs. This immediately raises a number of questions. Why are these other isoforms required? Do they have different functions and if so how are these specific roles achieved given the similarity of the isoforms? We are starting to understand the role of the Rac isoforms Rac1, 2, 3 and Rho isoforms RhoA and RhoC within the context of cell migration. Recent results from a number of different laboratories, including ours, are uncovering novel functions of these isoforms that could explain their expression in different cell types and also their roles in pathological processes such as metastasis.
In this review we will focus on the recent discoveries regarding Rac, Rho and Cdc42 isoforms in cell motility and how the use of FRET-biosensor imaging technologies are revealing unexpected roles for these small GTPases.
The ABCs of Rho GTPases
Since the identification in 1985 of the genes for the Rho proteinsCitation34 a vast amount of work has been done in order to understand the biology of RhoGTPases. The Rho family of GTPases encompasses three genes named RhoA, RhoB and RhoC. These different isoforms share 85% sequence homology.Citation35 The original studies from Allan HallCitation36 and Paul MonaghanCitation37 focused on understanding the cellular localizations of these proteins as well as on gathering the first insights into their biological function. These studies demonstrated that RhoB is localized in endosomes while RhoA and RhoC are found at the plasma membrane and associated with actin-rich cytoskeletal structures. Since then, a substantial amount of work has been done to understand the role of RhoB in intracellular trafficking and adhesionCitation35, Citation38-42 and the functions of RhoA and RhoC in regulating the actin cytoskeleton during migratory processes (reviewed inCitation40,43). These works emphasized that while highly related in sequence, the final output of the intracellular signaling by these isoforms is different. This is presumably due to the intricate way each RhoGTPase network is wired, whereby a different subset of the key GTPase regulators, GEFs, GAPs and GDIs, cooperate to trigger a unique physiological response. Given the similarities between these GTPases, how can we distinguish between the activities of these Rho isoforms within cells during migration and invasion?
A major breakthrough in the understanding of RhoGTPase biology came with the development of biosensors for Rho family GTPases in the early 2000s.Citation44,45 For the first time, these tools allowed the direct observation of the localization of RhoGTPase activation in real-time in living cells. These studies revealed the localized patterns of GTPase activation within subcellular compartments and during physiological processes, such as in the actin-rich protrusions that are critical for cell migration.Citation31,46,47 The observed activity patterns opened the window to a new field of GTPase biology that focuses on understanding the spatiotemporal regulation of GTPase activation during biological processes at subcellular compartments. More importantly, these tools provided new approaches for gaining insight into how upstream molecules, including GEFs, GAPs and GDI, regulate GTPase activity. The intricate activation dynamics of GTPases suggested that the regulation of upstream GEFs and GAPs are likely to confer specificity to the pattern and dynamics of Rho GTPase activation that we see in cellular processes.Citation22 Specifically, the RhoA biosensor revealed important aspects of its spatiotemporal regulation. Traditional biochemical and imaging approaches had shown a key role for RhoA in mediating cellular contraction during motility. Unexpectedly, biosensor imaging showed that RhoA activity localized to the leading edge of migrating cells,Citation44,48 suggesting a novel role in leading edge protrusion. By combining biosensor imaging with a computational approach, Machacek et al.Citation46 demonstrated that RhoA GTPase activation dynamics during leading edge protrusion are directly coordinated with edge movement immediately at the cell periphery. In tumor cells it was also shown that in the absence of a lamellipodium, RhoA displayed high activation levels in mDia1-dependent finger like protrusions that formed after EGF stimulation.Citation49 Furthermore, biosensor technology has shown how RhoA activity is regulated in other cellular processes such as cytokinesisCitation50 and T-cell transendothelial migration.Citation51 The RhoA biosensor design has evolved since the first versionCitation44,45 (A) to more recent forms,Citation44,52 which maintain an intact C-terminus and allowed the adaptation of the design to other isoforms, including RhoCCitation53 (B). This new biosensor has revealed some important aspects of the spatiotemporal regulation of RhoC in tumor cells and put in context the distinct roles of RhoA and RhoC proposed during tumor cell invasion.Citation54,55
Figure 1 (See previous page). Schematic showing the various designs used in GTPase Biosensors. (A) The single-chain Raichu design was used to generate genetically encoded biosensors for RhoA, Rac1, Cdc42 and TC10. In this design, the FRET pair (CFP/YFP) is placed on the N- and C-terminus of the molecule. Thus, the C-terminus of the GTPase is not free and these biosensors are insensitive to GDI. The CAAX box (membrane targeting) from K-Ras is encoded at the C-terminus of the YFP in order to place the probe constitutively in the membrane. Upon activation of the GTPase, it interacts with the GTPase binding domain (specific to each GTPases), resulting in a conformational change that brings the fluorescent proteins into close proximity leading to increased FRET signal. (B) The RhoA single chain biosensor places the donor and acceptor fluorescent proteins internally allowing the native C-terminus of RhoA to be maintained. High FRET occurs when the active GTPases interacts with the Rho binding domain (RBD) of Rhotekin. (C) The first bimolecular Rac1 biosensor in which Rac1 is linked to EGFP and the Pak binding domain (PBD) of PAK1 is coupled to the dye Alexa-546. FRET occurs when the interaction of Rac1 and the PBD bring EGFP and Alexa-546 into close proximity. This biosensor was later modified to be fully genetically encoded by replacing GFP and Alexa-546 with YFP and CFP respectively. (D) The newest version of the Rac1 biosensor is genetically encoded and uses mCerulean1 and mVenus as the FRET pair. This biosensor maintains the C-terminus of Rac1 in its native state. In addition, a tandem PBD module is utilized to regulate the interaction with Rac1 and further decrease the chances of spurious FRET. This design was recently adapted to generate a single chain Cdc42 biosensor (E) The Cdc42 GBD sensor detects active endogenous Cdc42. Upon binding of active endogenous Cdc42 to the GBD of WASP, a conformational change occurs that causes CFP and YFP to be forced apart. This results in low FRET signal where active Cdc42 is present in the cell. (F) In a modification of (E) the VCA domain of WASP is incorporated into the biosensor. This domain competes with Cdc42 for binding to the GBD and this may result in larger differences in the on and off states of the biosensor. (F) This biosensor was generated by adding Cdc42 to the N-terminus of the GBD/VCA sensor. Upon activation of Cdc42 by a GEF, Cdc42 binds to the GDB and results in decreased FRET. A caveat of this biosensor is that the rate of GTP hydrolysis is significantly slower than for wild type Cdc42 and in addition, the C-terminus of Cdc42 is occluded. (G) The MeroCBD biosensor allows measurements of the activation of endogenous Cdc42 by using a dye that changes fluorescence emission intensity as a function of the local solvent polarity. The dye is coupled to the GTPase effector and changes in fluorescence emmision occur upon interaction with the active GTPase. The dye/effector moiety is also labeled with GFP, which is used as reference for ratiometric intensity measurements.
