Abstract
eIF4E is over-expressed in many tumours, including a high proportion of breast cancers. eIF4E is an oncogene, and signalling pathways which promote eIF4E activity represent potential targets for therapeutic intervention in cancer. MNKs phosphorylate eIF4E on serine 209, a modification that can be required for eIF4E-dependent cell transformation. There is therefore a clear requirement to determine the role of MNKs in the proliferation and survival of cells from the major human tumours, such as breast cancer. Phosphorylated eIF4E protein was readily detectable in some breast tumour samples, but was below the limits of detection in others. Of 6 breast cancer cell lines representing the major sub-types of breast cancer, phosphorylated eIF4E was readily detectable in 5 of them, with MCF-7 cells displaying markedly lower levels. Long term colony forming assays demonstrated that all the five lines with high levels of phosphorylated eIF4E were highly sensitive to a MNK inhibitor. In short term assays, a range of responses was revealed. MCF-7 cells were insensitive in both assays. The anti-proliferative effects of the MNK inhibitor in breast cancer cells are primarily cytostatic, rather than cytotoxic, and are potentially due to the inhibition of cyclin D1 synthesis. Our data provide evidence that the sensitivity of breast cancer cells to MNK inhibition may correlate with baseline levels of eIF4E phosphorylation, and suggest that a proportion of breast cancers could be sensitive to inhibiting MNK kinase activity, and that the presence of phosphorylated eIF4E could provide a biomarker for the identification of responsive tumours.
Introduction
Breast cancer represents a major health problem with over 500,000 deaths annually world-wide. Substantial advances in the understanding of the biology underlying this disease have been made, and although the incidence continues to rise, mortality is falling in the western world. Despite this, in the face of metastatic disease, breast cancer remains almost universally fatal and, although cure rates have improved in early stage disease, patients continue to relapse despite maximal therapy.
Eukaryotic initiation factor 4E (eIF4E) has two distinct activities in the cell; in the nucleus it associates with a subset of mRNAs that contain ‘4E-sensitivity elements’ within their 3′UTRs and promotes their export through nuclear pores. In the cytoplasm it binds the 5′-7-methylguanosine cap of mRNAs and recruits the eIF4F complex (eIF4G and eIF4A, an RNA helicase) to promote translation, particularly of mRNAs with complex 5′-UTRs (e.g., containing inhibitory stem-loop structures).Citation1,Citation2 Through these two mechanisms eIF4E selectively enhances the synthesis of a variety of proteins involved in cell growth, proliferation and invasion, including the cell cycle regulatory protein cyclin D1,Citation3 the transcription factor c-Myc, growth factors such as VEGF and FGF2,Citation4 as well as the anti-apoptotic protein MCL-1.Citation5 eIF4E acts as an oncogene in experimental modelsCitation5–Citation7 and is overexpressed in a variety of human cancers.Citation8–Citation12 In breast cancer eIF4E overexpression correlates with cyclin D1, c-Myc and VEGF protein abundance,Citation13 and is associated with poor outcome in node negativeCitation14 and node positiveCitation15 disease with a higher rate of relapse. Low eIF4E protein following neoadjuvant therapy of breast tumors has been associated with improved outcome.Citation16 The mTOR signaling pathway, which promotes eIF4E activity, has already been validated as a therapeutic target in specific types of cancer.Citation17 Alternative strategies of targeting eIF4E are in development, including small molecule inhibitors of the eIF4E-eIF4G interaction and anti-sense oligonucleotides targeted to eIF4E.Citation2
Regulation of eIF4E activity forms a node of convergence of the PI3K/AKT and RAS/MAPK signaling pathways. Activated AKT phosphorylates and inactivates TSC2 resulting in the Rheb-dependent activation of mTORC1. mTORC1 in turn phosphorylates 4E-BP proteins, which reduces their ability to inhibit eIF4E binding to eIF4G.Citation7 ERK and p38 MAP kinases phosphorylate MNK1,Citation18 leading to phosphorylation of eIF4E at serine 209.Citation19 This phosphorylation event can promote the eIF4E-dependent export of mRNA such as cyclin D1,Citation20 and MDM2;Citation21 its effects on eIF4E-dependent translation remain unclear.Citation22 Critically, however, serine 209 phosphorylation is required for the transforming and oncogenic effects of eIF4E.Citation5,Citation20 Furthermore MNK kinases are dispensable for development and survival in mammalian modelsCitation23 making them potentially attractive as therapeutic targets for cancer.
