Abstract
Thiopeptides are sulfur containing highly modified macrocyclic antibiotics with a central pyridine/tetrapyridine/dehydropiperidine ring with up to three thiazole substituents on positions 2, 3 and 6. Thiazole antibiotics with central pyridine nucleus have a macrocyclic loop connecting thiazole rings at position 2 and 3 described as ring A. In addition antibiotics with central tetrahydropyridine nucleus have a quinaldic acid macrocycle also connected to thiazole on position 2 described as ring B. We have demonstrated before that thiazole antibiotics thiostrepton and Siomycin A act as proteasome inhibitors in mammalian tumor cells. Here we decided to test whether other known thiazole antibiotics such as berninamycin, micrococcin P1 and P2, thiocillin and YM-266183 (lacking the quinaldic acid ring B) demonstrate this activity. We found that none of them act as proteasome inhibitors. Moreover, structural modification of thiostrepton to thiostrepton methyl ester (with open B ring) also did not demonstrate this activity. These data suggest that B ring of thiostrepton and Siomycin A that is absent in other thiazole antibiotics determines the proteasome inhibitory activity of these drugs.
</br>See commentary: The fellowship of the ring
Introduction
An emerging class of naturally occurring thiopeptide antibiotics, characterized by highly complex sulfur-containing heterocyclic rings has demonstrated a variety of physiologic activities including antibacterial, antiparasitic activities. Thiazole peptide antibiotics are sulfur containing highly modified macrocyclic antibiotics that block protein synthesis in bacteria targeting ribosome (class 1) or elongation factor EF-Tu (class 2) ().Citation1,Citation2 The characteristic feature of thiazole antibiotics is a central pyridine/tetrapyridine/dehydropiperidine ring with up to three thiazole substituents on positions 2, 3 and 6.Citation3 We identified thiazole antibiotics Siomycin A and thiostrepton as inhibitors of FoxM1 and inducers of apoptosis in human cancer cells.Citation4–Citation6 Later, we found that exposure cells to Siomycin A, surprisingly led to the stabilization of a variety of proteins, such as p21, Mcl-1, p53 and hdm2,Citation7 suggesting that Siomycin A may act as a proteasome inhibitor.
The major site for proteolytic activity of intracellular proteins is the proteasome, a catalytic protein complex with multiple peptidase activities. The proteasome complex catalyzes the degradation of most proteins in eukaryotic cells in an ATP-dependent manner.Citation8,Citation9 The active site of the proteasome complex involves a 20S subunit (700 kDa) proteasome that functions through a mechanism involving a threonine active site. The 26S (2,000 kDa) complex, which degrades ubiquitinated proteins, contains in addition to the 20S proteasome a 19S regulatory complex composed of multiple ATPases and components necessary for binding protein substrates. Latest progress in the understanding of the proteasome function led to introduction of novel drugs against human cancer, proteasome inhibitors.Citation10 Bortezomib (Velcade) was the first proteasome inhibitor approved for the treatment of multiple myeloma with potential benefits against other types of cancer in the future. Since certain types of cancer may rely on a functional proteasome for growth, inhibition of proteasome activity would selectively kill these tumors. To directly test whether the Siomycin A and thiostrepton inhibit proteasome activity in vitro, thiazole antibiotics were compared with well-known proteasome inhibitors MG132 and lactacystin against the purified 20S proteasome.Citation7 Data from this experiment suggested that both thiostrepton and Siomycin A (in addition to their FoxM1 inhibitory activity) act as proteasome inhibitors in vitro, though weaker than bona-fide proteasome inhibitors, MG132 and lactacystin.Citation7
To understand the structural features of thiazole antibiotics that are critical for proteasome inhibitory effect, we decided to test whether other known thiazole antibiotics such as Berninamycin, Micrococcin P1 and P2, thiocillin and YM-266183 also demonstrate this activity. The thiazole antibiotics investigated varied in the B ring. We found that none of them act as proteasome inhibitors. Moreover, structural modification of thiostrepton to thiostrepton methyl ester (with open B ring) also did not demonstrate this activity.