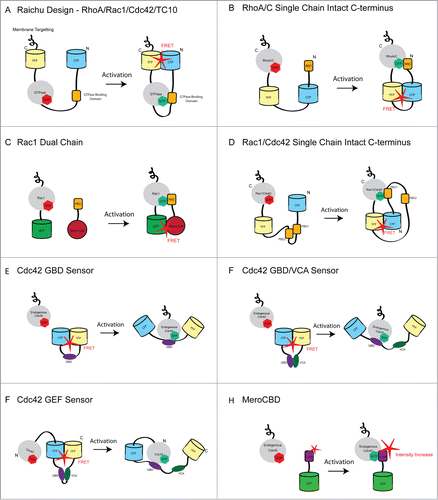
Depletion of RhoC in tumor cells inhibits invasionCitation56 and metastasis.Citation57 The RhoC biosensor was used to reveal the mechanism by which RhoC contributes to the invasion of tumor cells. Spatial activation of RhoC surrounding the core of invasive protrusions named invadopodiaCitation56 or within regions located behind the leading edgeCitation49 triggers a signaling pathway that phosphorylates cofilinCitation58 and confines cofilin activity and actin polymerization at the core of invadopodia or to the tip of the leading edge, a localization which is essential for efficient invasion. These activation zones are regulated by the spatial and geometric arrangements of the direct upstream regulators p190RhoGEF and p190RhoGAP.
This biosensor also allowed the direct comparison of the activation patterns of RhoA and RhoC at these subcellular structures.Citation49,53,56 At invadopodia, RhoC has a geometrically confined activation pattern, while RhoA displays a more random activation pattern.Citation56 At leading edge protrusions, RhoC activation is confined spatially behind RhoA activation.Citation49 Also, using computational approaches in fibroblasts, the activation of RhoC was quantitatively compared with that of RhoA.Citation53 This analysis showed that areas of RhoC activation appeared to be more diffuse and evenly distributed at the edge of the cell.Citation53 Furthermore, the authors showed that RhoA and RhoC activities have different kinetics during lamellipodial protrusions. These different activation dynamics suggest that the key determining factors that control subcellular GTPase activation dynamics is likely the spatial distribution of their upstream regulators.Citation49,56 Recent studies have also extended the use of Rho biosensors to in vivo systems. For example, Timpson et al.,Citation59 have shown how RhoA activity is modulated in an animal model of pancreatic cancer using the fluorescence lifetime imaging (FLIM)-FRET measurements of the Raichu-RhoA biosensor.
The development of new imaging tools capable of measuring the real-time spatial activation patterns of RhoA and RhoC has significantly extended our understanding of the differential regulation of these isoforms in the context of tumor cell invasion and migration. However, there is more to be done, for example, how is RhoB spatiotemporally regulated? The door is open for the development of biosensors for this particular isoform in order to answer this question.
Rac GTPases: Lighting Up the Lamellipodia
The Rac family of GTPases consists of three closely related isofoms Rac1, Rac2, Rac3, which share more than 90% homology. Differences in these isoforms are found in the C-terminal 15 amino acids, which is the region important for localization of the molecules.Citation60-63 In addition, this family of proteins includes the more distantly related RhoG, which has about 70% homology to Rac1.Citation64 Rac1 is the prototypical member of this family and its functions were first elucidated in the classical studies of Ridley and Hall. Microinjection of Rac1 into fibroblasts resulted in the formation of lamellipodia and membrane ruffles and also promotes the formation of focal contacts and E-cadherin dependent cell-cell contacts.Citation9,10,65 The importance of Rac1 was further highlighted by the generation of a knockout mouse that revealed that Rac1 is essential for embryonic development, as in its absence gastrulation fails to proceed, due to defects in lamellipodia formation and cell adhesion that compromise the ability of the embryonic cells to migrate.Citation66 Since these original studies, Rac1 has been established as a key regulator of cell motility (reviewed inCitation67,68).
As with RhoA and RhoC, over the last 10–15 years, much progress has been made on the development of FRET-based biosensors that show the localized activation status of Rac1 in live cells. Despite many studies highlighting the important function of Rac1 in actin polymerization and cell migration, it was only with the advent of fluorescent biosensors that specific insights into the high-resolution spatiotemporal regulation of Rac1 activity were elucidated.Citation31,46,47 Initial studies utilized a dual chain FRET biosensor (C) to demonstrate high levels of Rac1 activity in membrane ruffles in response to PDGF stimulation.Citation31 In keeping with the role of Rac1 in lamellipodia formation and cell migration, this study also showed high levels of Rac1 activity at the leading edge, which declined in a broad gradient. A subsequent study used an improved Rac1 biosensor combined with a computational multiplexing approach to compare the activity patterns of Rac1, RhoA and Cdc42 in migrating fibroblasts.Citation46 This study unexpectedly revealed that Rac1 activity is highest not at the protruding front of the cell but in fact peaks at a distance of approximately 2μm from the plasma membrane, directly behind a focused region of RhoA activation.Citation46 The activation of Rac1 was also delayed with respect to the protrusion of the membrane. This study shed new light on the function of Rac1 in cell migration and challenged the hypothesis that Rac1 initiates the protrusion of the leading edge in migrating cells.Citation9, Citation69-71 Furthermore this study gave support to the theory that Rac1 antagonizes RhoA activityCitation72,73 and may serve to restrict RhoA signaling to a narrow region at the leading edge.Citation46 Preliminary studies with the single chain Raichu biosensor (A), which eliminates the issues of equal expression of both biosensor components that is inherent in dual chain designs, also showed that Rac1 activity is highest directly behind the leading edge and further demonstrated that Rac1 activity decreases sharply with the plasma membrane retraction that occurs when the cell changes direction.Citation47
Recently, a new single chain Rac1 biosensor, which maintains the C-terminus of the protein in its native state, has been used to explore the role of Rac1 in tumor cell invasion (D).Citation74 This study found that Rac1 activity is suppressed during invadopodia formation but is subsequently activated to promote the disassembly of the invadopod structure. Unlike earlier versions of Rac1 biosensors, a key strength of this biosensor is that it is responsive to the full complement of endogenous GTPase regulatory mechanisms, including RhoGDI. Therefore this biosensor is likely to provide fresh insight into the roles and regulation of Rac1 in myriad physiological and pathological processes. Together the data from these biosensor studies demonstrated the clear importance of high spatial and temporal regulation of Rac1 activity in the control of cell migration and invasion. However, many questions still remain as to how this spatiotemporal activation is mediated. Rac1 biosensors have also been used to shed light on these questions. For example, the pathways regulating activation of Rac1 in the control of Drosophila border cell migration were elucidated in a number of studies using the Rac1 biosensor.Citation75,76 Furthermore, the importance of the Rac1 GEFs, PLCϵ,Tiam1 and Trio in fibroblast chemotaxis, T-cell migration and invasion and tumor cell invasion, respectively were determined in studies that employed Rac1 FRET biosensorsCitation74,77,78
