Abstract
Gene therapy protocols for the treatment of cancer often employ gene promoter sequences that are known to be over-expressed in specific tumor cell types relative to normal cells. These promoters, while specific, are often weakly active. It would be desirable to increase the activity of such promoters, while at the same time retain specificity, so that the therapeutic gene is more robustly expressed. Using a luciferase reporter DNA construct in both in vitro cell transfection assays and in vivo mouse tumor models, we have determined that in the absence of any other DNA sequence, a previously identified 18-base pair enhancer sequence called CanScript, lying upstream of the MSLN gene, has ~25% of the promoter activity of CAG, a very strong non-specific promoter/enhancer, in tumor cells in which MSLN is highly expressed. Furthermore, tandem repeat copies of CanScript enhance transcription in a dose-dependent manner and, when coupled with promoter sequences that are active in tumor cells, increase promoter activity. These findings suggest that the incorporation of CanScript into gene constructs may have application in enhancing activity of promoters used in cancer-targeting gene therapy strategies, thereby improving therapeutic efficacy.
Introduction
The MSLN gene, encoding mesothelin, is highly expressed in a third of human malignancies including ovarian, pancreatic, bronchial, gastroesophageal, cervical, endometrial and biliary carcinomas.Citation1–Citation3,Citation9,Citation12 In ovarian tumors that have little or no CA125 expression, MSLN is one of two proteins, the other being HE4, proven to be the most reliable biomarkers for ovarian cancer, as compared to a number of other candidate proteins, when expression in normal tissues was considered.Citation4,Citation11 The differential expression of MSLN between tumor and normal cells makes the promoter of the MSLN gene an attractive choice when designing gene therapy strategies for the treatment of ovarian and pancreatic cancers. In a recent study, we used a 1.8 kb sequence upstream of the human MSLN gene to regulate expression of a therapeutic diphtheria toxin (DT-A) gene and nanoparticles to deliver the MSLN/DT-A DNA to the peritoneal space of mice bearing ovarian tumors.Citation5 The growth rate of tumors in mice treated with MSLN/DT-A nanoparticles was significantly reduced compared to control mice, resulting in a significant increase in life span. Subsequently, we were able to duplicate this work in pancreatic cancer cell lines overexpressing MSLN.Citation13
The high cancer cell-specific activity of MSLN is attributable to an 18 bp upstream enhancer, termed CanScript, which selectively elevates transcription, in part, upon binding of TEF-1(TEAD1), a transcription enhancer factor, to a MCAT motif.Citation6 It has previously been shown that three concatemerized copies of CanScript produce a 30-fold enhancement of the SV40 minimal promoter activity in MSLN-expressing cancer cells in culture.Citation6 This finding suggested that the CanScript sequence may find utility in selectively enhancing the activity of mammalian promoter sequences in targeted gene therapy protocols. To test this possibility, we generated luciferase reporter gene constructs having different copy numbers of CanScript (1X, 2X, 3X, 6X) either alone or in combination with different tumor-specific promoters (MSLN, HE-4, PSA) and then assayed gene activity in various cell lines and in tumor-bearing mice. While the general use and molecular mechanism of CanScript to boost the activity of promoters that are overexpressed in tumors requires further analysis, the results of our study support incorporating CanScript into the design of gene therapy strategies when increased, tumor-specific gene expression is desired to improve therapeutic efficacy.
Results
Transcriptional efficacy of CanScripts 1X, 2X, 3X and 6X in cancer cells.
We generated luciferase reporter gene constructs, each having a single copy or multiple copies of the CanScript sequence in tandem (1X, 2X, 3X, 6X) (). The tandem repeats were sequenced for validation and to rule out a mutation induced by the sub-cloning process (data not shown). Multiple cell lines were transiently transfected with these constructs and luciferase activity was measured after 24 and 48 hrs to test CanScript promoter function and cell-type specificity (). As there was no difference in the results at the two time points, we only report our observations after 24 hrs. MSLN-expressing ovarian tumor cells (A2780) and pancreatic tumor cells (PL-5) transfected with CS1X/Luc (a single copy of the 18-base pair CanScript sequence linked to firefly luciferase coding sequence), expressed luciferase. In A2780 cells, luciferase activity increased in proportion to the number of copies of CanScript (e.g., CS6X/Luc has 6.8X more activity than CS1X/Luc). Three other MSLN-expressing ovarian cancer cell lines (OVCA420, OVCAR3 and ES-2) displayed similar CanScript dose-proportional luciferase activity (data not shown). Luciferase expression in transfected PL-5 cells did not display the same proportionality, although cells transfected with CS6X/Luc had 2.4X more activity than cells transfected with CS1X/Luc. Four other pancreatic cancer cell lines (CAPAN1, AsPc1, Hs766T and MiaPaca2) showed similar dose responsiveness (data not shown). Upon transfection of MSLN nonexpressing cells (HepG2, A549, PC3 and LNCaP), background luciferase expression was observed for constructs with 1, 2 and 3 copies of CanScript. Only when these cells were transfected with CS6X/Lluc was a very low level of expression observed, except for LNCaP cells which had no expression.