Results and Discussion
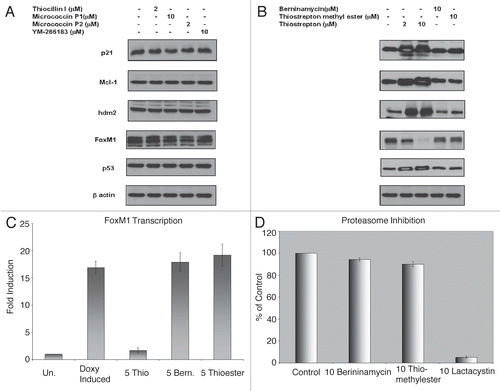
To determine whether several additional thiopetides may also act as proteasome inhibitors, we evaluated their proteasome inhibitory activity in mammalian cells. Six thiazole antibiotics, namely Berninamycin (comprising of atypical amino acids linked to thiazole and oxazoles), Micrococcin P1 and P2, thiocillin, YM-266183 (missing the B ring) and thiostrepton methyl ester (with open B ring) (kind gift from Albert Bowers and Christopher Walsh, Harvard Medical School) () were evaluated. First, in contrast to thiostrepton and Siomycin A, none of the other thiazole antibiotics demonstrated stabilization of proteins (Hdm2, Mcl-1, p21 and p53) that usually get stabilized following treatment with proteasome inhibitors.Citation7 The levels of the various proteins measured were similar to vehicle treated-control cells following treatment with thiazole antibiotics except thiostrepton ( and B). Thiostrepton was used as a control and demonstrated stabilization of multiple proteins at concentration as low as 2 µM ().
Secondly, six thiazole antibiotics that were investigated did not inhibit expression of FoxM1 protein by immunoblot analysis ( and B) or the transcriptional activity of FoxM1 (data not shown and ), while thiostrepton ( and C) and Siomycin A, as well as bortezomib and MG132, strongly inhibited FoxM1.Citation4–Citation7 Thiostrepton downregulated the expression of FoxM1 protein in a concentration-dependent manner. 2 µM Thiostrepton demonstrated a 30–40% reduction in FoxM1 level, while complete loss of FoxM1 expression occurred at 10 µM. Thiocillin and Micorcoccin were evaluated at a concentration of 2 µM due to poor solubility. Other thiazole antibiotics did not inhibit FoxM1 expression at 10 µM concentration. In contrast to thiazole antibiotics/proteasome inhibitors Siomycin A and thiostrepton,Citation4–Citation6 six studied thiazole antibiotics did not induce apoptosis in human osteosarcoma U-2OS C3 cells as evaluated by caspase-3 cleavage following treatment (data not shown). To directly evaluate the proteasome inhibitory activity of Berninamycin and thiostrepton methyl ester in vitro, we used the Proteasome Activity Assay Kit (Millipore/Chemicon) based on detection of free AMC following cleavage from labeled substrate LLVY-AMC by 20S proteasome. In contrast to Siomycin A and thiostrepton,Citation7 berninamycin and thiostrepton methyl ester demonstrated no proteasome inhibitory activity. Lactacystin, a known inhibitor of proteasome activity was used as a positive control (). All these data suggest that Berninamycin, Micrococcin P1 and P2, thiocillin, YM-266183 and thiostrepton methyl ester are not acting as proteasome inhibitors.
Based on the above described data we showed that none of thiazole antibiotics that were introduced in this paper have proteasome inhibitory activity. Berninamycin, Micrococcin P1 and P2, thiocillin and YM-266183 do not have ring B (), while thiostrepton methyl ester with open B ring demonstrates complete loss of proteasome inhibitory activity relatively to thiostrepton, suggesting that the macrocyclic B ring () is a crucial structural feature required for the proteasome inhibitory activity of thiazole antibiotics. It was demonstrated previously by using erythrocyte 20S proteasome that intact A/B ring of thiostrepton was required for proteasome inhibitory activity.Citation12 However, in this study we show for the first time that thiazole antibiotics with missing/open B ring do not possess proteasome inhibitory activity and cannot inhibit FoxM1. These data confirm our previous observations that proteasome inhibitors, but not all thiazole antibiotics have intrinsic ability to inhibit FoxM1 and induce apoptosis in cancer cells.Citation7 Bortezomib and other proteasome inhibitors bind mainly to chymotryptic-type active site located in the β5 subunit.Citation13 The binding site of thiostrepton is not yet elucidated. Determination of binding site of thiostrepton will provide insight into improving the proteasome inhibitory activity and structural specificity of thiostrepton analogs.