Inhibition of MNK kinase activity has shown antiproliferative effects in lung cancer24 and prostate cancer cell lines.Citation25 Approximately 25% of breast cancers are characterised by overexpression of the HER2 receptor, which is associated with poor prognosis.Citation26 HER2 promotes signaling through both the PI3K/AKT and RAS/MAPK signaling pathways and MNK inhibition has been shown to be antiproliferative in a single HER2 overexpressing breast cancer cell line.Citation27 Here we have investigated the effects of an inhibitor of MNK activity on proliferation and survival signaling in a panel of breast cancer cell lines, including cells which are oestrogen receptor positive, HER2 overexpressing as well as an oestrogen receptor α (ERα), progesterone receptor (PR) and HER2 triple negative line.
Results
eIF4E phosphorylation in breast cancer tissues and cell lines.
Using western blotting, we assessed cell lysates from 10 human breast tumor tissue samples for expression of total eIF4E protein, as well as the abundance of the serine 209 phosphorylated form (peIF4E) (). Some samples in which eIF4E was readily detectable, also had higher amounts of the phosphorylated protein (samples 3, 8–10). However other samples had measurable total eIF4E with barely detectable peIF4E (samples 2, 4–7). The relative ratios of phosphorylated eIF4E to total eIF4E between the samples were also calculated; these displayed a wide range which suggests clear differences in MNK kinase activity between the tumors. We then proceeded to perform a comparable analysis of a panel of six breast cancer cell lines () representing some of the main breast cancer subtypes (reviewed in ref. Citation28). MCF-7, ZR75.1 and T47D have a luminal phenotype and express oestrogen receptor α, BT474 and SKBr3 are HER2+ve and MDA-MB-231 has a basal phenotype and is ERα, PR and HER2 negative. eIF4E was present in all of the lines, though with variable abundance. Phosphorylated eIF4E was, in most of the lines, more readily detectable than in the tumor samples, presumably because cell lines were proliferating optimally in high concentrations of serum growth factors. It was highest in the two HER2-expressing cell lines, as well as the ERα+ve line ZR75.1. Moderate levels were also seen in T47D (ERα+ve) as well as the MDA-MB-231 line which harbours an activating k-RAS mutation.Citation28 eIF4E phosphorylation was markedly lower in MCF-7 cells in this assay, consistent with our previous findings.Citation21
Time and dose-dependent inhibition of eIF4E phosphorylation by CGP57380 in breast cancer cell lines.
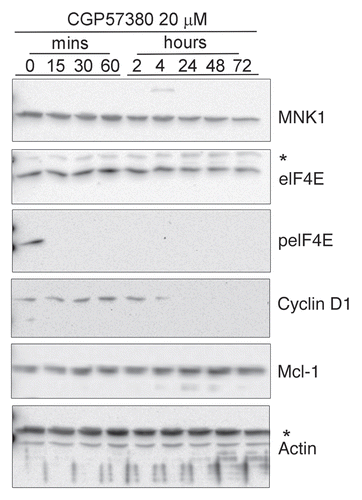
To investigate whether inhibition of eIF4E phosphorylation would have an effect on the proliferation of cell lines with higher basal levels of phosphorylated eIF4E we used a selective inhibitor of MNK kinase activity, CGP57380.Citation29,Citation30 We initially performed dose-response analysis at a single time point (24 h exposure to the compound) to determine the optimum concentration required to inhibit eIF4E phosphorylation. For this the lines SkBr3 and BT474 were used, as they exhibited the highest baseline phosphorylation at Ser209 (). In both cell lines there was a dose-dependent inhibition of eIF4E-phosphorylation, with maximal inhibition at 20 µM CGP57380, broadly consistent with findings from other investigators in different experimental models.Citation29,Citation30 Of the other lines (data not shown), the dose response in ZR75.1 closely matched that seen in BT474 and, as we have previously reported,Citation21 20 µM CGP57380 also inhibited eIF4E phosphorylation in T47D cells. Time course analysis () revealed some differences between the cell lines we examined. In BT474 cells, eIF4E phosphorylation was not detectable by the antibody after only 15 min exposure to CGP57380 and remained undetectable during 72 h continuous exposure to medium to which the compound had been added at the beginning of the experiment. The kinetics of inhibition were slightly slower in SKBr3 cells and phosphorylation could be detected again in the 72 h