To investigate whether the amount of TEF-1 in cells might account for differential CanScript function, we used real-time quantitative PCR (qPCR) to determine the amount of TEF-1 mRNA in each cell line used in this study (). We found no correlation between the amount of TEF-1 mRNA and CanScript transcriptional activation, confirming a previous report showing that TEF-1 presence is required for transcriptional activity, yet another cofactor is most likely involved.Citation6 For example, HepG2 and PC3 cells had very low CanScript activity, but these lines had high amounts of TEF-1 mRNA expression levels compared to the other cell lines ().
Transcriptional efficacy of MSLN-CanScript constructs.
Cell lines were also transfected with three other DNA constructs, MSLN/Luc, MSLN/CS3X/Luc and CS3X/MSLN/Luc (). These constructs contain the MSLN promoter region ± CS3X positioned 5′ or 3′ to the MSLN promoter. As expected, luciferase expression was observed in A2780 and PL-5 cells transfected with MSLN/Luc. A549 and PC3 cells also expressed luciferase at a low level, while there was only background expression in HepG2 and LNCaP cells (). MSLN/CS3X/Luc and CS3X/MSLN/Luc were equally effective at enhancing luciferase expression in A2780 and PL-5 cells as compared to cells transfected with MSLN/Luc (3.5X and 1.8X, respectively). Luciferase expression in A549 and PC3 cells transfected with either MSLN/CS3X/Luc or CS3X/MSLN/Luc was also enhanced as compared to MSLN/Luc transfected cells. In both these cell lines, however, enhancement was slightly higher when CS3X was positioned 5′ to the MSLN promoter.
Promoter specificity of canscript.
To test whether CanScript enhanced the activity of promoters other than its native MSLN promoter, we transfected cells with four additional reporter gene constructs: HE4/Luc, HE4/CS3X/Luc, PSES/Luc and PSES/CS3X/Luc (). HE4 is the promoter of the human epididymal 4 gene, HE4, a gene that is highly overexpressed in a majority of ovarian tumors.Citation4 PSES is a chimeric enhancer, composed of two modified regulatory elements controlling the human PSA and PSMA genes, which has multiple copies of the androgen receptor sequence.Citation8 The HE4 promoter was active in both A2780 and PL-5 cells and activity in these cells was enhanced with the addition of CS3X (5.8X and 2.0X, respectively) (). No luciferase activity was observed in the other cell lines transfected with HE4/Luc ± CS3X reporter constructs. The PSES promoter was highly active in androgen-sensitive LNCaP prostate cancer cells, and also had low activity in A2780 and PL-5 cells, but no activity in HepG2, A549 and PC3 cells. Transfection of A2780 and LNCaP cells with PSES/CS3X/Luc significantly enhanced luciferase expression as compared to PSES/Luc (5.8X and 1.5X, respectively), consistent with CS3X transcriptional activation, but did not significantly enhance expression in PL-5 cells, again consistent with the low activity associated with CS3X/Luc in these cells.
Naked CanScript sequence in the absence of vector sequences promotes transcription.
The fact that the CanScript sequence appeared to function as a gene promoter in the absence of any other promoter sequence was surprising. To ensure that its promoter activity was due to CanScript, and not some occult promoter activity associated with pGL4 vector sequences, we transfected A2780 cells with purified CS3X/Luc and CS6X/Luc insert sequences devoid of any vector sequences (). Luciferase activity in cells transfected with these constructs was significantly higher than in cells transfected with linearized pGL4 vector DNA (i.e., only luciferase sequence, no CanScript), confirming that the CanScript sequence does indeed have promoter function. Luciferase activity in cells transfected with CS6X/Luc was ∼4X higher than in cells transfected with CS3X/Luc.
