Abstract
1α,25-dihydroxyvitamin D3 (1,25D) is a powerful differentiation agent, which has potential for treatment of acute myeloid leukemia (AML), but induces severe hypercalcemia at pharmacologically active doses. We have previously shown that carnosic acid (CA), the polyphenolic antioxidant from rosemary plant, markedly potentiates differentiation induced by low concentrations of 1,25D in human AML cell lines. Here, we demonstrated similar enhanced differentiation responses to the 1,25D/CA combination in primary leukemic cells derived from patients with AML, and determined the role of the Nrf2/antioxidant response element (Nrf2/ARE) pathway in these effects using U937 human monoblastic leukemia cells as the model. CA strongly transactivated the ARE-luciferase reporter gene, induced the ARE-responsive genes, NADP(H)-quinone oxidoreductase and the γ-glutamylcysteine synthetase heavy subunit, and elevated cellular glutathione levels. Interestingly, 1,25D potentiated the effects of CA on these activities. Stable transfection of wild-type (wt) Nrf2 resulted in the enhancement, while transfection of dominant-negative (dn) Nrf2 produced suppression of differentiation induced by the 1,25D/CA combination and, surprisingly, by 1,25D alone. These opposite effects were associated with a corresponding increase or decrease in vitamin D receptor and retinoid X receptor-α protein levels, and in vitamin D responsive element transactivation. Cells transfected with wtNrf2 and dnNrf2 also displayed opposing changes in the levels of the AP-1 family proteins (c-Jun and ATF2) and AP-1 transcriptional activity. Pretreatment with AP-1 decoy oligodeoxynucleotide markedly attenuated the differentiation in wtNrf2-transfected cells, suggesting that the pro-differentiation action of Nrf2 is mediated by functional AP-1. Our findings suggest that the Nrf2/ARE pathway plays an important part in the cooperative induction of myeloid leukemia cell differentiation by 1,25D and a plant polyphenol.
Introduction
Acute myeloid leukemia (AML) is characterized by the uncontrolled proliferation of undifferentiated or poorly differentiated myeloid blasts. The standard approach for AML treatment is combination cytotoxic chemotherapy based on cytosine arabinoside (cytarabine) and anthracyclines, such as doxorubicin or idarubicin; however, despite initial responses to chemotherapy, the long-term survival rates continue to be modest. Furthermore, this treatment is poorly tolerated, particularly by older patients.Citation1 In recent years, several novel, targeted cytotoxic approaches to treat AML have been developed, e.g., the use of kinase and histone deacetylase inhibitors.Citation2 Differentiation therapy is an alternative AML treatment, based on the induction of leukemic blasts to mature beyond the differentiation block and, thus, to restore the normal cellular phenotype and cell cycle arrest. The differentiation inducer all-trans retinoic acid has proven extremely valuable in the treatment of one subtype of AML, the acute promyelocytic leukemia (APL), and this turned out to be the first effective targeted therapy of AML.Citation3,Citation4 However, APL accounts for only approximately 10% of AML, and no effective differentiation or other non-cytotoxic therapy is currently available for the other subtypes of AML.
Vitamin D, converted in the body to its hormonal form 1α, 25-dihydroxyvitamin D3 (1,25D), is a potent differentiation agent which can induce maturation and/or apoptosis in different types of cancer cells.Citation5 Although many laboratories have used 1,25D to differentiate various subtypes of AML cells in culture, the required concentrations would be lethal in vivo, principally due to hypercalcemia, with widespread vascular and kidney calcium deposits, incompatible with life. Many analogs of vitamin D with reduced calcemic activity have been synthesized,Citation6 but none has yet been found to be safe for human use at concentrations needed to induce differentiation of AML blasts. One solution to this challenge may be provided by the use of compounds that can potentiate the differentiation effect of low, non-toxic concentrations of 1,25D and its analogs.Citation7 We and others have previously shown that plant-derived polyphenolic antioxidants, such as carnosic acid (CA) isolated from the rosemary plant, curcumin from turmeric and silibinin from milk thistle, markedly enhance the differentiation effects of low nanomolar concentrations of 1,25D in AML cell lines, both human (HL60)Citation8–Citation12 and murine (WEHI-3B D−),Citation13,Citation14 as well as in leukemic blasts obtained from AML patients.Citation15 Furthermore, combined treatment with CA-rich extract of rosemary leaves and low-calcemic 1,25D analogs resulted in a strong cooperative antileukemic effect in syngeneic mouse AML models in vivo, without inducing hypercalcemia.Citation13,Citation14
The mechanisms by which phytochemicals potentiate 1,25D-induced differentiation of AML cells remain largely unclear. Some studies have suggested the role of inhibition of the transcription factor nuclear factor κB by these agents in overcoming the differentiation block in AML cells,Citation8,Citation16 and we found that the enhanced differentiation was associated with the ability of polyphenols to cooperate with 1,25D in the activation of MAPK pathways and of the transcription factor activator protein-1 (AP-1).Citation12,Citation17 Interestingly, the cooperative differentiation effects of 1,25D and CA were associated with a decrease in the intracellular levels of reactive oxygen species, elevation of the total cellular glutathione content,Citation12,Citation14 and an induction of the antioxidant enzyme NAD(P)H quinone oxidoreductase-1 (NQO1),Citation14 suggesting the role of redox-related mechanisms in the pro-differentiation activity of plant polyphenols. Furthermore, other redox-modulating agents, such as iron chelators,Citation18 have also been shown to potentiate 1,25D-induced differentiation of AML cells.
