Abstract
Commentary to: SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome Manoj Kumar Kashyap, H. C. Harsha, Santosh Renuse, Harsh Pawar, Nandini A. Sahasrabuddhe, Min-Sik Kim, Arivusudar Marimuthu, Shivakumar Keerthikumar, Babylakshmi Muthusamy, Kumaran Kandasamy, Yashwanth Subbannayya, Thottethodi Subrahmanya Keshava Prasad, Riaz Mahmood, Raghothama Chaerkady, Stephen Meltzer, Rekha V. Kumar, Anil K. Rustgi and Akhilesh Pandey
Esophageal squamous cell carcinoma (ESCC) is the sixth most frequent cause of cancer death worldwide. Human ESCC carcinogenesis is a multistage process involving multifactorial etiology and genetic-environment interactions.Citation1,Citation2 Patients with ESCC have a poor prognosis, with 5-year survival rates of less than 10%, because of its rapid spread and cancer-associated malnutrition due to dysphagia and cachexia.Citation3 The molecular mechanisms that underlie tumor formation and progression are still not completely understood, although several advances, based on alterations of gene expression and deregulated protein levels, have been reported. These studies identified candidate biomarker molecules, such as annexin I and tumor rejection antigen, via proteomic approaches.Citation4 Unfortunately, most of the newly described biomarkers have limited specificity, sensitivity or both.Citation5–Citation7 The discovery of new markers to discriminate normal from tumor cells is critically important for the early detection and diagnosis of ESCC. Among these, the identification of biomarkers secreted or shed by the tumor is the key step in the development of accessible and cost effective patient screening.
Most secreted proteins are predicted to have a 70 amino acid signal peptide located at the N-terminus of the nascent protein. This signal sequence is cleaved in the lumen of the endoplasmic reticulum and the protein is ultimately released outside of the plasma membrane through a tightly regulated multistage vesicle fusion event.Citation8 In this process, the secreted proteins are released in the blood stream or extracellular fluid, where they are diluted by six or more orders of magnitude and subjected to proteolysis.Citation9 As a consequence, cancer biomarkers are present at nanomolar concentrations in an abundant background of extracellular matrix and serum proteins. Conventional detection techniques may be limited by the complexity and broad dynamic range of such samples.Citation10 Currently, there is a growing consensus that a panel of markers, rather than individual molecules, would increase the efficacy and accuracy of early stage cancer detection. The “tumor secretome,” or group of proteins, secreted by the cancer cells,Citation11 can be analyzed to identify circulating molecules present at elevated levels in serum or plasma from cancer patients. These proteins have the potential to act as cancer derived marker candidates, which are distinct from hostresponsive marker candidates. In recent years, several groups have demonstrated the efficacy of secretome-based strategies in a variety of cancers including breast cancer, lung cancer and oral cancer.Citation12–Citation14 Similarly, a limited number of studies investigated differentially expressed proteins in ESCC versus non-lesional cells and identified a variety of candidate biomarkers including PRDX5 and HSP90.Citation15,Citation16 The secretome of cancer cells in these studies was resolved by one-or two-dimensional gel electrophoresis, subjected either to in-gel trypsic digestion and eventually analyzed by MALDI-TOF or LC-MS/MS or directly trypsinized in solution and run on the LC-MS/MS. Advances in both protein separation and detection resulted in the identification of more and more proteins and significant decrease of the false positive findings. In general, the LC-MS/MS methodology detected more proteins then the MALDI-TOF technique. Although the results from these proteomic-based approaches are quite encouraging, the progress of these studies has been hampered by the unresolved yet question how to accurately compare comprehensive proteomes and their subsets such as secretomes.
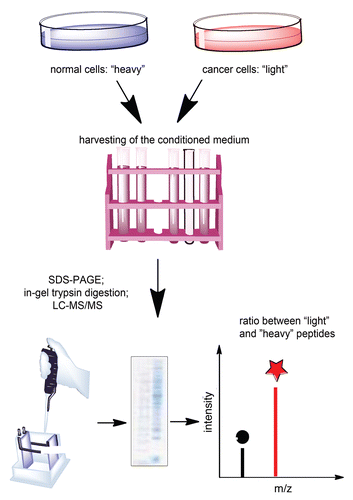
A possible solution of this problem may be the use of stable isotope labeling by amino acids in cell culture (SILAC). This is an emerging technology for quantitative proteomics that allows clear quantification of cellular aspects that differ between two phenotypes.Citation17–Citation19 In the current issue of Cancer Biology & Therapy,Citation20 Kashyap and colleagues successfully utilized this technique to identify potential biomarker panels for ESCC detection. SILAC uses the normal metabolic machinery of the cell to label proteins with “light” (normal) amino acid or “heavy” (isotope) amino acids (). The heavy amino acid can contain 2H instead of H, 13C instead of 12C or 15N instead of 14N. Within six doublings, the amino acids are fully incorporated into every peptide produced and secreted by the cell. Incorporation of the heavy amino acid into a peptide leads to a detectable (usually 2–6 Da) mass shift compared to the peptide that contains the light version of the amino acid, but no other chemical changes are introduced. In the study by Kashyap et al. the normal cells were labeled with the heavy amino acid and the cancer cells were labeled with the light amino acid.Citation20 Upon incorporation of the labeled amino acids, the conditioned media from both populations ware harvested, the proteomes were extracted, resolved on SDS-PAGE and analyzed by mass spectroscopy (MS). Metabolic labeling experiments are especially advantageous in identifying proteins secreted in culture because peptides naturally expressed appear as sequence-matched pairs separated by the fixed mass offset. If both cell populations secreted equal amounts of a given protein, recovered peptides appear at a 1:1 ratio. A higher intensity from the peptide that contains heavy amino acid indicates that the protein was more abundant in one of the populations (). Because there isn't any chemical difference between the light and the heavy amino acids, the ratio of the peak intensities in the mass spectrometer directly yields the ratio of the proteins in the respective cell population. Low signal-to noise ratio, as detected by Kashyap et al. ensures accuracy of the quantification. Based on that, the group identified not only several biomarkers previously known to be increased in ESCC, such as MMP1 and the transferrin receptor, but also a panel of novel candidate molecules.Citation20
Taken together, these results may lead to the discovery of novel diagnostic tests for ESCC. The approach used by the authors, however has considerable limitations. Whereas proteins in cultured cells can be readily labeled, those in living organisms cannot. Technologies have been developed to metabolically label worms, flies,Citation21 and even mice,Citation22 but human labeling has remained “unlikeable”. Moreover, the high-accuracy of SILAC-based quantification may be more than what is needed for proteomics-based biomarker discovery. The sensitivity of current mass spectroscopy-based detection methods suggests that subtle changes in the cancer secretome picked up by SILAC may be difficult if not impossible to accurately monitor in plasma levels at this time. Nevertheless, such studies offer valuable knowledge for selecting the right candidate molecules and pave the way for the development of more accessible and practical detection techniques.