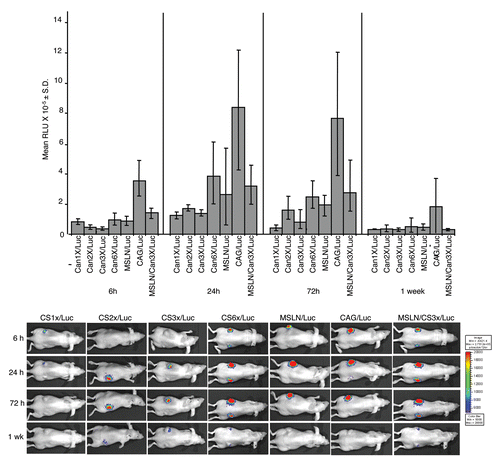
In vivo promoter-like activity of CanScripts (1X-6X).
To test whether CanScript promotes gene expression in vivo in a similar manner to what was observed in cells in culture, we generated A2780 xenografts in nude mice, and then injected the tumors with DNA/nanoparticles to deliver various luciferase reporter DNA constructs to tumor cells. The nanoparticles, generated by complexing a poly(β-amino ester), C32-117, with DNA, were previously shown to deliver DNA effectively to tumors in mice.Citation5 Mice were optically imaged at various times after injection (6, 24, 72 hr and 1 wk). As in the in vitro cell assays, CanScript had promoter function in the absence of other promoter sequences and luciferase activity was roughly proportional to the number of CanScript sequences (). Notably, six copies of CanScript yielded higher luciferase activity than that of the native MSLN promoter (containing 1 CanScript copy). A modest increase in luciferase activity (∼1.3-fold) was observed at 6, 24 and 72 hr time points in tumors injected with MSLN/CS3X/Luc nanoparticles as compared to tumors injected with MSLN/Luc nanoparticles. Maximal luciferase activity for each of the constructs tested was attained 24 hr after nanoparticle injection and remained near this level for at least another 48 hr. By 1 wk post-injection, luciferase activity was near baseline level for all constructs.
Discussion
We have discovered that a previously identified 18-base pair enhancer sequence, CanScript,Citation6 that lies 40 base pairs upstream of one of three consensus transcriptional initiation sequences in the native promoter region of the MSLN gene, has promoter function, even in the absence of any other DNA sequence. This finding was unexpected, as there is no evidence of a transcriptional start site within the CanScript sequence in its normal chromosomal context. In the absence of other promoter sequences, the isolated CanScript sequence displays cell-type specificity and activates transcription only in cell lines in which the MSLN gene is overexpressed, i.e., ovarian and pancreatic cancer cells. While binding of TEF-1 transcription factor to a MCAT motif in the CanScript sequence has previously been shown to elevate MSLN selectively in tumor cells, there is also evidence that a modulatory co-factor may bind to a SP1-like site present in the CanScript sequence, thereby facilitating transcription in cancer cells, but not in normal cells.Citation6 The identity of this factor remains to be determined.Citation9
In agreement with similar observations made by others,Citation9,Citation15 we found no correlation in the amount of TEF-1 mRNA with the ability of CanScript to enhance promoter activity. It seems likely that CanScript function is regulated by a delicate balance of TEF-1 and at least one other co-factor.Citation6 In this regard, the likelihood that CanScript enhances transcription in normal cells is remote given that mesothelin (MSLN) expression in normal tissues is very limited, and when it is expressed, expression is low. MSLN expression can serve as a barometer with which to gauge those cells that have the right mix of TEF-1 and its co-factor(s) to activate transcription. Since MSLN is overexpressed in nearly one third of human malignancies,Citation6 the expectation is that CanScript will function in these tumors, but not in MSLN non-expressing normal tissues. The fact that a search of the human genome sequence did not identify the CanScript sequence at any additional sites other than in the MSLN gene supports the likelihood that CanScript function is tumor specific (our unpublished data).