sample. MDA-MB-231 and T47D (not shown) demonstrated similar kinetics of inhibition to SKBr3 cells. The reasons for the differential time courses of eIF4E phosphorylation can only be speculated upon at present; differential rates of loss of phospho-eIF4E after addition of CGP57380 may indicate the activity of serine 209 phosphatases in some cell lines, though to our knowledge no such enzyme has yet been described. Differential rates of recovery of phosphorylation most likely reflect variations in the metabolism or export of the compound in different cell types, though the upregulation of a CGP57380-insensitive kinase is another possibility. Overall, the data demonstrate that eIF4E phosphorylated at serine 209 is either dephosphorylated or possibly degraded, in proliferating cells and MNK activity is continuously required to maintain eIF4E phosphorylation.
Inhibition of breast cancer cell line proliferation by the MN K inhibitor CGP57380.
Chrestensen et al.Citation27 reported that basal phosphorylation of eIF4E was elevated in breast cancer cells that overexpress HER2, compared to those that did not. However their analysis of the effects of CGP57380 on cell proliferation was restricted to a single cell line, AU565. This line overexpresses HER2 and its proliferation in soft agar is CGP57380-sensitive. We therefore examined the effects of CGP57380 on the proliferation of each of the breast cancer cell lines in our panel. Long-term colony-forming assays were used as they provide a sensitive readout irrespective of whether any antiproliferative effects are due to cell cycle inhibition or induction of apoptosis (). CGP57380 had marked antiproliferative effects on both of the two HER2 overexpressing lines SKBr3 and BT474, which exhibited high levels of phosphorylation of eIF4E. Proliferation was also clearly inhibited in ZR75.1, T47D and MDA-MB-231 cells, which all had either high or moderate levels of eIF4E phosphorylation. The only breast cancer cell line in which we did not observe substantial effects on proliferation in this assay were MCF-7, in which eIF4E phosphorylation is low or undetectable.
Colony forming assays can be highly sensitive, as cells are assayed under conditions of stress induced by reduced survival signals from neighbouring cells. We therefore used an MTS cell viability assays to determine whether the anti-proliferative effects of CGP57380 were detectable in short term (72 h) assays of cells cultured at higher (40–95%) cell densities (). Moderate inhibition of proliferation was seen in the two HER2 overexpressing cell lines, BT474 and SKBr3 and, to a lesser extent, MDA-MB-231 cells. MCF-7 cells were resistant to the compound, as they had been in the colony forming assays. T47D and ZR75.1 cells had shown a clear inhibitory response to CGP57380 in the colony forming assays, however in the MTS assays T47D showed no response and ZR75.1 appeared to show an increase in proliferation in response to the MNK inhibitor. This latter result needs interpreting with caution, as the MTS assay is only an indirect measure of cell viability; it determines the reductive capacity of poorly defined mitochondrial and cytoplasmic dehydrogenase enzymes. Thus an increase in the readout from this assay can be caused by, for example, a change in cellular metabolism that results in an increased cytoplasmic NADH:NAD+ ratio, rather than a genuine increase in cell proliferation.Citation31 The apparent proliferative response in ZR75.1 cells was not observed using an assay which measures cellular DNA content (CyQUANT, Invitrogen) (data not shown). In summary therefore, whilst all five cell lines that display moderate or high eIF4E phosphorylation are clearly responsive to CGP57380 in the sensitive colony forming assays, the two cell lines in which cell proliferation is known to be strictly dependent on signaling from the HER2 receptor are the most responsive in short term cell viability assays, the line containing an activating k-RAS mutation being partially responsive.
Mechanism of inhibition of breast cancer cell proliferation by CGP57380.