Different phytochemicals, such as polyphenols, carotenoids and isothiocyanates, have been shown to activate cellular antioxidant mechanisms, e.g., induction of antioxidant enzymes and de novo synthesis of glutathione, through the activation of the NF-E2-related factor-2/antioxidant response element (Nrf2/ARE) transcription pathway (reviewed in ref. Citation19–Citation21), which constitutes a major detoxification system in many cell types and is implicated in cancer prevention (reviewed in ref. Citation22 and Citation23). Nrf2 belongs to the “cap ‘n’ collar” subfamily of the basic region-leucine zipper transcription factors and is sequestered in the cytosol by its inhibitor, Kelch-like ECH-associated protein 1 (Keap-1), which carries multiple reactive SH-groups and, thus, can function as a sensor for various electrophilic agents. Upon stimulation by such agents, Nrf2 is released from Keap-1, translocates to the nucleus where it forms heterodimers with small Maf proteins and transactivates the ARE sequences found in the promoters of genes encoding phase II detoxifying and antioxidant enzymes [e.g., NQO1, heme oxygenase-1, glutathione peroxidase and glutathione S-transferase and the rate-limiting enzyme in glutathione synthesis γ-glutamylcysteine synthetase (γGCS)] as well as proteins with reducing capability, such as thioredoxin, thioredoxin reductase and peroxiredoxin.Citation22,Citation23
Recent reports indicate that Nrf2 may contribute to the induction of differentiation in contrasting ways, depending on the cell type. For instance, Nrf2 was found to positively regulate differentiation of neuronsCitation24,Citation25 and adipocytes,Citation26 but to inhibit differentiation of osteoblastsCitation27 and chondrocytes.Citation28 The role of Nrf2 in the differentiation of hematopoietic cells remains unclear. While Nrf2-/- mice have been shown to develop normally, without megakaryocytic and erythroid cell lineage dysfunction or anemia,Citation29,Citation30 Nrf2 was found to be upregulated during megakaryocytic and erythroid differentiation of human CD34+ cord blood cells.Citation31 Recently, Nrf2 has been reported to regulate the expression of CD36 scavenger receptor in subset of macrophages;Citation32 however, the involvement of this transcription factor in the induction of myeloid differentiation has not as yet been investigated.
In this study we determined the role of Nrf2 in the enhancing effect of CA on 1,25D-induced differentiation in human AML cells, using the U937 cell line as the model. We show that CA activates the Nrf2/ARE transcription pathway and that 1,25D cooperates with CA at least in some aspects of this activation. Furthermore, ectopic expression of wild-type (wt) Nrf2 or its dominant-negative mutant (dnNrf2) results in a marked enhancement or attenuation, respectively, of the differentiation efficacy of the 1,25D/CA combination and, surprisingly, of 1,25D alone. These opposite changes in cell differentiation were accompanied by respective upregulation and downregulation of protein components and functional activity of both AP-1 and vitamin D receptor. Collectively, these results suggest that Nrf2 can mediate, probably via upregulation and activation of AP-1 and vitamin D receptor, the differentiation-potentiating effect of CA, and is involved, at least in part, in the basic mechanism of differentiation of AML cells by 1,25D.
Results
Carnosic acid potentiates 1,25D-induced differentiation of AML cells in vitro and ex vivo.
Treatment of U937 cells for 96 h with 10 µM CA, a concentration which can be achieved in plasma,Citation39 had almost no effect on the expression of the monocytic (CD14) and general myeloid (CD11b) surface differentiation markers but significantly potentiated the induction of both markers by a low concentration (2.5 nM) of 1,25D ( and C). This was associated with characteristic changes in cell morphology consistent with the more mature monocyte-macrophage phenotypeCitation40,Citation41 (). In contrast to untreated or CA alone-treated cells, which had large nuclei and basophilic cytoplasm, 1,25D/CA-treated cells displayed decreased nucleus-to-cytoplasm ratio, less basophilic cytoplasm as well as phagosome-like vacuoles and external protrusions (). Similar morphologic changes were induced by high-dose 1,25D (100 nM) while a lower concentration of 1,25D alone (2.5 nM) had a moderate effect ().
The enhanced differentiation effect of 1,25D/CA combination was accompanied by a marked reduction in cell numbers (), without significant cytotoxicity (). The lack of cytotoxicity of 1,25D/CA treatment in U937 cells is consistent with the previous reports that in contrast to breastCitation42 and prostateCitation43 cancer cells or squamous cell carcinoma cells,Citation44 1,25D (alone or together with CA) acts as an anti-proliferative, though pro-survival agent in AML cells.Citation10,Citation45,Citation46
Similar, though less pronounced and varying potentiation of 1,25D-induced differentiation by CA was observed in primary cultures of leukemic blasts obtained from six patients with AML. As exemplified in , treatment with the 1,25D/CA combination resulted in enhanced expression of CD11b and CD14 as well as TPA-stimulated superoxide generation (a functional marker of differentiated myeloid cells) in AML-M1, -M2 and -M4 samples, compared with untreated controls or single treatments. These effects were comparable to those produced by a high concentration of 1,25D (100 µM) and were accompanied by differentiation-related changes in cell morphology (data not shown), but to a lesser extent compared with 1,25D/CA-treated U937 cells ().
Enhancement of 1,25D-induced differentiation by carnosic acid is accompanied by the activation of the Nrf2/ARE transcription pathway.
To determine whether the differentiation-enhancing effect of CA is associated with its ability to activate the Nrf2/ARE pathway, we first utilized the ARE-Luc reporter gene assay in transiently transfected U937 cells using tBHQ, the classical Nrf2/ARE activator,Citation22 as the positive control. As shown in , CA (1–15 µM) dose-dependently transactivated both the 4xARE-Luc and γGCSh-ARE4-Luc reporters, containing the ARE sequences from glutathione-S-transferase Ya subunit and γGCS heavy subunit (γGCSh) promoter regions, respectively. This is consistent with data showing Nrf2/ARE activation by CA in neuronal cells.Citation20 The addition of 2.5 nM 1,25D markedly enhanced CA (10 µM)-induced transactivation of both ARE-Luc reporters (), even though 1,25D alone had an insignificant effect even at a high (100 nM) concentration. These effects were apparently ARE-specific because CA or the 1,25D/CA combination failed to induce luciferase expression from the mutant γGCSh-AREm-Luc construct (). CA did not significantly affect mRNA expression of Nrf2 in untransfected U937 cells (). However, a marked increase in Nrf2 protein levels was evident as early as 1 h post-incubation with CA () and persisted for at least 24 h (not shown). This is consistent with the accumulating evidence that by interacting with Keap-1, different Nrf2/ARE activators stabilize Nrf2 protein via preventing its degradation.Citation23,Citation47 1,25D did not significantly alter Nrf2 mRNA () and protein () levels when added alone, nor did it potentiate CA-induced Nrf2 upregulation ().