Materials and Methods
Cell lines, media and chemical compounds.
The development of U-2OS C3 cell line with doxycyclin-inducible FoxM1-green fluorescent protein (GFP) fusion protein has been described before.Citation11 U-2OS C3 cell line was transfected with a plasmid expressing firefly luciferase under the control of 6x FoxM1 responsive promoterCitation11 along with pcDNA3.1 (Invitrogen, Carlsbad, CA) plasmid that expresses neomycin phosphotransferase. The resultant cells U-2OS C3-Luc showed severalfold doxycycline-dependent induction in firefly luciferase activity.Citation4 Cells were grown in DMEM medium (Invitrogen). The media was supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 1% penicillin-streptomycin (GIBCO) and the cell lines were kept at 37°C in 5% CO2. Thiazole antibiotics Siomycin A (NCI) and thiostrepton (Sigma), Micrococcin P1 (A), Micrococcin P2 (B), thiocillin I (C), YM-266183 (D), berninamycin (E), thiostepton methyl ester were dissolved in DMSO (dimethylsulfoxide).
Immunoblot analysis.
U-2OS C3 cancer cells treated as indicated for 24 hours, harvested and lysed by using IP buffer (20 mM HEPES, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 100 mM NaF, 10 mM Na4P2O7, 1 mM sodium othrovanadate, 0.2 mM PMSF supplemented with protease inhibitor tablet [Roche Applied Sciences]). Protein concentration was determined by the Bio-Rad Protein Assay reagent (BIO-RAD). Isolated proteins were separated on 8–15% gradient SDS-PAGE and transferred to PVDF membrane (Millipore). Immunoblotting was carried out with antibodies specific for FoxM1 (a gift from Dr. Costa's lab), p21 (BD-Pharmingen), hdm2 (Santa-Cruz Biotechnology), Mcl-1 (LabVision) and β-actin (Sigma).
20S proteasome activity assay.
The inhibition of proteasome activity by thiazole antibiotics in vitro was compared with that of known proteasome inhibitor, lactacystin using 20S Proteasome Activity Assay Kit (Millipore) according to the instructions of the manufacturer. The assay is based on the detection of the fluorophore 7-amino-4-methylcoumarin (AMC) after cleavage from the labeled substrate LLVY-AMC in the presence and the absence of tested compounds. Indicated concentrations of the compounds were pre-incubated with the proteasome samples for 15 minutes at room temperature and then the proteasome substrate was added. Samples were incubated for 1 hour at 37°C and the free AMC fluorescence was quantified at 380/460 nm in a fluorometer (Spectra Max GeminiXS, Molecular Devices).
Transcriptional activity assay.
U-2OS C3-Luc cells were treated with combination of 1 µg/ml doxycycline and the indicated concentrations of the thiazole antibiotics, thiostrepton methyl ester and berninamycin. The luciferase activity was determined by using the Luciferase Assay System (Promega) according to the manufacturer's instructions and data were normalized on the amount of protein in the samples.
Figures and Tables
Figure 1 Structures of Micrococcin P1 (A), Micrococcin P2 (B), Thiocillin I (C), YM-266183 (D), Berninamycin (E), Thiostrepton methyl ester (F), Thiostrepton (G) and Siomycin A (H). A and B rings of these compounds are marked.