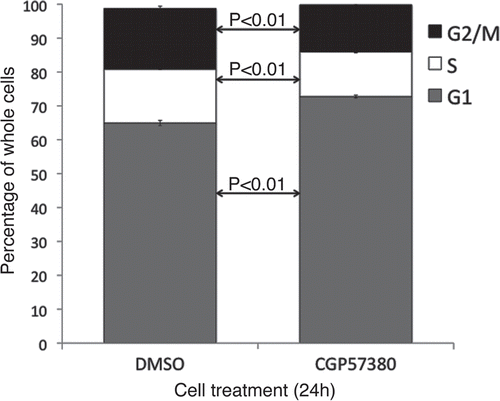
Live cell imaging was used to assess mitotic and apoptotic events in SKBr3 and MDA-MB-231 cells following 24 h treatment with CGP57380 (). During the subsequent 24 h period, control SKBr3 cells underwent 33.3 ± 1.9 mitotic events per 100 cells present at the beginning of the imaging period. In the CGP57380-treated cultures, this was reduced to 13.4 ± 1.5 mitotic events per 100 initial cells. Apoptotic events were low in both control and treated samples, with 0.4 ± 0.3 and 2.1 ± 0.5 apoptotic events per 100 initial cells in control and CGP57380 treated cells respectively. Whilst this does represent a ≈5 fold increase in the apoptotic rate, the overall contribution of apoptosis to the anti-proliferative effects of CGP57380 in SKBr3 cells is clearly minor compared to its more marked inhibitory effect on cell cycle progression. Essentially the same effects on cell cycle progression were seen in MDA-MB-231 cells, although in this cell line CGP57380 did not have any effect on the rate of apoptosis. We also assessed the effects of CGP57380 on cell cycle distribution using flow cytometry (). Consistent with the live cell imaging data, MDA-MB-231 cells treated with CGP57380 accumulated in G1 phase, with decreased numbers of cells in S and G2/M.
To further investigate the mechanism of the effects of CGP57380 on proliferation we assessed the impact of CGP57380 on known targets of phosphorylated eIF4E, i.e., cyclin D1 and MCL-1. BT474 cells were analysed as they showed the most profound changes in eIF4E phosphorylation and proliferation in response to the compound. CGP57380 reduced the abundance of cyclin D1 to below detectable levels within 4 h of treatment (). This is likely to be a cause of the inhibition of proliferation induced by the compound. In contrast the abundance of the pro-apoptotic protein MCL-1 was unaffected throughout the 72 h time course.
Discussion
We have shown that of a panel of six breast cancer cell lines representing a spectrum of phenotypes, five show a reduction in proliferation in response to an inhibitor of MNK kinase activity, CGP57380. This confirms the findings of Chrestensen et al.Citation27 who demonstrated that the proliferation of a single HER2 overexpressing breast cancer line is inhibited by CGP57380, but also extends this finding. Specifically, in the lines we have examined, all those which demonstrated readily detectable baseline amounts of phosphorylated eIF4E were sensitive to the compound. These included both ERα+ve lines as well as a HER2, ERα, PR negative line that harbours an activating k-RAS mutation. Importantly, no inhibition of proliferation was seen in the one cell line in which basal eIF4E phosphorylation was not readily detectable (MCF7). The breast tumor samples which we assessed showed considerable variation in the levels of eIF4E phosphorylation. Based on this, as well as the cell line data, it may therefore be that eIF4E phosphorylation could provide a biomarker for response to inhibition of the MNKs should this be developed as a therapeutic strategy in breast cancer.