The ability of CA and its combination with 1,25D to transactivate the ARE, particularly in the γGCSh-ARE-Luc construct, well correlated with the capacity of these agents to cooperatively induce γGCSh expression in untransfected U937 cells at both the mRNA level (), which was most noticeable following 1 h incubation, and protein level (). Interestingly, a similar γGCSh induction was observed in the presence of 100 nM 1,25D alone ( and E). Since γGCS is a rate-limiting enzyme in the de novo synthesis of glutathione, we determined the effects of CA and 1,25D on the total glutathione levels in non-transfected U937 cells. demonstrates that when added alone, CA, but not 2.5 nM 1,25D, significantly elevated glutathione content; however, their combination produced a marked cooperative increase, which correlated with the combinatorial effects of CA and 1,25D on both ARE-Luc transactivation () and γGCSh induction ( and E). In analogy with its effect on γGCSh expression, treatment with the high concentration of 1,25D alone (100 nM) resulted in a moderate increase in the total glutathione levels (). Treatment of U937 cells with CA resulted in a significant increase in the expression of the other classical Nrf2/ARE-responsive gene, NQO1, at both the mRNA (not shown) and protein ( and C) levels. Taken together, the above results indicate that CA is capable of activating the Nrf2/ARE pathway in U937 cells, and that 1,25D can potentiate this activation leading to enhanced cellular glutathione production.
Development of stably transfected AML cells ectopically expressing wtNrf2 or dnNrf2.
To explore the role of the Nrf2 in 1,25D/CA-induced differentiation, U937 cells were stably transfected with expression vectors for wtNrf2 or its dominant-negative mutant (dnNrf2). The transfected cells were then cloned by limiting dilution followed by characterization of the clones by determining protein levels of Nrf2 (), NQO1 and γGCSh ( and C) under both the basal and Nrf2/ARE-activating conditions. The empty vector-transfected cells (clone pEF#3 and mixed pEF transfectants) as well as untransfected U937 cells were used as the controls.
Low levels of a ∼97 kDa Nrf2 polypeptide were found in untransfected U937 cells as well as in the pEF and dnNrf2 transfectants under basal conditions, whereas the wtNrf2 clones had relatively higher Nrf2 levels (). The wtNrf2 #18 clone exhibited a slightly higher Nrf2 expression than the wtNrf2 #20 clone. In dnNrf2-transfected cells, the appearance of a truncated dnNrf2 form (∼22.5 kDa) was detected (). Stimulation with tBHQ resulted in a further increase in Nrf2 levels, particularly, in the wtNrf2-transfected cells, wtNrf2#18 being more responsive than wtNrf2#20. In contrast, tBHQ did not significantly increase Nrf2 expression in dnNrf2 clones (). The enhancement or attenuation of the Nrf2 expression due to transfection of wtNrf2 or dnNrf2 was accompanied by corresponding changes in the levels of NQO1 and γGCSh ( and C). Thus, wtNrf2 overexpression resulted in an increase in both the basal and tBHQ- or CA-induced NQO1 and, particularly, γGCSh levels, as compared to control cells. On the other hand, dnNrf2 expression almost completely abrogated NQO1 and γGCSh induction ( and C). In addition, 4xARE-Luc reporter gene assays in transiently transfected wtNrf2- or dnNrf2-expressing stable U937 clones demonstrated substantially increased or reduced luciferase expression in response to both CA and tBHQ, as compared to empty vector-expressing cells (not shown). In summary, manipulation of Nrf2 expression/activity resulted in cells with a functionally upregulated or downregulated Nrf2/ARE pathway.
Functional Nrf2 is involved in the differentiation of AML cells induced by 1,25D and carnosic acid.
The role of Nrf2 in the differentiation-enhancing effects of CA was evaluated using two pairs of U937 cell clones transfected with either wtNrf2 (wtNrf2#18 & #20) or dnNrf2 (dnNrf2#13 and #17) in comparison with pEF#3 and untransfected cells. The wtNrf2 and dnNrf2 clones demonstrated markedly increased or almost abrogated CD11b expression in response to the 1,25D/CA combination, respectively (). Comparable, though somewhat less pronounced, changes in CD14 expression were observed as well (). Surprisingly, the differentiation induced by 1,25D alone, at both a low (2.5 nM) and a high (100 nM) concentration, was also enhanced or reduced in the respective transfectants, the changes in CD11b induction being more pronounced, as compared to CD14 ( and B). Since dnNrf2, which lacks the transactivation domain of the intact Nrf2,Citation35 opposes the positive influence of wtNrf2 on cell differentiation, the above results strongly suggest that the transcriptionally active Nrf2 largely mediates the enhancing effect of CA on 1,25D-induced differentiation and, participates in the basic mechanism of the differentiation effect of 1,25D on AML cells.
Modulation of the AP-1 protein levels and transcriptional activity by wtNrf2 and dnNrf2 expression.
AP-1 has been recognized as one of the critical transcription factors implicated in myeloid differentiation,Citation48 and has a role in 1,25D-induced differentiation of AML cells.Citation38 Therefore, we asked whether transfection of wtNrf2 or dnNrf2 affected the expression of cJun, cFos and ATF-2 protein components of AP-1 complex and its transcriptional activity in U937 cells. As shown in and B, in pEF#3 cells CA markedly increased the levels of cJun and ATF2, as compared to untreated controls, whereas 1,25D alone at either 2.5 or 100 nM had no appreciable effect and did not augment the CA-induced increases ( and B). Nrf2 overexpression resulted in a substantial elevation of both the basal and CA-induced levels of both cJun and ATF2 while the effects of 1,25D remained minor (). On the other hand, the expression of dnNrf2 markedly reduced the ability of CA to elevate cJun and ATF2 levels, as compared to the empty vector-transfected cells, ATF-2 being mostly affected (). In contrast to cJun and ATF2, the expression of cFos was practically unaffected by CA or 1,25D in the stable U937 transfectants tested here ( and B). The overall cFos levels mildly increased following wtNrf2 transfection while remaining unaltered in dnNrf2-expressing cells, as compared to the empty vector-transfected cells ( and B).