In addition to probes that monitor the activation status of Rac1 in cells, optical tools have also been developed that allow the direct, localized manipulation of Rac1 activity. A genetically encoded photoactivatable Rac1 was engineered in which Rac1 is fused to the LOV domain, which sterically hinders the ability of Rac1 to interact with downstream effectors.Citation79 Upon irradiation with 458 or 473nm light, a conformational change occurs that removes this hindrance and allows Rac1 to signal. This tool has been effectively employed both in cellular and multicellular models to demonstrate that activation of Rac1 is required for directional cell migration.Citation75, Citation79-82 Recently a number of studies have combined the use of biosensors to monitor the activity of Rac1 with the manipulation of this protein by specific photoactivation have been used to determine the functions of Rac1 in Drosophila border cell migration and tumor cell invasion.Citation74,83 Studies such as these which combine different microscopy and optical approaches to dissect the mechanism of GTPase signaling in different contexts will pave the way for a complete understanding of the biology of these proteins.
Since the original cell-based studies using Rac1 FRET biosensors, much progress has been made in imaging Rac1 activity in multicellular organisms. The first in vivo analysis of Rac1 activity was performed in migrating neural crest cells in xenopus embryos.Citation84 More detailed in vivo analysis of Rac1 activity was performed in migrating border cells in Drosophila. In this study, a combination of PA-Rac1 and Raichu-Rac1 biosensor showed that Rac1 activation regulates the directionality of border cell migration in Drosophila embryos.Citation80 It was found that the directionality of the cells depends on the asymmetric localization of Rac and that the establishment of asymmetry is dependent on the guidance receptors, PVR and EGFR. Furthermore, Rac1 activity has been observed in migrating germ cells in developing zebrafish embryos.Citation85 A recent study also showed that the activity of Rac1 is high in invasive glioblastoma cells that are engrafted into rat brains.Citation86 In addition, a transgenic mouse that ubiquitously expresses the Raichu-Rac1 biosensor has recently been generated.Citation87 In this study Johnsson et al., imaged Rac1 activity in primary MEFs and chemotaxing neutrophils, as well as in intestinal crypt cultures, where it was found that Rac1 activity is higher in the cells at the base of intestinal crypts compared with distally located cells.Citation87 Furthermore, these mice were crossed with two cancer mouse models to demonstrate that Rac1 activity is higher both in pancreatic and mammary tumors compared with non-cancerous tissues.Citation87 These studies have given great insights into the importance of Rac1 signaling in vivo, however much remains to be done to fully understand the patterns and regulation of Rac1 signaling and its physiological significance in mammals.
Other Rac Isoforms: Shaping the Immune System and the Brain
Insights into the roles of the other Rac GTPases have revealed both distinct and overlapping functions for these isoforms. Rac2 expression is restricted to hematopoietic cells where it plays an important role in regulating the migration and chemotaxis of neutrophils, macrophages and B- and T-lymphocytes.Citation88-91 Deletion of Rac2 in mice revealed numerous other defects in the immune system as actin polymerization, superoxide production and phagocytosis are impaired in the absence of Rac2.Citation92 Combined deletion of both Rac1 and Rac2 resulted in more severe migratory defects than are observed in Rac2−/− cells alone indicating that there is some synergy in the functioning of these GTPases.Citation93,94 A dual chain biosensor approach was used to demonstrate that active Rac2 is localized at both the leading edge and in the following tail of chemotaxing neutrophils.Citation95 This study showed similar results using the dual chain Rac1 biosensor and was the first report of a role for Rac signaling in tail retraction in migrating cells.Citation95 Rac2 also exhibits isoform specific functions in cell migration, as in its absence neutrophils cannot chemotax in response to fMLP. In contrast, loss of Rac1 in neutrophils does not result in chemotactic deficits in response to fMLP.Citation96
Rac3 has also been linked to cell migration as well as the invasion of metastatic cancer cells. Gest et al. demonstrate that depletion of Rac3 in the breast carcinoma cell line MB-MDA-231 leads to decreased migration in wound healing assays, however the ability of glioblastoma cells to migrate in a similar assay was only slightly affected by the loss of Rac3.Citation97,98 Conversely, depletion of Rac1 strongly affected the ability of the glioblastoma cells to migrate in two dimensions.Citation97 Similarly, the loss of Rac1 but not Rac3 was found to impair the migration of non-tumor primary fibroblasts in Dunn chamber chemotaxis assays.Citation99 In contrast, Rac3 and Rac1 appear to perform synergistic roles in the development of the nervous system as loss of both isoforms results in defects in the migration and differentiation of a number of brain cell types.Citation100-103 Loss of either isoform in the brain does not lead to obvious defects.Citation100 These results highlight that each of the Rac isoforms had been specialized for a very particular function and displays its own specific biology.
It is clear that much is known about the distinct roles that the Rac family isoforms play in cell motility, which contribute to a wide range of physiological functions including the normal action of the immune system and neuronal development as well as the pathogenesis of cancer. However many questions remain, for example, the activation pattern of Rac2 has only been characterized using a dual chain approach while that of Rac3 has not yet been studied at all. Furthermore the upstream signaling pathways that control the differential regulation of these isoforms remain unclear. Thus the challenge ahead is to design biosensors that can be used to visualize the activation dynamics of Rac2 and Rac3 in order to shed light on these pertinent questions. The generation of specific tools to observe the activation of the Rac GTPases will enable a greater understanding of the functions of these isoforms as well as the regulatory mechanisms that enable such closely related molecules to play distinct roles.