The restriction of anti-proliferative effects of CGP57380 to cell lines showing basal levels of phosphorylated eIF4E is also an important finding when one considers the target specificity of the MNK inhibitor. It has been shown, at least in vitro, that CGP57380 is able to inhibit protein kinases in addition to the MNKs,Citation32 raising the possibility that some of the effects of CGP57380 in cells could be through its effects on these enzymes. Suggested criteria for assessing the specificity of the effects of chemical inhibitors of protein function in cell regulation have recently been published.Citation33 With respect to these criteria, we have determined (data not shown) that the effects of CGP57380 on cell proliferation occur in the same concentration range that inhibits eIF4E phosphorylation in the breast cancer cells. We also show a concordance between the presence of phosphorylated eIF4E in cells and their sensitivity to inhibition of proliferation by the compound. That MNKs are the relevant target of CGP57380 with respect to their antiproliferative effects is further supported by downregulation of cyclin D1 and cell cycle arrest and being the dominant response to the inhibitor, the synthesis of cyclin D1 being known to be increased by eIF4E phosphorylation.Citation20,Citation34
High expression of eIF4E has been established by several groups to be a poor prognostic marker in breast cancer.Citation14–Citation16 Phosphorylated eIF4E can independently predict prognosis in lung cancer in a multivariate analysis of prognostic factors,Citation35 and is also elevated in gastric and colorectal cancers.Citation36 Thus the possibility of targeting MNK kinases in tumors in which overexpression of phosphorylated eIF4E is seen may well be considered as a novel therapeutic strategy worthy of further consideration.
Materials and Methods
Culture of the panel of human breast cancer lines has been described previously.Citation37 Primary human breast cancer material was obtained from the Southampton Cancer Research UK tumor bank and assayed with local ethics committee approval. The MNK 1 and 2 inhibitor N3-(4-fluorophenyl)-1H-pyrazolo-[3,4-d]pyrimidine-3,4-diamine (CGP57380, Tocris Bioscience) was used at concentrations as indicated, in dimethyl sulfoxide vehicle to a final concentration of 0.5% (v/v) in culture medium. For proliferation assays, cells were plated to 96 well plates at 1,000 cells/well with triplicate wells for each condition. 24 h later CGP57380 was added at concentrations as indicated. After 72 h MTS reagent (CellTitre Aqueous One Cell Proliferation assay, Promega) was added as per the manufacturers instructions. Absorbance at 490 nm was measured after 2 h using a varioskan flash plate reader (Thermo Scientific). Mean of triplicate values from each of three experiments were combined for each cell line.
For colony assays, cells were treated for 24 h before being trypsinised and counted. 1,250 cells were re-plated in medium containing CGP57380 or DMSO and incubated for 9–12 d before being fixed in methanol and stained with Giemsa stain (Sigma). For live cell imaging, cells were treated for 24 h before being imaged for 24 h using an Olympus IX81 microscope with CO2 and temperature controlled environmental chamber, controlled by SIS Cell P software. At least 30 cells per field were counted from three fields from each of three independent wells. Image J software was used for analysis. For cell cycle analysis, cells were treated for 24 h before being trypsinised with all washes retained and pelleted by centrifugation (1,000x g 4 min 4°C). Cells were washed in PBS and fixed with ethanol. Fixed cells were further washed in PBS and stained with propidium iodide (20 µg/ml propidium iodide, 0.2 mg/ml RNAse A, 0.1% (v/v) Triton X-100 in PBS) for 30 min at room temperature. Flow cytometry was performed using a FACSCalibur (Becton Dickinson) and analysed with CellQuest software (Becton Dickinson).
Proteins were analysed by immunoblotting as previously described.Citation37 Images of blots were captured on a Bio-Rad Fluoro-S-Max imager and quantified using Bio-Rad Quantity One software. The following antibodies were used: MNK1 #2196, eIF4E #9742, peIF4E (serine 209) #9741 (Cell Signalling Technology), Cyclin D1 Ab-3 (Oncogene Research Products), MCl-1 sc-819 (Santa Cruz Biotechnology), actin (20-33) A5060 (Sigma).
Abbreviations
| CGP57380 | = | N3-(4-fluorophenyl)-1H-pyrazolo-[3,4-d]pyrimidine-3,4-diamine |
| ERα | = | oestrogen receptor alpha |
| peIF4E | = | eIF4E phosphorylated at serine 209 |
| PR | = | progesterone receptor |
Figures and Tables
Figure 1 eIF4E expression and phosphorylation in breast cancer tissue. Equal protein concentrations from 10 breast cancer tissue samples were separated by SDS-PAGE and western blots probed with antibodies to eIF4E and serine 209 phosphorylated eIF4E. Figure representative of two independent blots. Quantitation shows the relative ratios of phosphorylated- to total eIF4E between the samples (*indicates total eIF4E is too low for a reliable analysis).