The wtNrf2- and dnNrf2-induced changes in cJun and ATF-2 levels were associated with corresponding changes in the functional activity of AP-1, as measured by the TRE-Luc reporter assay in transiently transfected cells. In untransfected U937 and pEF#3 cells, CA caused only a modest TRE-Luc transactivation while 1,25D had no effect, either alone or in combination with CA (). Consistent with the AP-1 protein expression data ( and B), U937 cells stably expressing wtNrf2 showed both the increased basal TRE-Luc activity and dramatically enhanced response to CA, as compared to untransfected or pEF#3 cells. Also, in wtNrf2 cells 1,25D was capable of significantly potentiating the CA-induced TRE transactivation even though it alone was still ineffective (). Conversely, in the dnNrf2 cells CA and/or 1,25D failed to significantly transactivate TRE-Luc (not shown). These results suggest that in U937 cells Nrf2 can function as an upstream regulator of AP-1.
AP-1 can mediate the Nrf2-regulated differentiation of AML cells.
To determine whether AP-1 is functionally involved in Nrf2/ARE-regulated differentiation of U937 cells we employed the transcription factor “decoy” strategyCitation38 using a double-stranded ODN carrying the TRE consensus sequence (TRE-ODN) derived from the hVDR promoter. Empty vector-transfected cells (pEF#3 and, pEF-mix) and wtNrf2-transfected clones #18 and #20 were preincubated for 24 h either without ODN or with 7.5 µM TRE-ODN, followed by treatment with 2.5 nM 1,25D, 10 µM CA, their combination or 100 nM 1,25D (a positive control) for an additional 60 h. Cell differentiation was then measured by double labeling for CD11b and CD14. To evaluate the specificity of TRE-ODN effects, parallel cell cultures were preincubated with a mutated TRE sequence (mTRE-ODN) under the same conditions. The data, exemplified in and averaged in , demonstrate that pretreatment with TRE-ODN resulted in a specific decrease in 1,25D/CA-induced expression of both CD11b and CD14 differentiation markers to approximately similar residual levels in both moderately differentiated pEF transfectants and markedly more differentiated wtNrf2 transfectants. Consistent with the previous data,Citation38 the differentiation effects of 1,25D alone were also reduced by TRE-ODN but to a lower extent than the effects of the 1,25D/CA combination (). Thus, the almost complete abrogation of the differentiation response to the 1,25D/CA combination in TRE-ODN-treated wtNrf2 overexpressing cells suggests that wtNrf2-dependent increment in cell differentiation is largely mediated by the functional AP-1.
Modulation of VDR and RXRα protein levels and VDRE transcriptional activity by wtNrf2 and dnNrf2 expression.
Since AP-1 decoy based on the hVDR-derived TRE sequence was capable of markedly suppressing the differentiation of wtNrf2-transfected U937 cells () and has also been shown to reduce VDR protein levels in HL60 cells,Citation38 we hypothesized that Nrf2 may regulate the expression and transcriptional activity of the nuclear VDR and hence the sensitivity of AML cells to 1,25D. We, thus, measured the effects of 1,25D and CA on the protein levels of VDR and its heterodimeric partner, RXRα,and on VDRE-Luc transactivation in wtNrf2 and dnNrf2 clones in comparison with untransfected U937 and pEF#3 cells. As shown in and B, treatment of pEF#3 cells with CA for 24 h increased, to various extents, both VDR and RXRα protein levels, as compared to untreated control cells. On the other hand, 1,25D elevated the VDR content but had almost no effect on RXRα levels. The 1,25D/CA combination was slightly more effective than single agents ( and B). Importantly, wtNrf2 overexpression resulted in a marked increase in both VDR and, particularly, RXRα levels both under basal and treatment conditions (). In contrast, transfection of dnNrf2 strongly reduced both the basal and treatment-induced VDR and RXRα expression (). These changes in protein expression correlated with the corresponding changes in VDRE transactivation, as determined by the VDREx6-Luc reporter assay in transiently transfected U937 cells (). In pEF#3 and untransfected cells, CA, which alone was practically ineffective, moderately increased VDRE activation by a low concentration of 1,25D (2.5 nM) to the level comparable to that induced by a 100 nM 1,25D (). wtNrf2 overexpression strongly augmented this potentiating effect of CA, whereas dnNrf2 expression abrogated it (). Importantly, the VDRE activation induced by 100 nM 1,25D3 alone was also strongly enhanced in the wtNrf2 clones or reduced in the dnNrf2 clones, as compared to untransfected or empty vector-transfected cells (). Collectively, these results suggest that the active Nrf2 can positively control the VDR transcriptional activity in U937 cells, likely via upregulating the VDR and RXRα protein levels.
Discussion
In this study, we demonstrate for the first time that the transcription factor Nrf2 largely mediates the enhancing effect of a plant polyphenol on 1,25D-induced differentiation and is involved in the differentiation effect of 1,25D alone in human AML cells. This pro-differentiation role of Nrf2 was supported by the following lines of evidence.
First, the differentiation-enhancing effect of CA in U937 cells correlated with its ability to activate the Nrf2/ARE transcription system and to increase de novo synthesis of cellular glutathione in these cells. Moreover, 1,25D was able to augment these Nrf2/ARE-related CA activities. Since in our experiments 1,25D did not affect Nrf2 levels, its potentiation of CA-stimulated ARE-Luc reporter activity in U937 cells could result from post-translational modification of Nrf2. For instance, different signaling protein kinases () which are known to be activated by 1,25D in AML cells (reviewed in ref. Citation5 and Citation49), have also been shown to phosphorylate and activate Nrf2.Citation50–Citation52 Interestingly, despite the fact that 1,25D alone, even at a high concentration, had only a slight effect on the ARE-transactivation, including γGCSh-ARE4-Luc activation, it was capable of significantly inducing γGCSh expression and moderately elevating glutathione levels. This suggests that in addition to the cooperation with CA in the Nrf2/ARE-dependent γGCSh induction, 1,25D may also activate its expression by an independent mechanism, thereby increasing the glutathione generating capacity of the 1,25D/CA system.
Second, ectopic expression of wtNrf2 or dnNrf2 in U937 cells resulted in a strong enhancement or attenuation, respectively, of the differentiation effects of both the 1,25D/CA combination and, unexpectedly, 1,25D alone. The latter finding implies that while 1,25D alone had minimal effects on Nrf2/ARE, this transcriptional pathway or Nrf2 itself plays an important regulatory role in the differentiation-inducing activity of 1,25D in at least some subtypes of AML cells.