The Cdc42 Family—Not just Filopodia
As with Rho and Rac, the identification of Cdc42 as a key regulator of cytoskeletal dynamics was identified in classical microinjection experiments. Activation of Cdc42 in Swiss 3T3 fibroblasts results in the rapid induction of filopodia, which subsequently develop into lamellipodia if Rac is present.Citation104 The same study also identified a role for Cdc42 in promoting focal complex formation.Citation104 Since then, key roles for Cdc42 in chemotaxis and directed cell migration as well as in the establishment of epithelial cell polarity have been identified (reviewed inCitation67). The first report showing the activation dynamics of Cdc42 in live cells came from the Matsuda group, which developed the Raichu-Cdc42 biosensor based on a similar design as the Raichu-Rac1 biosensorCitation47 (A). This study showed that, like Rac1, activation of Cdc42 increases in a gradient toward the leading edge of motile cells but that in contrast to Rac1, the activation of Cdc42 is highest at the tip of the leading edge. A subsequent study resulted in the development of three sensors that can be used collectively to report on the activation status of Cdc42.Citation105 Two of these sensors involve either the GTPase binding domain (GBD) of WASP or the GBD and the VCA region of WASP sandwiched between CFP and YFP (E,F). Upon the specific interaction of the GBD or GBD/VCA with active Cdc42, a conformational change occurs that results in a decrease in FRET efficiency. Unlike the majority of FRET sensors, these probes can detect the activity of the endogenous GTPase. The third sensor consists of Cdc42 coupled at its C-terminus to the CFP-GBD-VCA-YFP sensor (G). This acts as a sensor of Cdc42 specific GEFs but again has the caveat that the C-terminus of Cdc42 is occluded which is likely to affect the correct localization of the sensor. Detailed biochemical analyses revealed that this sensor reports the timing of Cdc42 activation accurately but that there is 16-fold slower rate of GTP hydrolysis by the GEF sensor than by wild type Cdc42. Thus this sensor may not be optimal for monitoring processes in which the rapid turnover of Cdc42 activation is important. Imaging studies showed that these Cdc42 sensors can be expressed in cells and respond to the presence of GEFs and GAPs, however a detailed analysis of Cdc42 activation using these probes have not been performed.Citation105
Greater insight into the function of Cdc42 came from a study in which another novel biosensor capable of reporting the activity of endogenous Cdc42 was developed.Citation106 This biosensor, termed MeroCBD, comprises the CRIB (GBD) domain of the Cdc42-specific effector protein WASP, covalently labeled with a fluorescent dye (H). Upon binding to active GTP-bound Cdc42, the dye undergoes a measurable change in fluorescence.Citation106,107 Microinjection of this biosensor into fibroblasts showed that after attachment, Cdc42 is activated in a thin band in areas of the plasma membrane that are extending filopodia, although no activity is observed within the filopodia themselves. At later timepoints after adhesion, Cdc42 activity is localized in larger dynamic protrusions. This study also showed that the indicated fluctuations in Cdc42 activity are correlated with extension and retraction of cellular protrusions and suggested that Cdc42 is important for the initiation but not the maintenance of these protrusions.Citation106 More recently, a new genetically encoded single chain Cdc42 biosensor has been developedCitation108 (D). Like the new RhoA, RhoC and Rac1 biosensors (B,D), this biosensor maintains the native C-terminus of Cdc42 and thus undergoes correct membrane targeting and interaction with GDI. As well as confirming the previously documented activation of Cdc42 at the leading edge of motile cells, the authors of this study also extended their observations to macrophages and showed that Cdc42 is activated during phagocytosis and after cytokine stimulation. Furthermore, Cdc42 was found to be transiently activated during the formation of podosomes, which are adhesive and degradative structures involved in extracellular matrix remodeling and cell migration.Citation109 While these papers collectively reveal the patterns of Cdc42 activation in different contexts, studies utilizing Cdc42 biosensors to examine the regulation of its activation are still lacking.
Like the Rho and Rac families, different isoforms of Cdc42 have also been identified. These are termed TC10 (RhoQ) and TCL (RhoJ). This family of proteins shares 70–85% homology.Citation110,111 TC10 has been shown to play a role in the regulation of glucose transport, while TCL is highly expressed in endothelial cells and is important for vascular morphogenesis.Citation110,112,113 Like Cdc42, expression of these GTPases in cells results in modulation of the morphology of the actin cytoskeleton.Citation110,111 The activity of TC10 has been visualized using a modified version of the Raichu biosensorsCitation114 (A). This study showed that TC10 is highly activated on exocytosing vesicles and recycling endosomes, but not on early or late endosomes in Hela cells. Furthermore, hydrolysis of GTP by TC10 was shown to be necessary to promote the fusion of vesicles with the plasma membrane.Citation114 This function of TC10 was found to be conserved in neurites, suggesting that TC10 activity is broadly important for exocytosis.Citation115
Conclusion
FRET-biosensors have come in multiple flavors since their early development, but technology has vastly improved and we now have powerful tools that allow the precise analysis of GTPase activation in the time scale of seconds and at subcellular resolution. The expansion in the use of FRET-biosensor imaging to address biological question is changing the way we understand and study GTPase biology. Computational analysis combined with FRET imaging is revealing unpredicted roles for GTPases in different subcellular compartments. The field is wide open, we must now expand our use of biosensor technology to analyze the activity of upstream GEF and GAP regulators and GDIs in order to complete the entire picture of GTPase regulation.
FRET-biosensor imaging technology has been widely used to address the functions of RhoGTPases during motility but there are many other processes in which these proteins play a major role including cell division, membrane trafficking, polarity and proliferation. The application of FRET biosensor technology to these fields will greatly improve our understanding of these critical physiological processes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
This work was supported by NIH GM093121 (LH, SKD, JJBC).