We also determined that 1 to 6 tandem repeats of CanScript enhance cell-specific transcription in a dose-dependent manner in MSLN-expressing ovarian tumor cells in culture as well as in xenografts. Apparently, TEF-1 and its co-factor are overexpressed in tumor cells in sufficient amounts to initiate transcription through the CanScript motif. We are currently investigating whether higher numbers of CanScript repeats can result in even higher, cancer-specific gene expression. We also observed that addition of CanScript sequence to the non-CanScript-containing HE4 and PSES promoters enhances transcriptional activity in ovarian tumor A2780 cells, and to a lesser degree in pancreatic PL-5 cells (), presumably because both TEF-1 and its cofactor are expressed in these cells. Curiously, addition of CanScript also enhanced PSES promoter activity in LNCaP prostate cancer cells. The fact that neither the MSLN promoter, CanScript 1X-6X, or the MSLN promoter + CanScript3X displayed activity in LNCaP cells suggests that the addition of CanScript to the PSES promoter may aid RNA polymerase II binding to an active promoter sequence in some, as yet, undefined way. While we did not observe enhancement of transcription by CanScript in all cell types, our data support the idea that if a promoter is active in cancer cells, the addition of CanScript to the promoter will enhance its activity. Importantly, CanScript transcriptional enhancement of MSLN promoter activity also occurred in ovarian tumors in tumor-bearing mice. Gene promoters which are highly active in target organs or in tumors are increasingly used in gene therapy strategies to direct therapeutic gene activity to specific populations of cells. Unfortunately, these “tissuespecific” promoters often have weak activity. Our data suggest that, for therapies targeting ovarian tumors, and perhaps for other tumor types as well, addition of at least one CanScript sequence to the promoter sequence may boost gene expression while retaining the desired specificity.
This work lays the groundwork for the possibility that there may be other, as yet undiscovered, enhancer sequences that activate gene expression specifically in cancer cells similar to CanScript. Identification of such sequences would provide additional options for optimizing gene therapy outcomes. In the future, personalized cancer therapy may include selecting patients, based on MSLN and TEF-1-related biomarkers, for gene therapy strategies utilizing CanScript technology.
Materials and Methods
RNA isolation and real-time quantitative PCR (qPCR).
Total RNA was isolated from the indicated cancer cell lines using RNeasy RNA isolation kit (Qiagen, Valencia, CA). RT-PCR was performed on equal amounts of RNA (measured via optical density) from each cell line to generate cDNA. Optical Density of cDNA was assessed and then real-time quantitative PCR (qPCR) was performed on an ABI 7500 machine against generated TaqMan probes (Applied Biosystems, Foster City, CA) for TEF-1 and a control gene (GAPDH) using 75 ng of cDNA template per reaction to determine relative expression of TEF-1 in cancer cell lines.
DNA constructs.
pGL4-CS3X/Luc. To construct pGL4-Can3X/Luc, a 3X CS fragment (∼66 bp) was excised by digestion of pGL3-CS3X/SV40/Luc (donated to us by Dr. Scott E. Kern)Citation6 with BglII and XhoI, then ligated with a (BglII + XhoI) digest of pGL4.10(luc2) (Promega).
pGL4-CS1X/Luc. A 1X CanScript sequence with a 5′ XhoI and 3′ BglII was PCR amplified from pZero2 (Integrated DNA Technologies), containing the CS1X duplex 5′-CTC GAG CTC CAC CCA CAC ATT CCT GGA GAT CT-3′ made by Integrated DNA Technologies (forward primer: 5′-TAT CTC GAG CTC CAC CCA CA-3′; reverse primer: 5′-ATA AGA TCT CCA GGA ATG TG-3′). The PCR product was digested with XhoI and BglII and then ligated with a (XhoI + BglII) digest of pGL4-CS3X/Luc.
pGL4-CS2X/Luc. A 2X CanScript sequence with a 5′ XhoI and 3′ BglII was PCR amplified from pZero2, containing the CS2X duplex 5′-CTC GAG CTC CAC CCA CAC ATT CCT GGC TCC ACC CAC ACA TTC CTG GAG ATC T-3′ made by Integrated DNA Technologies (same PCR primers as for pGL4-Luc/CS1X). The PCR product was digested with XhoI and BglII and then ligated with a (XhoI + BglII) digest of pGL4-CS3X/Luc.