Third, the opposite consequences of wtNrf2 or dnNrf2 transfection on differentiation responses in U937 cells were associated with concomitant changes in the AP-1 and VDR transcription factor modules, the critical regulators of differentiation in AML cells. Moreover, the fact that AP-1 decoy ODN inhibited the wtNrf2-augmented differentiation responses further suggests that AP-1 can mediate, at least in part, the prodifferentiation activity of Nrf2.
The possible modes of Nrf2 involvement in the differentiation of human AML cells are represented in . Our observations that transfection of dnNrf2, which lacks the transactivation domain of the intact Nrf2,Citation35 produced negative effects on the induction of differentiation markers and on AP-1 and VDR/RXRα expression and activity indicates that the transcriptionally active Nrf2 is required for the upregulation of these systems. In support of the involvement of the functional Nrf2/ARE transcription pathway in the differentiation of AML cells, we have previously shown that treatment of HL60 cells with buthionine sulfoximine, the specific inhibitor of γGCS, resulted in a marked inhibition of 1,25D/CA- and 1,25D alone-induced differentiation which paralleled glutathione depletion.Citation12 Another glutathione-depleting compound, diethyl maleate, has been shown to block TPA-induced differentiation in HL60 cells and KG-1 human AML cells.Citation53
We have previously shown that CA and 1,25D, alone and in combination, decrease the levels or reactive oxygen species in human and murine AML cell lines.Citation12,Citation14 This supports the hypothesis that generation of reducing conditions through elevation of intracellular glutathione and the induction of other Nrf2/ARE-responsive cellular antioxidants, such as thioredoxin (), may represent one possible mechanism by which CA and, probably, other Nrf2-activating agents can potentiate 1,25D-differentiation of AML cells. Reducing intracellular environment can lead to increased activity of AP-1 through maintaining the cysteine residues in the DNA-binding sites of Jun/Fos proteins in their active reduced form.Citation54,Citation55 However, as indicated by our data, activated Nrf2 may also contribute to AP-1 activation by upregulating the expression of at least some of the AP-1 family proteins, such as cJun and ATF-2 in AML cells. This is supported by the earlier published resultsCitation56 showing that fibroblasts isolated from Nrf2-/- mice had much lower cJun and cFos levels compared to their wild-type counterparts and that treatment with the Nrf2 activator tBHQ resulted in a marked increase in cJun and cFos levels in the wild-type cells but not in Nrf2-/- cells.
AP-1 has been found to play an important role in myeloid differentiation of hematopoietic progenitors and different types of AML cells induced by various agents,Citation48,Citation57,Citation58 including 1,25D,Citation38 and downregulation of the Jun family proteins has been associated with the pathogenesis of myeloproliferative disorders, chronic myelogenous leukemia and AML.Citation58,Citation59 Although the precise mechanism of the pro-differentiation activity of AP-1 in AML cells remains unclear, one possibility is that AP-1 may transcriptionally enhance VDR expression. The VDR promoter contains two AP-1 response elements.Citation60 The putative role of AP-1 in the regulation of VDR expression is supported by our previous finding that the enhancement of 1,25D-induced differentiation by CA in HL60 cells is accompanied by increased DNA-binding capacity of nuclear proteins to both AP-1 motifs present in VDR promoter.Citation12 Furthermore, treatment of HL60 cells with AP-1 decoy ODN resulted in a decrease in VDR protein content.Citation38 AP-1-mediated regulation of VDR expression was also demonstrated in breast cancer cells by application of dominant negative c-Jun and by site-directed mutagenesis of the TRE in the VDR promoter.Citation60 Taken together, these data suggest that by activating AP-1, the Nrf2/ARE pathway can upregulate the expression of VDR and RXRα in AML cells, thereby increasing VDR availability and, hence, cell responsiveness to low concentrations of 1,25D.
In conclusion, this study demonstrates that Nrf2 has a novel pro-differentiation role in AML cells, which may provide the mechanistic basis for the development of new 1,25D-based strategies of differentiation therapy of at least some subtypes of AML. Although the differentiation responses of patient-derived blasts to 1,25D/CA () were less pronounced compared to U937 () or HL60,Citation10,Citation61 cell lines, we believe that these and earlierCitation15 findings demonstrate the proof of concept that at least some plant polyphenols may act as enhancers of antileukemic actions of 1,25D in the human disease. Furthermore, since vitamin D (reviewed in ref. Citation62) and phytochemicals (reviewed in ref. Citation22 and Citation63) have been implicated in cancer prevention, our evidence for the cooperation between 1,25D and a plant polyphenol in the activation of the Nrf2/ARE pathway may provide a basis for the mechanism-based clinical and epidemiological chemoprevention studies of these combinations in AML and other neoplastic diseases.
Materials and Methods
Chemicals and antibodies.
Carnosic acid was purchased from Alexis Biochemicals (Läufenfingen, Switzerland). 1,25D was a gift from Dr. Milan Uskokovic (BioXell, Inc., Nutley, NJ). 12-O-tetradecanoylphorbol 13-acetate (TPA), cytochrome c, tert-butylhydroquinone (tBHQ) and DMSO were from Sigma Chemical Co., (St. Louis, MO). The antibodies against Nrf2 (C-20 and H-300), NQO1 (C-19), VDR (C-20), RXRα (D-20), c-Jun (H-79), c-Fos (4-10G) and ATF2 (C-19X), were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). γGCS heavy subunit (γGCSh) antibody (Ab-1; RB-1697-P) was from NeoMarkers (Fremont, CA). Calreticulin antibody (PA3-900) was from Affinity BioReagents (Goden, CO). Peroxidase-conjugated donkey anti-rabbit and donkey antigoat IgG were from Jackson ImmunoResearch Laboratories Inc. (West Grove, PA). Tissue culture media and reagents were from Biological Industries (Beit Ha'Emek, Israel). Stock solutions of CA (10 mM), 1,25D (0.25 mM) and tBHQ (30 mM) were prepared in absolute ethanol. The precise concentrations of 1,25D in stock solutions were verified spectrophotometrically at 264 nm (ε = 19,000).
Reporter and expression constructs.