References
- Shaw TJ, Martin P. Wound repair at a glance. J Cell Sci 2009; 122:3209-13; PMID:19726630; http://dx.doi.org/10.1242/jcs.031187
- Abreu-Blanco MT, Watts JJ, Verboon JM, Parkhurst SM. Cytoskeleton responses in wound repair. Cell Mol Life Sci 2012; 69:2469-83; PMID:22349211; http://dx.doi.org/10.1007/s00018-012-0928-2
- Fort P, Théveneau E. PleiotRHOpic: Rho pathways are essential for all stages of Neural Crest development. Small GTPases 2014; 5:e27975; PMID:24614304; http://dx.doi.org/10.4161/sgtp.27975
- Walck-Shannon E, Hardin J. Cell intercalation from top to bottom. Nat Rev Mol Cell Biol 2014; 15:34-48; PMID:24355988; http://dx.doi.org/10.1038/nrm3723
- Wiesner C, Le-Cabec V, El Azzouzi K, Maridonneau-Parini I, Linder S. Podosomes in space: Macrophage migration and matrix degradation in 2D and 3D settings. Cell Adh Migr 2014; 8:•••; PMID:24713854; http://dx.doi.org/10.4161/cam.28116
- Lam PY, Huttenlocher A. Interstitial leukocyte migration in vivo. Curr Opin Cell Biol 2013; 25:650-8; PMID:23797028; http://dx.doi.org/10.1016/j.ceb.2013.05.007
- Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol 2012; 24:277-83; PMID:22209238; http://dx.doi.org/10.1016/j.ceb.2011.12.004
- Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol 2013; 13:709-21; PMID:23995626; http://dx.doi.org/10.1038/nri3520
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995; 81:53-62; PMID:7536630; http://dx.doi.org/10.1016/0092-8674(95)90370-4
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 1992; 70:401-10; PMID:1643658; http://dx.doi.org/10.1016/0092-8674(92)90164-8
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 1992; 70:389-99; PMID:1643657; http://dx.doi.org/10.1016/0092-8674(92)90163-7
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 2006; 16:522-9; PMID:16949823; http://dx.doi.org/10.1016/j.tcb.2006.08.006
- Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr 2011; 5:170-80; PMID:21178402; http://dx.doi.org/10.4161/cam.5.2.14403
- Narumiya S, Yasuda S. Rho GTPases in animal cell mitosis. Curr Opin Cell Biol 2006; 18:199-205; PMID:16487696; http://dx.doi.org/10.1016/j.ceb.2006.02.002
- Rajakylä EK, Vartiainen MK. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases 2014; 5:e27539; PMID:24603113; http://dx.doi.org/10.4161/sgtp.27539
- Infante E, Ridley AJ. Roles of Rho GTPases in leucocyte and leukaemia cell transendothelial migration. Philos Trans R Soc Lond B Biol Sci 2013; 368:20130013; PMID:24062583; http://dx.doi.org/10.1098/rstb.2013.0013
- Hanna S, El-Sibai M. Signaling networks of Rho GTPases in cell motility. Cell Signal 2013; 25:1955-61; PMID:23669310; http://dx.doi.org/10.1016/j.cellsig.2013.04.009
- Hakoshima T, Shimizu T, Maesaki R. Structural basis of the Rho GTPase signaling. J Biochem 2003; 134:327-31; PMID:14561717; http://dx.doi.org/10.1093/jb/mvg149
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 2001; 294:1299-304; PMID:11701921; http://dx.doi.org/10.1126/science.1062023
- Michaelson D, Silletti J, Murphy G, D’Eustachio P, Rush M, Philips MR. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol 2001; 152:111-26; PMID:11149925; http://dx.doi.org/10.1083/jcb.152.1.111
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93:269-309; PMID:23303910; http://dx.doi.org/10.1152/physrev.00003.2012
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 2005; 6:167-80; PMID:15688002; http://dx.doi.org/10.1038/nrm1587
- Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol 2003; 13:13-22; PMID:12480336; http://dx.doi.org/10.1016/S0962-8924(02)00004-1
- Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol 2011; 12:493-504; PMID:21779026; http://dx.doi.org/10.1038/nrm3153
- Forster T. Intermolecular energy migration and fluorescence. Ann Phys 1948; 2:55-75
- Lakowicz JR. in Principles of Fluorescence Spectroscopy 305-341 (Plenum Press, New York, 1983).
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 2002; 9:789-98; PMID:11983170; http://dx.doi.org/10.1016/S1097-2765(02)00496-3
- Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc 2006; 1:1278-86; PMID:17406412; http://dx.doi.org/10.1038/nprot.2006.201
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 2006; 3:995-1000; PMID:17072308; http://dx.doi.org/10.1038/nmeth947
- Söderberg O, Leuchowius KJ, Kamali-Moghaddam M, Jarvius M, Gustafsdottir S, Schallmeiner E, Gullberg M, Jarvius J, Landegren U. Proximity ligation: a specific and versatile tool for the proteomic era. Genet Eng (N Y) 2007; 28:85-93; PMID:17153934
- Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science 2000; 290:333-7; PMID:11030651; http://dx.doi.org/10.1126/science.290.5490.333
- Chamberlain CE, Kraynov VS, Hahn KM. Imaging spatiotemporal dynamics of Rac activation in vivo with FLAIR. Methods Enzymol 2000; 325:389-400; PMID:11036621; http://dx.doi.org/10.1016/S0076-6879(00)25460-8
- Pertz O, Hahn KM. Designing biosensors for Rho family proteins–deciphering the dynamics of Rho family GTPase activation in living cells. J Cell Sci 2004; 117:1313-8; PMID:15020671; http://dx.doi.org/10.1242/jcs.01117
- Madaule P, Axel R. A novel ras-related gene family. Cell 1985; 41:31-40; PMID:3888408; http://dx.doi.org/10.1016/0092-8674(85)90058-3
- Wheeler AP, Ridley AJ. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp Cell Res 2004; 301:43-9; PMID:15501444; http://dx.doi.org/10.1016/j.yexcr.2004.08.012
- Adamson P, Paterson HF, Hall A. Intracellular localization of the P21rho proteins. J Cell Biol 1992; 119:617-27; PMID:1383236; http://dx.doi.org/10.1083/jcb.119.3.617