Acknowledgements
This work was supported in part by NIH Grant CA140615.
Commentary to:
References
- Bird-Lieberman EL, Fitzgerald RC. Early diagnosis of oesophageal cancer. Br J Cancer 2009; 101:1 - 6
- Endo M, Kawano T. Detection and classification of early squamous cell esophageal cancer. Dis Esophagus 1997; 10:155 - 158
- Kranzfelder M, Buchler P, Lange K, Friess H. Treatment options for squamous cell cancer of the esophagus: a systematic review of the literature. J Am Coll Surg 2010; 210:351 - 359
- Zhou G, Li H, DeCamp D, Chen S, Shu H, Gong Y, et al. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol Cell Proteomics 2002; 1:117 - 124
- Yang YF, Li H, Xu XQ, Diao YT, Fang XQ, Wang Y, et al. An expression of squamous cell carcinoma antigen 2 in peripheral blood within the different stages of esophageal carcinogenesis. Dis Esophagus 2008; 21:395 - 401
- Kubota M, Yamana H, Sueyoshi S, Fujita H, Shirouzu K. The significance of telomerase activity in cancer lesions and the noncancerous epithelium of the esophagus. Int J Clin Oncol 2002; 7:32 - 37
- Bacus JW, Bacus JV, Stoner GD, Moon RC, Kelloff GJ, Boone CW, et al. Quantitation of preinvasive neoplastic progression in animal models of chemical carcinogenesis. J Cell Biochem Suppl 1997; 28:21 - 38
- Walter P, Gilmore R, Blobel G. Protein translocation across the endoplasmic reticulum. Cell 1984; 38:5 - 8
- Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics 2003; 2:1096 - 1103
- Issaq HJ, Conrads TP, Prieto DA, Tirumalai R, Veenstra TD. SELDI-TOF MS for diagnostic proteomics. Anal Chem 2003; 75:148 - 155
- Volmer MW, Radacz Y, Hahn SA, Klein-Scory S, Stühler K, Zapatka M, et al. Tumor suppressor Smad4 mediates downregulation of the anti-adhesive invasion-promoting matricellular protein SPARC: Landscaping activity of Smad4 as revealed by a “secretome” analysis. Proteomics 2004; 4:1324 - 1334
- Kulasingam V, Diamandis EP. Proteomics analysis of conditioned media from three breast cancer cell lines: a mine for biomarkers and therapeutic targets. Mol Cell Proteomics 2007; 6:1997 - 2011
- Lou X, Xiao T, Zhao K, Wang H, Zheng H, Lin D, et al. Cathepsin D is secreted from M-BE cells: its potential role as a biomarker of lung cancer. J Proteome Res 2007; 6:1083 - 1092
- Weng LP, Wu CC, Hsu BL, Chi LM, Liang Y, Tseng CP, et al. Secretome-based identification of Mac-2 binding protein as a potential oral cancer marker involved in cell growth and motility. J Proteome Res 2008; 7:3765 - 3775
- Fujita Y, Nakanishi T, Hiramatsu M, Mabuchi H, Miyamoto Y, Miyamoto A, et al. Proteomics-based approach identifying autoantibody against peroxiredoxin VI as a novel serum marker in esophageal squamous cell carcinoma. Clin Cancer Res 2006; 12:6415 - 6420
- Fujita Y, Nakanishi T, Miyamoto Y, Hiramatsu M, Mabuchi H, Miyamoto A, et al. Proteomics-based identification of autoantibody against heat shock protein 70 as a diagnostic marker in esophageal squamous cell carcinoma. Cancer Lett 2008; 263:280 - 290
- Pimienta G, Chaerkady R, Pandey A. SILAC for global phosphoproteomic analysis. Methods Mol Biol 2009; 527:107 - 116
- Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods 2003; 29:124 - 130
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002; 1:376 - 386
- Kashyap MK. SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome. Cancer Biol Ther 2010; 10:796 - 810
- Krijgsveld J, Ketting RF, Mahmoudi T, Johansen J, Artal-Sanz M, Verrijzer CP, et al. Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat Biotechnol 2003; 21:927 - 931
- Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 2008; 134:353 - 364