γGCSh-ARE4-Luc reporter construct containing the ARE4 sequence (5′-GTG ACT CAG C-3′) identified in the 5′-flanking region of the human γGSCh gene, and its mutated counterpart (γGCSh-ARE4m-Luc; 5′-GGG ACT CAG C-3′), were kindly provided by Dr. J.J. Gipp (University of Wisconsin, Madison, WI).Citation33 The construct 4xARE-Luc containing four tandem repeats of the ARE sequence from the glutathione S-transferase Ya subunit (5′-TGA CAA AGC ACC C-3′) was a gift from Dr. M. Hannink (University of Missouri, Columbia, MO).Citation34 The construct 3xTRE-Luc containing three tandem copies of the cis-acting TPA response element (TRE; 5′-TGA CTC A-3′) was provided from Dr. E. Shaulian (Hebrew University School of Medicine, Jerusalem, Israel). VDRE-Luc reporter construct containing two similar direct repeats separated by three nucleotides (DR-3; 5′-AGG TCA CAG AGG TCA-3′) was gifted by Dr. David G. Garner (University of California, San Francisco, CA). Expression constructs for wtNrf2 and its dominant-negative mutant Nrf2M (dnNrf2), which lacks the transactivation domain residues 1–392 in the NH2-terminal portion of the protein,Citation35 as well as the empty vector (pEF) were kindly provided by Dr. J. Alam (Louisiana State University Medical Center, New Orleans, LA).
In vitro and ex vivo cell culture and treatment.
U937 human myelomonocytic leukemia cells (CRL-1593.2) were obtained from American Type Culture Collection (Rockville, MD) and were used within 6 months of resuscitation from frozen stocks stored in liquid N2. Peripheral blood specimens were obtained from six newly diagnosed patients with AML who presented to Soroka University Medical Center (Beer-Sheva, Israel) and Rabin Medical Cenetr (Petakh Tikva, Israel) and gave informed consent according to the institutional IRB protocol. One patient had M1, two had M2 and three had M4 subtype of AML by the French-American-British (FAB) classification.Citation36 Blasts were isolated from these specimens on Ficoll gradients, as described previously.Citation15 Cell cultures were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (0.1 mg/ml) and 10 mM Hepes (pH = 7.4), and treated with test agents, as described in references Citation10, Citation12 and Citation15.
Determination of differentiation markers.
The expression of CD11b and CD14 surface antigens was determined by flow cytometry, as described in references Citation10 and Citation12. One- or two-parameter analysis was performed using Cytomics FC500 flow cytometer equipped with CXP software (Beckman Coulter). The TPA-stimulated production of superoxide (O2−) was measured in a 96-multiwell format by the reduction of cytochrome c, as described in reference Citation12, in a VERSAmax microplate spectrophotometer (Molecular Devices, Menlo Park, CA). The maximal rates of O2− generation were determined and expressed as nmol O2−/106 cells/min using the extinction coefficient (E550) of 21 mM−1/cm−1. Cell maturation was also assessed by light microscopy of Wright-Giemsa-stained cytospin preparations of control and treated cells.
Total glutathione assay.
The total glutathione (GSH+GSSG) concentration was determined in cells lysed by sulfosalicylic acid by the glutathione reductase recycling assay,Citation37 as described in reference Citation12, using VERSAmax microplate spectrophotometer (Molecular Devices).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
Total RNA was extracted from cells with the YRB Kit (RBC Bioscience, Chung Ho, Taiwan) and cDNA was prepared with M-MuLV Reverse Transcriptase (Promega, Madison, WI), according to the manufacturer's instructions, using oligo-dT as a primer. Nrf2, NQO1, γGCSh mRNA was determined by qRT-PCR and the results were normalized by ARP0 mRNA content. The primers used in this study are given in .
cDNA samples (7 µl) were diluted nine-fold, mixed with the specific primers (0.2 mM) and Thermo-Start master mix (ABgene, Surrey, UK). SYBR green I dye (Amresco, Cleveland) was then added to the reaction mixture. Reactions were carried out in the Rotor-Gene Real-Time PCR machine (Corbett-Research, Northlake, Australia). Standard cycling conditions for this instrument were: 15 min initial enzyme activation at 95°C, then 35 cycles as follows: 10 sec at 95°C, 15 sec at the annealing temperature (as detailed above) and 20 sec at 72°C.
Western blotting.
Whole cell lysates (30 µg of protein) were subjected to SDS-PAGE and western blot analysis, as described in references Citation10, Citation12 and Citation17. Protein band absorbance was quantitated using an ImageMaster VDS-CL imaging system (Amersham Parmacia Biotech, Piscataway, NJ) and Image Gauge 3.11 software (Fuji Photo Film Co., Tokyo, Japan). Optical density values for each protein were normalized to calreticulin and are displayed beneath each protein band.
Transient transfection and reporter gene assay.
Cells were transiently co-transfected with the indicated reporter plasmids (0.8 µg) and Renilla luciferase (pRL-null) plasmid (0.2 µg) using JetPEI Reagent (Polyplus-transfection, New York, NY), as described in reference Citation17, with minor modifications. Transfected cells were exposed to the indicated treatments for 24 h followed by measurement of firefly and Renilla luciferase using the Dual Luciferase Reporter Assay system (Promega). The data are presented as the normalized ratios of firefly luciferase to Renilla luciferase activity (RLU).
Nucleoporation of cells.
U937 cells (1–2 × 107 cells/ml) were suspended in the Cell Line Nucleofector Kit V electroporation buffer with either 1 µg of wtNrf2, dnNrf2 or pEF expression vector carrying the neomycin (neoR) resistance gene and nucleofected using T-19 and V-01 programs, respectively, in an Amaxa Nucleofector™ (AMAXA, Inc., Gaithersburg, MD) according to the manufacturer's protocol. Cells were then selected in the growth medium containing 1 mg/ml G418 (Calbiochem, Darmstadt, Germany). Neomycin-resistant cells were established after a 2-week period and then cloned by a limited dilution.
Cell treatment with AP-1 decoy oligodeoxynucleotides.
Cells were preincubated with 7.5 µM double-stranded oligodeoxynucleotide containing the proximal binding site for AP-1 (TPA-response element; TRE) from the promoter region of hVDR (-77 to -97 relative to the transcription start site; 5′-CTG GCA AGA GAG GaC TGG ACC-3′) or its mutant (5′-CTG GCA AGA GAG tGC TGG ACC-3′) in the culture medium, for 24 h, followed by treatment with test agents for an additional 60 h, essentially as described in reference Citation38.
Statistical analysis.