- Robertson D, Paterson HF, Adamson P, Hall A, Monaghan P. Ultrastructural localization of ras-related proteins using epitope-tagged plasmids. J Histochem Cytochem 1995; 43:471-80; PMID:7537292; http://dx.doi.org/10.1177/43.5.7537292
- Alfano D, Ragno P, Stoppelli MP, Ridley AJ. RhoB regulates uPAR signalling. J Cell Sci 2012; 125:2369-80; PMID:22366462; http://dx.doi.org/10.1242/jcs.091579
- Ellis S, Mellor H. Regulation of endocytic traffic by rho family GTPases. Trends Cell Biol 2000; 10:85-8; PMID:10675900; http://dx.doi.org/10.1016/S0962-8924(99)01710-9
- Ridley AJ. RhoA, RhoB and RhoC have different roles in cancer cell migration. J Microsc 2013; 251:242-9; PMID:23488932; http://dx.doi.org/10.1111/jmi.12025
- Vega FM, Colomba A, Reymond N, Thomas M, Ridley AJ. RhoB regulates cell migration through altered focal adhesion dynamics. Open Biol 2012; 2:120076; PMID:22724071; http://dx.doi.org/10.1098/rsob.120076
- Wheeler AP, Ridley AJ. RhoB affects macrophage adhesion, integrin expression and migration. Exp Cell Res 2007; 313:3505-16; PMID:17692842; http://dx.doi.org/10.1016/j.yexcr.2007.07.014
- Ridley AJ. Rho GTPases and cell migration. J Cell Sci 2001; 114:2713-22; PMID:11683406
- Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 2006; 440:1069-72; PMID:16547516; http://dx.doi.org/10.1038/nature04665
- Yoshizaki H, Ohba Y, Kurokawa K, Itoh RE, Nakamura T, Mochizuki N, Nagashima K, Matsuda M. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol 2003; 162:223-32; PMID:12860967; http://dx.doi.org/10.1083/jcb.200212049
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature 2009; 461:99-103; PMID:19693013; http://dx.doi.org/10.1038/nature08242
- Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol 2002; 22:6582-91; PMID:12192056; http://dx.doi.org/10.1128/MCB.22.18.6582-6591.2002
- Kurokawa K, Matsuda M. Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol Biol Cell 2005; 16:4294-303; PMID:15987744; http://dx.doi.org/10.1091/mbc.E04-12-1076
- Bravo-Cordero JJ, Sharma VP, Roh-Johnson M, Chen X, Eddy R, Condeelis J, Hodgson L. Spatial regulation of RhoC activity defines protrusion formation in migrating cells. J Cell Sci 2013; 126:3356-69; PMID:23704350; http://dx.doi.org/10.1242/jcs.123547
- Birkenfeld J, Nalbant P, Bohl BP, Pertz O, Hahn KM, Bokoch GM. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev Cell 2007; 12:699-712; PMID:17488622; http://dx.doi.org/10.1016/j.devcel.2007.03.014
- Heasman SJ, Carlin LM, Cox S, Ng T, Ridley AJ. Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J Cell Biol 2010; 190:553-63; PMID:20733052; http://dx.doi.org/10.1083/jcb.201002067
- Fritz RD, Letzelter M, Reimann A, Martin K, Fusco L, Ritsma L, Ponsioen B, Fluri E, Schulte-Merker S, van Rheenen J, et al. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci Signal 2013; 6:rs12; PMID:23882122; http://dx.doi.org/10.1126/scisignal.2004135
- Zawistowski JS, Sabouri-Ghomi M, Danuser G, Hahn KM, Hodgson L. A RhoC biosensor reveals differences in the activation kinetics of RhoA and RhoC in migrating cells. PLoS One 2013; 8:e79877; PMID:24224016; http://dx.doi.org/10.1371/journal.pone.0079877
- Vega FM, Fruhwirth G, Ng T, Ridley AJ. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J Cell Biol 2011; 193:655-65; PMID:21576392; http://dx.doi.org/10.1083/jcb.201011038
- Simpson KJ, Dugan AS, Mercurio AM. Functional analysis of the contribution of RhoA and RhoC GTPases to invasive breast carcinoma. Cancer Res 2004; 64:8694-701; PMID:15574779; http://dx.doi.org/10.1158/0008-5472.CAN-04-2247
- Bravo-Cordero JJ, Oser M, Chen X, Eddy R, Hodgson L, Condeelis J. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr Biol 2011; 21:635-44; PMID:21474314; http://dx.doi.org/10.1016/j.cub.2011.03.039
- Hakem A, Sanchez-Sweatman O, You-Ten A, Duncan G, Wakeham A, Khokha R, Mak TW. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev 2005; 19:1974-9; PMID:16107613; http://dx.doi.org/10.1101/gad.1310805
- Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol 2013; 14:405-15; PMID:23778968; http://dx.doi.org/10.1038/nrm3609
- Timpson P, McGhee EJ, Morton JP, von Kriegsheim A, Schwarz JP, Karim SA, Doyle B, Quinn JA, Carragher NO, Edward M, et al. Spatial regulation of RhoA activity during pancreatic cancer cell invasion driven by mutant p53. Cancer Res 2011; 71:747-57; PMID:21266354; http://dx.doi.org/10.1158/0008-5472.CAN-10-2267
- Tao W, Filippi MD, Bailey JR, Atkinson SJ, Connors B, Evan A, Williams DA. The TRQQKRP motif located near the C-terminus of Rac2 is essential for Rac2 biologic functions and intracellular localization. Blood 2002; 100:1679-88; PMID:12176888
- Shirsat NV, Pignolo RJ, Kreider BL, Rovera G. A member of the ras gene superfamily is expressed specifically in T, B and myeloid hemopoietic cells. Oncogene 1990; 5:769-72; PMID:2189110
- Haataja L, Groffen J, Heisterkamp N. Characterization of RAC3, a novel member of the Rho family. J Biol Chem 1997; 272:20384-8; PMID:9252344; http://dx.doi.org/10.1074/jbc.272.33.20384
- Didsbury J, Weber RF, Bokoch GM, Evans T, Snyderman R. rac, a novel ras-related family of proteins that are botulinum toxin substrates. J Biol Chem 1989; 264:16378-82; PMID:2674130
- Vincent S, Jeanteur P, Fort P. Growth-regulated expression of rhoG, a new member of the ras homolog gene family. Mol Cell Biol 1992; 12:3138-48; PMID:1620121
- Braga VM, Machesky LM, Hall A, Hotchin NA. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J Cell Biol 1997; 137:1421-31; PMID:9182672; http://dx.doi.org/10.1083/jcb.137.6.1421
- Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, Sakagami H, Kondo H, Nozawa S, Aiba A, et al. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene 1998; 17:3427-33; PMID:10030666; http://dx.doi.org/10.1038/sj.onc.1202595
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 2008; 9:690-701; PMID:18719708; http://dx.doi.org/10.1038/nrm2476
- Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal 2010; 8:23; PMID:20822528; http://dx.doi.org/10.1186/1478-811X-8-23
- Ridley AJ, Hall A. Distinct patterns of actin organization regulated by the small GTP-binding proteins Rac and Rho. Cold Spring Harb Symp Quant Biol 1992; 57:661-71; PMID:1339704; http://dx.doi.org/10.1101/SQB.1992.057.01.072
- Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol 1999; 1:45-50; PMID:10559863; http://dx.doi.org/10.1038/9018
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol 2004; 265:23-32; PMID:14697350; http://dx.doi.org/10.1016/j.ydbio.2003.06.003
- Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat Cell Biol 2003; 5:236-41; PMID:12598902; http://dx.doi.org/10.1038/ncb938
- Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol 1999; 147:1009-22; PMID:10579721; http://dx.doi.org/10.1083/jcb.147.5.1009
- Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol 2014; 16:574-86; PMID:24859002; http://dx.doi.org/10.1038/ncb2972
- Ramel D, Wang X, Laflamme C, Montell DJ, Emery G. Rab11 regulates cell-cell communication during collective cell movements. Nat Cell Biol 2013; 15:317-24; PMID:23376974; http://dx.doi.org/10.1038/ncb2681
- Fernández-Espartero CH, Ramel D, Farago M, Malartre M, Luque CM, Limanovich S, Katzav S, Emery G, Martín-Bermudo MD. GTP exchange factor Vav regulates guided cell migration by coupling guidance receptor signalling to local Rac activation. J Cell Sci 2013; 126:2285-93; PMID:23525006; http://dx.doi.org/10.1242/jcs.124438
- Martins M, Warren S, Kimberley C, Margineanu A, Peschard P, McCarthy A, Yeo M, Marshall CJ, Dunsby C, French PM, et al. Activity of PLCϵ contributes to chemotaxis of fibroblasts towards PDGF. J Cell Sci 2012; 125:5758-69; PMID:22992460; http://dx.doi.org/10.1242/jcs.110007
- Rajagopal S, Ji Y, Xu K, Li Y, Wicks K, Liu J, Wong KW, Herman IM, Isberg RR, Buchsbaum RJ. Scaffold proteins IRSp53 and spinophilin regulate localized Rac activation by T-lymphocyte invasion and metastasis protein 1 (TIAM1). J Biol Chem 2010; 285:18060-71; PMID:20360004; http://dx.doi.org/10.1074/jbc.M109.051490
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 2009; 461:104-8; PMID:19693014; http://dx.doi.org/10.1038/nature08241
- Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol 2010; 12:591-7; PMID:20473296; http://dx.doi.org/10.1038/ncb2061
- Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell 2010; 18:226-36; PMID:20159593; http://dx.doi.org/10.1016/j.devcel.2009.11.015
- Schwechter B, Rosenmund C, Tolias KF. RasGRF2 Rac-GEF activity couples NMDA receptor calcium flux to enhanced synaptic transmission. Proc Natl Acad Sci U S A 2013; 110:14462-7; PMID:23940355; http://dx.doi.org/10.1073/pnas.1304340110
- Wang H, Zhao G, Liu X, Sui A, Yang K, Yao R, Wang Z, Shi Q. Silencing of RhoA and RhoC expression by RNA interference suppresses human colorectal carcinoma growth in vivo. J Exp Clin Cancer Res 2010; 29:123; PMID:20828398; http://dx.doi.org/10.1186/1756-9966-29-123
- Matthews HK, Marchant L, Carmona-Fontaine C, Kuriyama S, Larraín J, Holt MR, Parsons M, Mayor R. Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Development 2008; 135:1771-80; PMID:18403410; http://dx.doi.org/10.1242/dev.017350
- Kardash, E. et al. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat Cell Biol 12, 47-53; sup pp 1-11 (2010).
- Hirata E, Yukinaga H, Kamioka Y, Arakawa Y, Miyamoto S, Okada T, Sahai E, Matsuda M. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J Cell Sci 2012; 125:858-68; PMID:22399802; http://dx.doi.org/10.1242/jcs.089995
- Johnsson AK, Dai Y, Nobis M, Baker MJ, McGhee EJ, Walker S, Schwarz JP, Kadir S, Morton JP, Myant KB, et al. The Rac-FRET mouse reveals tight spatiotemporal control of Rac activity in primary cells and tissues. Cell Rep 2014; 6:1153-64; PMID:24630994; http://dx.doi.org/10.1016/j.celrep.2014.02.024
- Troeger A, Williams DA. Hematopoietic-specific Rho GTPases Rac2 and RhoH and human blood disorders. Exp Cell Res 2013; 319:2375-83; PMID:23850828; http://dx.doi.org/10.1016/j.yexcr.2013.07.002
- Carstanjen D, Yamauchi A, Koornneef A, Zang H, Filippi MD, Harris C, Towe J, Atkinson S, Zheng Y, Dinauer MC, et al. Rac2 regulates neutrophil chemotaxis, superoxide production, and myeloid colony formation through multiple distinct effector pathways. J Immunol 2005; 174:4613-20; PMID:15814684; http://dx.doi.org/10.4049/jimmunol.174.8.4613
- Faroudi M, Hons M, Zachacz A, Dumont C, Lyck R, Stein JV, Tybulewicz VL. Critical roles for Rac GTPases in T-cell migration to and within lymph nodes. Blood 2010; 116:5536-47; PMID:20870900; http://dx.doi.org/10.1182/blood-2010-08-299438
- Henderson RB, Grys K, Vehlow A, de Bettignies C, Zachacz A, Henley T, Turner M, Batista F, Tybulewicz VL. A novel Rac-dependent checkpoint in B cell development controls entry into the splenic white pulp and cell survival. J Exp Med 2010; 207:837-53; PMID:20308364; http://dx.doi.org/10.1084/jem.20091489
- Roberts AW, Kim C, Zhen L, Lowe JB, Kapur R, Petryniak B, Spaetti A, Pollock JD, Borneo JB, Bradford GB, et al. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity 1999; 10:183-96; PMID:10072071; http://dx.doi.org/10.1016/S1074-7613(00)80019-9