pGL4-CS6X/Luc. CS3X, a 66 bp (XhoI + HindII) digestion fragment of pGL4-CS3X/Luc, was ligated to a (SalI + HindII) digest of pEGFP-1 (Clontech) to create pCS3X/EGFP. To insert one more CS3X fragment into pGL4-CS3X/Luc, a released Can3X fragment from digestion of pCS3X/EGFP with KpnI and XhoI was ligated to a (KpnI + XhoI) digest of pGL4-CS3X/Luc.
pGL4-MSLN/luc and pGL4-HE4/Luc were constructed as previously described.Citation5
pGL4-MSLN/CS3X/Luc. A CS3X fragment, released by digestion of pGL3-CS3X/SV40/Luc with BglII and XhoI was ligated to a (BglII + XhoI) digest of pMECACitation14 to create the plasmid pMECA-CS3X. Following BamHI and XbaI digestion of pMECA-CS3X, the released CS3X sequence was ligated to a (NheI + BamH1) digest of pDC312 (Microbix Biosystems, Inc., Toronto, Canada) to create pDC312-CS3X. To insert a CS3X fragment into pGL4-MSLN/Luc, a CS3X fragment released from digestion of pDC312-CS3X with BamHI and HindIII, was ligated to a (BglII + HindIII) digestion of pGL4-MSLN/Luc.
pGL4-HE4/CS3X/Luc. A CS3X fragment, released by digestion of pGL3-CS3X/SV40/Luc with BglII and XhoI, was ligated to a (BglII + XhoI) digest of pMECA to create pMECA-CS3X. Following BamHI and XbaI digestion of pMECA-CS3X, then released CS3X was ligated to a (NheI + BamHI) digestion of pDC312 to create pDC312-CS3X. To insert a CS3X fragment into pGL4-HE4/Luc, a released CS3X fragment resulting from digestion of pDC312-CS3X with BamHI and HindIII, was ligated to a (BglII + HindIII) digest of pGL4-HE4/Luc.
pGL4-PSES/Luc. A 468 bp PSES fragement, released by (KpnI + SmaI) digestion of pGL3-AREc3-PSME(DEL2)-TATA (gift of L. Wu, UCLA), was ligated to a (KpnI + SmaI) digest of pDSRed-monomer (Clontech) to create pDsRed/PSES. A PSES fragment, released by (KpnI + BamHI) digestion of pDsRed/PSES, was ligated to a (KpnI + BamHI) digest of pMECA to create pMECA-PSES. Following (KpnI + XhoI) digestion of pMECA-PSES, the released PSES was ligated to a (KpnI + XhoI) digest of pGL4.10(luc2) (Promega).
pGL4-PSES/CS3X/Luc. A PSES fragment, released by (KpnI + XhoI) digestion of pGL4-PSES/Luc, was ligated to a (KpnI + XhoI) digest of pGL4-CS3X/Luc.
pGL4-CS3X/Luc and pGL4-CS6X/Luc were digested with KpnI and BamHI to isolate CS3X/Luc and CS6X/Luc vector-free fragments.
The DNA sequence of each construct was confirmed using an ABI 3730 DNA Analyzer and BigDye chemistry, version 3.1.
Cell culture and transfection.
Sources for the cell lines A2780, PL-5, HepG2, A549, PC3 and LNCaP and the conditions for their culture were as previously described.Citation1,Citation5,Citation13 All cell lines were transiently transfected using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's directions.
Luciferase assay.
We used the Luciferase Assay System (Promega) and a Monolight 2010 luminometer to measure luciferase activity in cell extracts that were prepared according to the manufacturer's instructions.
In vivo experiments.
To generate xenografts, 2 × 106 A2780 cells in 100 ml phosphate buffered saline containing 20% cold Matrigel (BD Biosciences) were injected subcutaneously into the flank of 8 week-old female athymic nude-Foxn1nu/Foxn1+ mice (Harlan).
Nanoparticles, resulting from complexation of plasmid DNA with a poly(β-amino ester), were prepared as previously described.Citation1 For administration of nanoparticles to xenografts, 50 µg complexed DNA/60 µL total volume were delivered directly to the tumor by injection using a 30G needle.
Optical imaging to detect luciferase activity in mice was performed using an IVIS 100 series Bioluminescence Imaging System (Caliper Life Sciences) as described previously with a 5 min integration time for image acquisition.Citation10
All procedures performed on mice in this study were done in accordance with protocols approved by the Lankenau Institutional Animal Care and Use Committee.