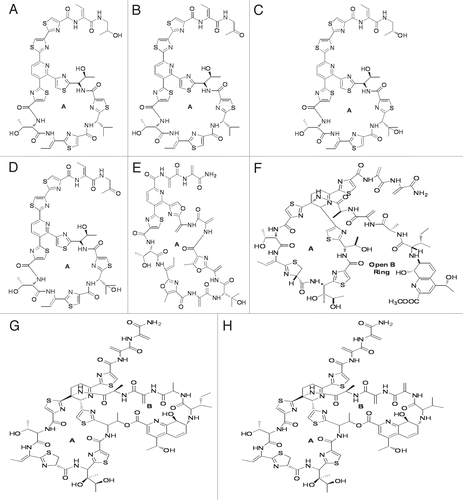
Figure 2 Thiocillin I, Micrococcin P1, Micrococcin P2, YM-266183, Berninamycin and Thiostrepton methyl ester are not proteasome inhibitors. (A) Thiocillin, Micrococcin P1, P2 and YM266183 do not stabilize expression of multiple cellular proteins and suppress FoxM1 expression. U-2OS C3 osteosarcoma cells treated as indicated were harvested and immunoblotting was carried out with antibodies specific for p21, p53, hdm2, Mcl-1, FoxM1 and β-actin. (B) Thiostrepton, but not berninamycin and thistrepton methyl ester stabilize expression of multiple cellular proteins and suppress FoxM1 expression. U-2OS C3 osteosarcoma cells treated as indicated were harvested and immunoblotting was carried out with antibodies specific for p21, p53, hdm2, Mcl-1, FoxM1 and β-actin. (C) Thiostrepton, but not berninamycin or thiostepton methyl ester inhibits FoxM1 transcriptional activity. U-2OS C3-Luc cells were treated with a combination of 1 µg/ml doxycycline and the indicated concentrations of the thiazole antibiotics and FoxM1 transcriptional activity measured by luciferase assay. (D) Berninamycin and thiostrepton methyl ester do not inhibit proteasome activity in vitro. The inhibition of proteasome activity by the thiazole antibiotics in vitro was negligible compared with proteasome inhibitor lactacystin as detected by using 20S Proteasome activity kit (Millipore).
Acknowledgements
We would like to thank Dr. Albert Bowers and Dr. Christopher Walsh for kindly providing the thiazole antibiotics used in this paper. This work was supported by NIH grants 1RO1CA1294414 and 1R21CA134615 to A.L.G.
References
- Jonker HR, Ilin S, Grimm SK, Wohnert J, Schwalbe H. L11 domain rearrangement upon binding to RNA and thiostrepton studied by NMR spectroscopy. Nucleic Acids Res 2007; 35:441 - 454
- Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, et al. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell 2008; 30:26 - 38
- Bowers AA, Acker MG, Koglin A, Walsh CT. Manipulation of thiocillin variants by prepeptide gene replacement: structure, conformation and activity of heterocycle substitution mutants. J Am Chem Soc 2010; 132:7519 - 7527
- Radhakrishnan SK, Bhat UG, Hughes DE, Wang IC, Costa RH, Gartel AL. Identification of a chemical inhibitor of the oncogenic transcription factor forkhead box M1. Cancer Res 2006; 66:9731 - 9735
- Bhat UG, Zipfel PA, Tyler DS, Gartel AL. Novel anticancer compounds induce apoptosis in melanoma cells. Cell Cycle 2008; 7:1851 - 1855
- Bhat UG, Halasi M, Gartel AL. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS ONE 2009; 4:5592
- Bhat UG, Halasi M, Gartel AL. FoxM1 is a general target for proteasome inhibitors. PLoS ONE 2009; 4:6593
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994; 78:761 - 771
- Craiu A, Gaczynska M, Akopian T, Gramm CF, Fenteany G, Goldberg AL, et al. Lactacystin and clastolactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J Biol Chem 1997; 272:13437 - 13445
- Nencioni A, Grunebach F, Patrone F, Ballestrero A, Brossart P. Proteasome inhibitors: antitumor effects and beyond. Leukemia 2007; 21:30 - 36
- Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev 2004; 18:830 - 850
- Schoof S, Pradel G, Aminake MN, Ellinger B, Baumann S, Potowski M, et al. Antiplasmodial thiostrepton derivatives: proteasome inhibitors with a dual mode of action. Angewandte Chemie International ed 2010; 49:3317 - 3321
- Groll M, Berkers CR, Ploegh HL, Ovaa H. Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure 2006; 14:451 - 456