- Zhang H, Sun C, Glogauer M, Bokoch GM. Human neutrophils coordinate chemotaxis by differential activation of Rac1 and Rac2. J Immunol 2009; 183:2718-28; PMID:19625648; http://dx.doi.org/10.4049/jimmunol.0900849
- Niggli V, Schlicht D, Affentranger S. Specific roles of Rac1 and Rac2 in motile functions of HT1080 fibrosarcoma cells. Biochem Biophys Res Commun 2009; 386:688-92; PMID:19555660; http://dx.doi.org/10.1016/j.bbrc.2009.06.098
- Gardiner EM, Pestonjamasp KN, Bohl BP, Chamberlain C, Hahn KM, Bokoch GM. Spatial and temporal analysis of Rac activation during live neutrophil chemotaxis. Curr Biol 2002; 12:2029-34; PMID:12477392; http://dx.doi.org/10.1016/S0960-9822(02)01334-9
- Filippi MD, Harris CE, Meller J, Gu Y, Zheng Y, Williams DA. Localization of Rac2 via the C terminus and aspartic acid 150 specifies superoxide generation, actin polarity and chemotaxis in neutrophils. Nat Immunol 2004; 5:744-51; PMID:15170212; http://dx.doi.org/10.1038/ni1081
- Chan AY, Coniglio SJ, Chuang YY, Michaelson D, Knaus UG, Philips MR, Symons M. Roles of the Rac1 and Rac3 GTPases in human tumor cell invasion. Oncogene 2005; 24:7821-9; PMID:16027728; http://dx.doi.org/10.1038/sj.onc.1208909
- Gest C, Joimel U, Huang L, Pritchard LL, Petit A, Dulong C, Buquet C, Hu CQ, Mirshahi P, Laurent M, et al. Rac3 induces a molecular pathway triggering breast cancer cell aggressiveness: differences in MDA-MB-231 and MCF-7 breast cancer cell lines. BMC Cancer 2013; 13:63; PMID:23388133; http://dx.doi.org/10.1186/1471-2407-13-63
- Monypenny J, Zicha D, Higashida C, Oceguera-Yanez F, Narumiya S, Watanabe N. Cdc42 and Rac family GTPases regulate mode and speed but not direction of primary fibroblast migration during platelet-derived growth factor-dependent chemotaxis. Mol Cell Biol 2009; 29:2730-47; PMID:19273601; http://dx.doi.org/10.1128/MCB.01285-08
- Corbetta S, Gualdoni S, Ciceri G, Monari M, Zuccaro E, Tybulewicz VL, de Curtis I. Essential role of Rac1 and Rac3 GTPases in neuronal development. FASEB J 2009; 23:1347-57; PMID:19126596; http://dx.doi.org/10.1096/fj.08-121574
- Pennucci R, Tavano S, Tonoli D, Gualdoni S, de Curtis I. Rac1 and Rac3 GTPases regulate the development of hilar mossy cells by affecting the migration of their precursors to the hilus. PLoS One 2011; 6:e24819; PMID:21949760; http://dx.doi.org/10.1371/journal.pone.0024819
- Vaghi V, et al. Rac1 and Rac3 GTPases Control Synergistically the Development of Cortical and Hippocampal GABAergic Interneurons. Cereb Cortex 2012; PMID:23258346
- Dimidschstein J, Passante L, Dufour A, van den Ameele J, Tiberi L, Hrechdakian T, Adams R, Klein R, Lie DC, Jossin Y, et al. Ephrin-B1 controls the columnar distribution of cortical pyramidal neurons by restricting their tangential migration. Neuron 2013; 79:1123-35; PMID:24050402; http://dx.doi.org/10.1016/j.neuron.2013.07.015
- Nobes CD, Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem Soc Trans 1995; 23:456-9; PMID:8566347
- Seth A, Otomo T, Yin HL, Rosen MK. Rational design of genetically encoded fluorescence resonance energy transfer-based sensors of cellular Cdc42 signaling. Biochemistry 2003; 42:3997-4008; PMID:12680752; http://dx.doi.org/10.1021/bi026881z
- Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science 2004; 305:1615-9; PMID:15361624; http://dx.doi.org/10.1126/science.1100367
- Toutchkine A, Kraynov V, Hahn K. Solvent-sensitive dyes to report protein conformational changes in living cells. J Am Chem Soc 2003; 125:4132-45; PMID:12670235; http://dx.doi.org/10.1021/ja0290882
- Hanna S, Miskolci V, Cox D, Hodgson L. A new genetically encoded single-chain biosensor for Cdc42 based on FRET, useful for live-cell imaging. PLoS One 2014; 9:e96469; PMID:24798463; http://dx.doi.org/10.1371/journal.pone.0096469
- Dovas A, Cox D. Signaling networks regulating leukocyte podosome dynamics and function. Cell Signal 2011; 23:1225-34; PMID:21342664; http://dx.doi.org/10.1016/j.cellsig.2011.02.004
- Vignal E, De Toledo M, Comunale F, Ladopoulou A, Gauthier-Rouvière C, Blangy A, Fort P. Characterization of TCL, a new GTPase of the rho family related to TC10 andCcdc42. J Biol Chem 2000; 275:36457-64; PMID:10967094; http://dx.doi.org/10.1074/jbc.M003487200
- Murphy GA, Solski PA, Jillian SA, Pérez de la Ossa P, D’Eustachio P, Der CJ, Rush MG. Cellular functions of TC10, a Rho family GTPase: regulation of morphology, signal transduction and cell growth. Oncogene 1999; 18:3831-45; PMID:10445846; http://dx.doi.org/10.1038/sj.onc.1202758
- Chiang SH, Baumann CA, Kanzaki M, Thurmond DC, Watson RT, Neudauer CL, Macara IG, Pessin JE, Saltiel AR. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature 2001; 410:944-8; PMID:11309621; http://dx.doi.org/10.1038/35073608
- Yuan L, Sacharidou A, Stratman AN, Le Bras A, Zwiers PJ, Spokes K, Bhasin M, Shih SC, Nagy JA, Molema G, et al. RhoJ is an endothelial cell-restricted Rho GTPase that mediates vascular morphogenesis and is regulated by the transcription factor ERG. Blood 2011; 118:1145-53; PMID:21628409; http://dx.doi.org/10.1182/blood-2010-10-315275
- Kawase K, Nakamura T, Takaya A, Aoki K, Namikawa K, Kiyama H, Inagaki S, Takemoto H, Saltiel AR, Matsuda M. GTP hydrolysis by the Rho family GTPase TC10 promotes exocytic vesicle fusion. Dev Cell 2006; 11:411-21; PMID:16950130; http://dx.doi.org/10.1016/j.devcel.2006.07.008
- Fujita A, Koinuma S, Yasuda S, Nagai H, Kamiguchi H, Wada N, Nakamura T. GTP hydrolysis of TC10 promotes neurite outgrowth through exocytic fusion of Rab11- and L1-containing vesicles by releasing exocyst component Exo70. PLoS One 2013; 8:e79689; PMID:24223996; http://dx.doi.org/10.1371/journal.pone.0079689