Abbreviations
| CS | = | canscript |
| DT-A | = | diphtheria toxin, A chain |
| MSLN | = | mesothelin |
Figures and Tables
Figure 1 Schematics of DNA constructs used in this study (not drawn to scale). CAG is a ubiquitously active regulatory sequence consisting of the CMV enhancer and the chicken β-actin promoter.Citation7 All constructs are housed in the plasmid vector, pGL4.10(luc2).
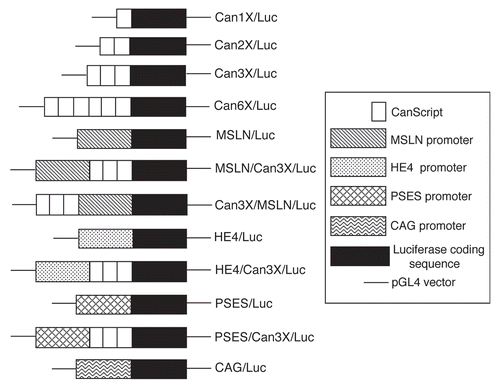
Figure 2 CanScript promoter function as measured by luciferase activity following transient transfection of different cancer cell lines with various plasmid DNA luciferase reporter constructs. Luciferase activity was measured 24 hr after transfection and is normalized to activity in cells transfected with CAG/Luc plasmid DNA. (A) Transfection with plasmids containing single or tandem repeats of CanScript sequence and MSLN promoter ± CS3X. (B) Transfection with plasmids containing HE 4 and PSES promoters ± CS3X. A2780 (ovarian), PL-5 (pancreatic), HepG2 (liver), A549 (lung), PC3 (prostate), LNCaP (prostate).
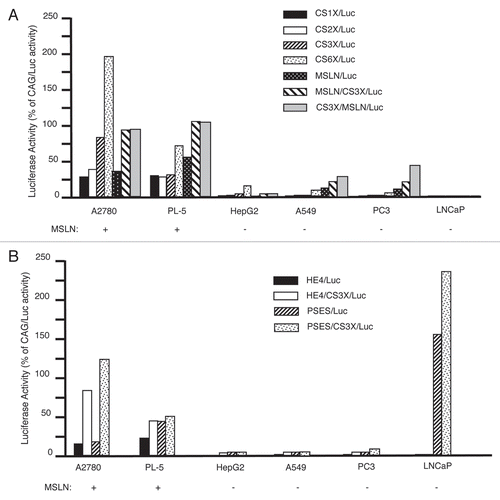
Figure 3 TEF-1 expression in cancer cell lines. mRNAs isolated from the indicated cell lines were reverse transcribed and TEF-1 expression levels were measured by qPCR. Values were mass normalized to GAP DH expression. The A2780 ovarian cancer cell line was chosen as a calibrator. Values represent the means from three independent data points.
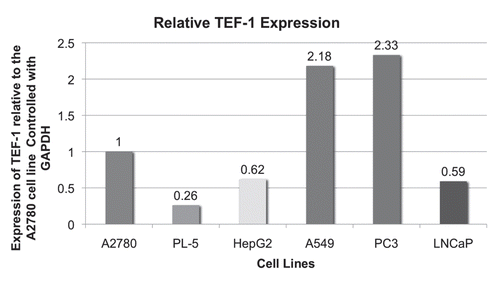
Figure 4 CanScript promoter function as measured by luciferase activity following transient transfection of A2780 cells with purified insert DNA (i.e., no vector sequence) or with pGL4.10(luc2) vector. Cells were transfected with 0.5 µg DNA/well in a 24-well plate. Each construct was assayed three times.
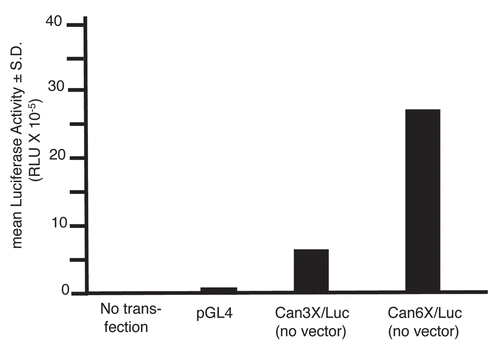
Figure 5 In vivo CanScript promoter function. (A) Relative light units (RLU) emitted by A2780 xenografts in nude mice at various times after intratumoral injection of DNA-nanoparticles prepared with different CanScript constructs. Eight to 10 tumors were injected for each construct. (B) Representative longitudinal optical images of mice following intratumoral injection of DNA-nanoparticles.