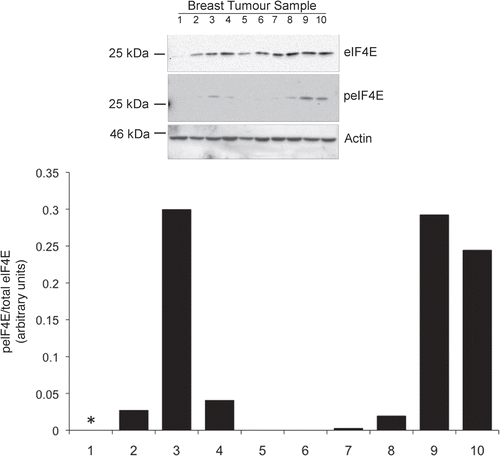
Figure 2 eIF4E expression and phosphorylation in breast cancer tissue. Cells in exponential growth phase were lysed 24 h after the application of fresh medium. Proteins were separated by SDS-PAGE and western blots probed with antibodies to MNK1, eIF4E and serine 209 phosphorylated eIF4E. Figure representative of two independent blots. Quantitation is as per .
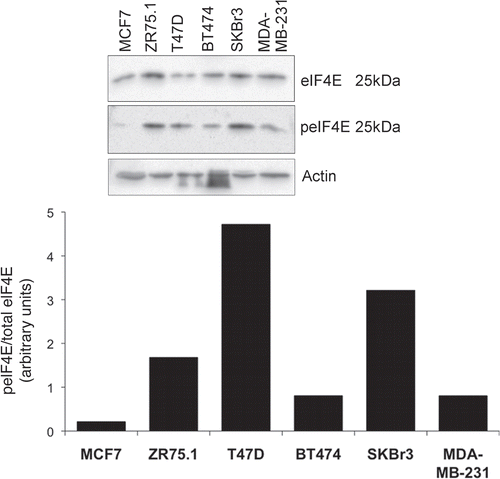
Figure 3 Dose response analysis of the inhibition of eIF4E phosphorylation by CGP57380. BT474 and SKBr3 cells were treated with the indicated concentrations of CGP57380 for 24 h prior to analysis of cell lysates by western blotting.
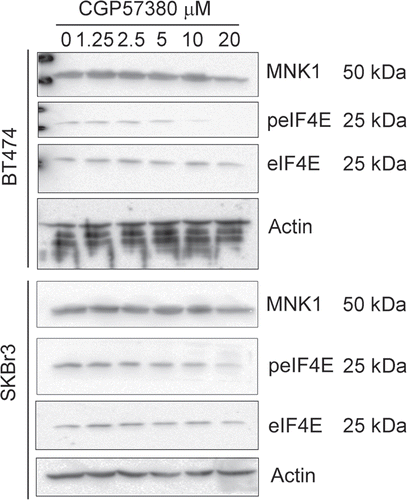
Figure 4 Time course analysis of the inhibition of eIF4E phosphorylation by CGP57380. Cells were treated with 20 µM CGP57380 for the times as indicated. Proteins were analysed by western blotting. *, cross reacting band.
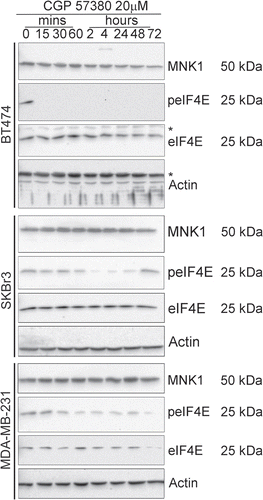
Figure 5 Effects of the MNK inhibitor, CGP57380, on colony formation by breast cancer cell lines. Cells were treated with CGP57380 20 µM or DMSO vehicle control for 24 h before being trypsinised, counted and re-plated at equal number in CGP57380 or DMSO. Colonies were fixed with methanol and stained with Giemsa 9-12 days later. Representative of three replicate plates.