The significance of the differences between the means of the various subgroups was assessed by one-way ANOVA followed by Newmans-Keuls multiple comparison test. Data are presented as the mean ± SE. A p value less than 0.05 was considered statistically significant. The computations were performed using GraghPad Prism 4.0 program (GraphPad Software, La Jolla, CA).
Abbreviations
| AML | = | acute myeloid leukemia |
| APL | = | acute promyelocytic leukemia |
| AP-1 | = | activator protein-1 |
| ARE | = | antioxidant response element |
| 1,25D | = | 1α,25-dihydroxyvitamin D3 |
| CA | = | carnosic acid |
| FAB | = | the French-American-British classification of leukemia |
| γGCS | = | γ-glutamylcysteine synthetase |
| Keap-1 | = | kelch-like ECH-associated protein 1 |
| NQO1 | = | NAD(P)H quinone oxidoreductase-1 |
| Nrf2 | = | NF-E2-related factor-2 |
| dnNrf2 | = | dominant-negative Nrf2 |
| wtNrf2 | = | wild type Nrf2 |
| tBHQ | = | tertbutylhydroquinone |
| TPA | = | 12-O-tetradecanoylphorbol 13-acetate |
| TRE | = | TPA response element |
Figures and Tables
Figure 1 Carnosic acid (CA) and 1,25D (D3) cooperate in the induction of differentiation and growth inhibition in U937 cells without inducing cytotoxicity. Cells (4 × 104/ml) were incubated with 0.1% ethanol (vehicle control) or the indicated test agents for 96 h. (A and C) CD11b and CD14 expression was determined by flow cytometry. (B) Wright-Giemsa-stained cytospin smears of the indicated cell samples (magnification, 1,000x). Arrows indicate relatively enlarged cytoplasmic areas of more mature cells with phagosome-like vacuoles and external protrusions. Cell numbers (D) and viability (E) were assessed by the trypan blue exclusion assay. 1,25D at 100 nM was used as a positive control. (A) demonstrates representative bivariate flow cytometric measurements, and (C–E) summarize the data of five independent experiments. *p < 0.05 and $p < 0.001 versus 2.5 nM 1,25D alone. ¶p < 0.01 versus control.
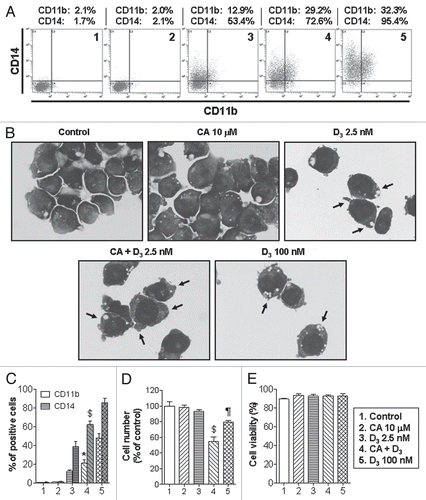
Figure 2 Enhanced differentiation of leukemic blasts from patients with AML following combined treatment with 1,25D and CA ex vivo. Mononuclear cells were isolated from blood specimens of patients with AML-M1 (A), M2 (B) and M4 (C) and incubated at 2 × 105 cell/ml with the indicated agents for 144 h, as described in Materials and Methods. CD11b and CD14 expression was determined by flow cytometry. TPA-stimulated superoxide production was measured in triplicate by the cytochrome c reduction assay (means ± SD).
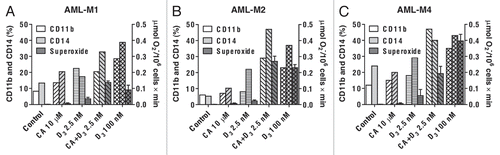
Figure 3 Carnosic acid (CA) and 1,25D (D3) cooperate in the activation of the Nrf2/ARE transcription system and glutathione synthesis in U937 cells. (A) Dose-dependent transactivation of the 4xARE-Luc and γGCSh-ARE4-Luc by CA. Luciferase activity was measured in transiently transfected cells following 24 h incubation with CA, as described in Material and Methods. (B) CA and D3 cooperatively transactivate the 4xARE-Luc and γGCSh-ARE4-Luc, but not the γGCSh-ARE4m-Luc containing mutated ARE4 sequence. Luciferase activity was measured following 24 h incubation with the indicated agents, as in (A). tBHQ was used as the positive control. #p < 0.01 and $p < 0.001 versus CA alone. ‡p < 0.001 versus control. (C) Treatment with CA and D3 does not affect Nrf2 mRNA expression. Cells were incubated with the indicated agents for 1 or 4 h followed by isolation of total mRNA and qRT-PCR analysis, as described in Material and Methods. (D) CA, but not D3, induces elevation of Nrf2 protein levels while the two agents cooperate in the induction of γGCSh. Cells were incubated with the indicated agents for 1 and 6 h (for Nrf2 detection) or 24 h (for γGCSh detection) followed by lysis and western blot analysis, as described in Material and Methods. Calreticulin (CRN) was used as the protein loading control. A representative of three similar blots is shown. (E) CA and D3 cooperatively activate the γGCS mRNA expression. Cells were treated with the indicated agents for 1 and 4 h followed by qRT-PCR, as described above. #p < 0.01 versus CA alone. §p < 0.05 and ¶p < 0.01 versus control. (F) CA and D3 cooperatively elevate the total glutathione levels. Cells were incubated with the indicated agents for 24 h followed by the glutathione reductase recycling assay, as described in the Material and Methods. *p < 0.05 versus CA alone. ¶p < 0.01 and versus control. The treatment groups in (B, C, E and F) are designated “1” to “6” as indicated in (B). (A–C, E and F) The data are the means ± SE of at least three independent experiments performed in duplicate or triplicate.