Acknowledgements
We wish to thank Jiping Chen for technical assistance. We also thank Scott E. Kern (Johns Hopkins University, Baltimore, MD) for kindly donating, and allowing the use of, the original CanScript clone, and for helpful suggestions and discussion. This work was supported by the National Institutes of Health (CA132091 to J.A.S., EB000244 to R.L.); the Teal Ribbon Ovarian Cancer Research Foundation, Inc. to J.A.S.; Wawa, Inc. Corporate Charities to J.A.S.; and the Red Bra Ladies to J.B. and J.A.S.; Supported by Grant #IRG-08-060-01 from the American Cancer Society (J.B.) and a PanCAN Career development Award, American Association of Cancer Research (J.B.).
References
- Anderson DG, Peng W, Akinc AB, Houssain N, Kohn A, Padera R, et al. A polymer library approach to suicide gene therapy for cancer. Proc Natl Acad Sci USA 2004; 101:16028 - 16033
- Chang K, Pastan I. Molecular cloning and expression of a cDNA encoding a protein detected by the K1 antibody from an ovarian carcinoma (OVCAR-3) cell line. Int J Cancer 2004; 57:90 - 97
- Frierson HF Jr, Moskaluc CA, Powell SM, Zhang H, Cerilli LA, Stoler MH, et al. Large-scale molecular and tissue microarray analysis of mesothelin expression in common human carcinomas. Hum Pathol 2003; 34:605 - 609
- Hellstrom I, Hellstrom KE. SMRP and HE4 as biomarkers for ovarian carcinoma when used alone and in combination with CA125 and/or each other. Adv Exp Med Biol 2008; 622:15 - 21
- Huang YH, Zugates GT, Peng W, Holtz D, Dunton C, Green JJ, et al. Nanoparticle-delivered suicide gene therapy effectively reduces ovarian tumor burden in mice. Cancer Res 2009; 69:6184 - 6191
- Hucl T, Brody JR, Gallmeier E, Iacobuzio-Donahue CA, FarranceI K, Kern SE. High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor-dependent MCAT motif. Cancer Res 2007; 67:9055 - 9065
- Ikawa M, Kominami K, Yoshimura Y, Tanaka K, Nishimune Y, Okabe M. Green fluorescent protein as a marker in transgenic mice. Develop Growth Differ 1995; 37:455 - 459
- Lee SJ, Kim HS, Yu R, Lee K, Gardner TA, Jung C, et al. Novel prostate-specific promoter derived from PSA and PSMA enhancers. Mol Ther 2002; 6:415 - 421
- Ordonez NG. Application of mesothelin immunostaining in tumor diagnosis. Am J Sug Pathol 2003; 27:1418 - 1428
- Peng W, Chen J, Huang YH, Sawicki JA. Tightly-regulated suicide gene expression kills PSA-expressing prostate tumor cells. Gene Ther 2005; 12:1573 - 1580
- Rosen DG, Wang L, Atkinson JN, Yu Y, Lu KH, Diamandis EP, et al. Potential markers that complement expression of CA125 in epithelial ovarian cancer. Gynecol Oncol 2005; 99:267 - 277
- Scholler N, Fu N, Yang Y, Ye Z, Goodman GE, Hellstrom KE, et al. Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in sera from patients with ovarian carcinoma. Proc Natl Acad Sci USA 1999; 96:11531 - 11536
- Showalter SL, Huang YH, Witkiewicz A, Costantino CL, Yeo CJ, Green JJ, et al. Nanoparticulate delivery of diphtheria toxin DNA effectively kills mesothelin expressing pancreatic cancer cells. Cancer Biol Ther 2008; 7:1584 - 1590
- Thomson JM, Parrott WW. pMECA: A cloning plasmid with 44 unique restriction sites that allows selection of recombinants based on colony size. BioTechniques 1998; 24:922 - 928
- Xiao J, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression and transcriptional properties of the human enhancer factor TEF-1. Cell 1991; 65:551 - 568