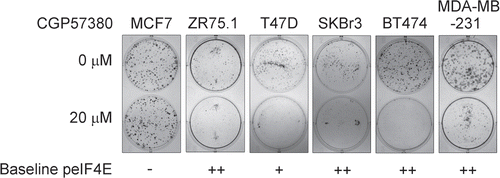
Figure 6 Effects of 72 h exposure to CGP57380 on breast cancer cell line proliferation. Cells for the experiment were plated at a density which resulted in them being in exponential growth phase at the point of assay. Data represents the results from three independent experiments for each cell line. (Bars = mean + SEM, X axis labels represent CGP57380 concentration (µM) 0 = 0.5% DMSO carrier control).
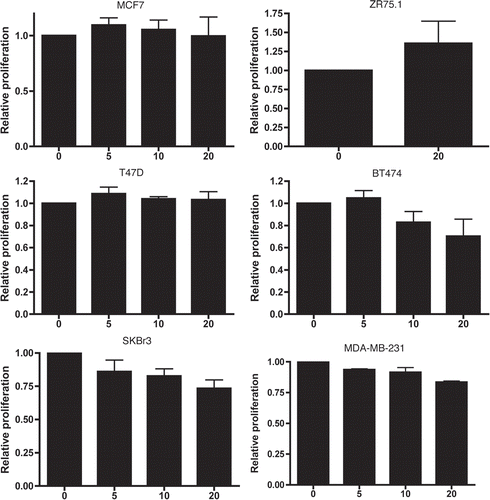
Figure 7 Live cell imaging analysis of CGP57380 treated breast cancer cells. MDA-MB-231 cells and SKBr3 cells were treated with CGP57380 20 µM or DMSO vehicle control before undergoing time lapse photomicroscopy. Graphs show the number of mitotic (black bars) and apoptotic events (white bars) per 100 initial cells during the 24 h imaging period. Results are mean of three experiments + SEM.
Figure 8 Cell cycle analysis of MDA-MB-231 cell line following CGP57380 treatment. MDA-MB-231 cells were treated with CGP57380 20 µM or DMSO (vehicle control); for 24 h prior to analysis of DNA content by propidium iodide staining and flow cytometry. Data shown is the means of three independently repeated experiments, each comprising three replicates. Student's t-test was used for statistical analysis.
Acknowledgements
We are grateful to Matthew Darley for technical support and the Southampton School of Medicine Bioimaging Unit for assistance with live cell imaging. We thank Prof. Chris. Proud, University of Southampton, for helpful discussion. M.W. is supported by Cancer Research UK.
References
- Silva RL, Wendel HG MNK. EIF4E and targeting translation for therapy. Cell Cycle 2008; 7:553 - 555
- Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res 2008; 68:631 - 634
- Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci USA 1996; 93:1065 - 1070
- De Benedetti A, Harris AL. eIF4E expression in tumors: its possible role in progression of malignancies. Int J Biochem Cell Biol 1999; 31:59 - 72
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev 2007; 21:3232 - 3237
- Lazaris-Karatzas A, Sonenberg N. The mRNA 5′ capbinding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol Cell Biol 1992; 12:1234 - 1238
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E—from translation to transformation. Oncogene 2004; 23:3172 - 3179
- Sorrells DL, Black DR, Meschonat C, Rhoads R, De Benedetti A, Gao M, et al. Detection of eIF4E gene amplification in breast cancer by competitive PCR. Ann Surg Oncol 1998; 5:232 - 237
- Sorrells DL, Ghali GE, Meschonat C, DeFatta RJ, Black D, Liu L, et al. Competitive PCR to detect eIF4E gene amplification in head and neck cancer. Head Neck 1999; 21:60 - 65
- Berkel HJ, Turbat-Herrera EA, Shi R, de Benedetti A. Expression of the translation initiation factor eIF4E in the polyp-cancer sequence in the colon. Cancer Epidemiol Biomarkers Prev 2001; 10:663 - 666
- Matthews-Greer J, Caldito G, de Benedetti A, Herrera GA, Dominguez-Malagon H, Chanona-Vilchis J, et al. eIF4E as a marker for cervical neoplasia. Appl Immunohistochem Mol Morphol 2005; 13:367 - 370
- Wang R, Geng J, Wang JH, Chu XY, Geng HC, Chen LB. Overexpression of eukaryotic initiation factor 4E (eIF4E) and its clinical significance in lung adenocarcinoma. Lung Cancer 2009; 66:237 - 244
- Kleiner HE, Krishnan P, Tubbs J, Smith M, Meschonat C, Shi R, et al. Tissue microarray analysis of eIF4E and its downstream effector proteins in human breast cancer. J Exp Clin Cancer Res 2009; 28:5
- Flowers A, Chu QD, Panu L, Meschonat C, Caldito G, Lowery-Nordberg M, et al. Eukaryotic initiation factor 4E overexpression in triple-negative breast cancer predicts a worse outcome. Surgery 2009; 146:220 - 226
- McClusky DR, Chu Q, Yu H, Debenedetti A, Johnson LW, Meschonat C, et al. A prospective trial on initiation factor 4E (eIF4E) overexpression and cancer recurrence in node-positive breast cancer. Ann Surg 2005; 242:584 - 590
- Hiller DJ, Chu Q, Meschonat C, Panu L, Burton G, Li BD. Predictive value of eIF4E reduction after neoadjuvant therapy in breast cancer. J Surg Res 2009; 156:265 - 269
- Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, et al. Temsirolimus, interferon alfa or both for advanced renal-cell carcinoma. N Engl J Med 2007; 356:2271 - 2281
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J 1997; 16:1909 - 1290
- Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the capbinding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol 1999; 19:1871 - 1880
- Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res 2004; 64:8639 - 8642
- Phillips A, Blaydes JP. MNK1 and EIF4E are downstream effectors of MEKs in the regulation of the nuclear export of HDM2 mRNA. Oncogene 2008; 27:1645 - 1649
- Buxade M, Parra-Palau JL, Proud CG. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front Biosci 2008; 13:5359 - 5373
- Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol 2004; 24:6539 - 6549
- Wang X, Yue P, Chan CB, Ye K, Ueda T, Watanabe-Fukunaga R, et al. Inhibition of mammalian target of rapamycin induces phosphatidylinositol 3-kinase-dependent and Mnk-mediated eukaryotic translation initiation factor 4E phosphorylation. Mol Cell Biol 2007; 27:7405 - 7413
- Bianchini A, Loiarro M, Bielli P, Busa R, Paronetto MP, Loreni F, et al. Phosphorylation of eIF4E by MNKs supports protein synthesis, cell cycle progression and proliferation in prostate cancer cells. Carcinogenesis 2008; 29:2279 - 2288
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235:177 - 182
- Chrestensen CA, Shuman JK, Eschenroeder A, Worthington M, Gram H, Sturgill TW. MNK1 and MNK2 regulation in HER2-overexpressing breast cancer lines. J Biol Chem 2007; 282:4243 - 4252
- Hollestelle A, Nagel JH, Smid M, Lam S, Elstrodt F, Wasielewski M, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat 2009; 121:53 - 64
- Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol 2001; 21:5500 - 5511
- Buxade M, Parra JL, Rousseau S, Shpiro N, Marquez R, Morrice N, et al. The Mnks are novel components in the control of TNFalpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 2005; 23:177 - 189
- Dhanjaland P, Fry JR. Determinants of MTT reduction in rat hepatocytes. Biomarkers 1997; 2:111 - 116
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J 2007; 408:297 - 315
- Cohen P. Guidelines for the effective use of chemical inhibitors of protein function to understand their roles in cell regulation. Biochemical Journal 2009; 425:53 - 54
- Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle 2007; 6:65 - 69
- Yoshizawa A, Fukuoka J, Shimizu S, Shilo K, Franks TJ, Hewitt SM, et al. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in nonsmall cell lung cancer. Clin Cancer Res 16:240 - 248
- Fan S, Ramalingam SS, Kauh J, Xu Z, Khuri FR, Sun SY. Phosphorylated eukaryotic translation initiation factor 4 (eIF4E) is elevated in human cancer tissues. Cancer Biol Ther 2009; 8:1463 - 1469
- Phelps M, Darley M, Primrose JN, Blaydes JP. p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor alpha positive breast cancer cells. Cancer Res 2003; 63:2616 - 262