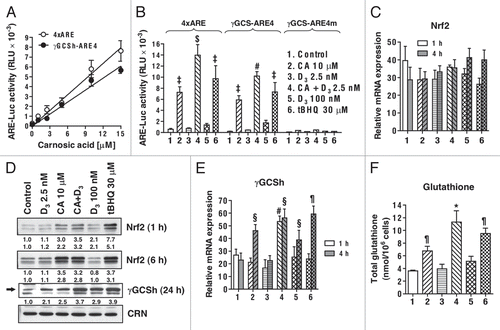
Figure 4 Effects of stable transfection with wtNrf2 or dnNrf2 on the basal and induced levels of Nrf2, NQO1 and γGCSh proteins in U937 cells. (A and B) The indicated wtNrf2, dnNrf2 or empty vector (pEF) transfected clones, mixed pEF clone culture (mix) or untransfected cells (U937) were incubated at 1 × 105 cells/ml with 0.1% ethanol (control) or tBHQ, for 24 h, followed by determination of Nrf2 (A) or NQO1 (B) protein levels by western blotting. (C) The indicated wtNrf2, dnNrf2 or empty vector (pEF) transfected clones or untransfected cells (U937) were incubated, as described above, with ethanol (control) or in the presence of CA or tBHQ, for 24 h, followed by determination of γ-GCS protein levels by western blotting. Calreticulin (CRN) was used as a protein loading control. Representative blots of three similar experiments are shown.
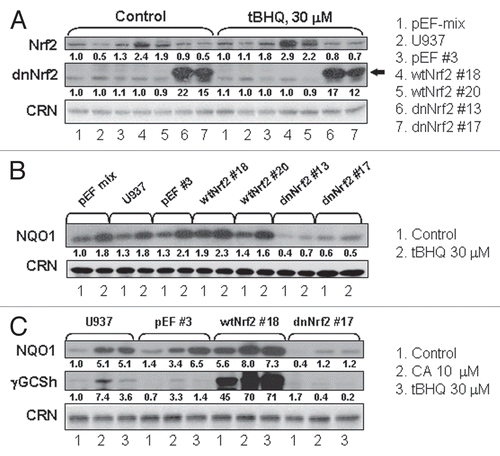
Figure 5 Stable expression of wtNrf2 or dnNrf2 modulates differentiation responses of U937 cells to the 1,25D/CA combination and 1,25D alone. The indicated wtNrf2-, dnNrf2- and empty vector (pEF)-transfected U937 clones and untransfected (U937) cells were incubated with 0.1% ethanol (control), CA, 1,25D (D3) or D3/CA combination for 96 h followed by analysis of CD11b (A) and CD14 (B) expression. Data are the means ± SE of five independent experiments. §p < 0.05; ¶p < 0.01 and ‡p < 0.001 versus corresponding responses of the p-EF#3 cells.
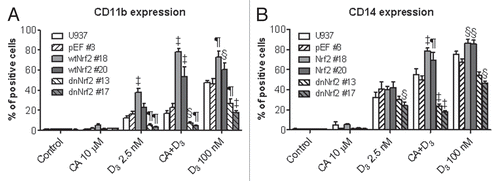
Figure 6 Stable expression of wtNrf2 or dnNrf2 modulates c-Jun, c-Fos and ATF2 protein levels and AP-1 transactivation in U937 cells. The indicated clones of U937 cells (1 × 105 cells/ml) stably transfected with empty vector (pEF#3; A and B), wtNrf2 (A) or dnNrf2 (B) were treated with vehicle (control) or the agents indicated in (C), for 24 h. Whole cell lysates were analyzed by western blotting. Calreticulin (CRN) was used as a protein loading control. Representative blots of three similar experiments are shown. The identification numbers of wtNrf2 and dnNrf2 clones are indicated next to the corresponding blots. (C) TRE × 3-Luc reporter activity was determined in untransfected U937 cells (U937) and in the indicated stable clones following transient transfection with TREx3-Luc and Renilla luciferase and treatment with vehicle (control) or indicated test agent for 24 h. The relative TRE-Luc activity (means ± SE) was calculated from the data of three individual experiments performed in triplicate. The treatment groups in (A and B) are designated “1” to “5” as indicated in (C). §p < 0.05; ¶p < 0.01 and ‡p < 0.001 versus corresponding responses of the pEF#3 cells.
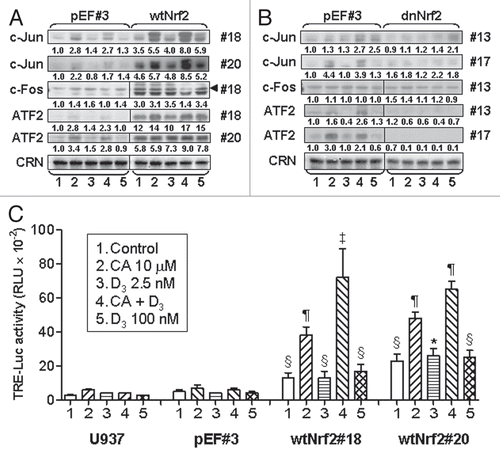
Figure 7 Treatment with AP-1 decoy oligonucleotide blocks the differentiation responses to the 1,25D/CA combination, most robustly in wtNrf2-transfected cells. U937 cells stably expressing empty vector (pEF) or wtNrf2 were preincubated with 7.5 µM TRE-ODN or the mutant mTRE-ODN for 24 h followed by treatment with the indicated agents for an additional 60 h. The expression of CD11b and CD14 was then measured by the bivariate flow cytometric analysis. (A) Comparative read-outs of CD11b and CD14 cell surface expression obtained from ODN-treated pEF #3 and wtNrf2 #18 cells. (B) Averaged CD11b and CD14 expression data from ODN-treated pEF #3 and pEF-mix cells (pEF-CD11b and pEF-CD14, respectively) and from wtNrf2 #18 and wtNrf2 #20 cells (wtNrf2-CD11b and wtNrf2-CD14, respectively). Data are the means ± SE of three independent experiments *p < 0.05 and #p < 0.01 versus response to mTRE-ODN in the same treatment group.
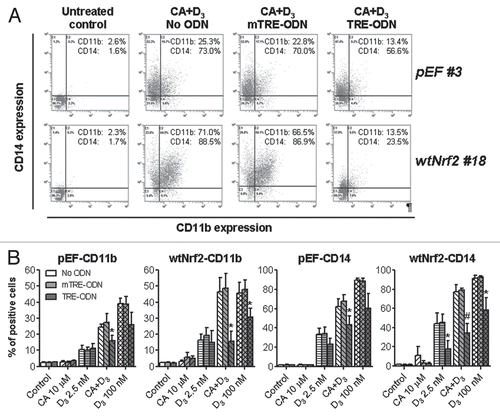
Figure 8 Modulation of VDR and RXRα protein levels and VDRE transactivation by stable expression of wtNrf2 or dnNrf2. The indicated clones of U937 cells (1 × 105 cells/ml) stably transfected with empty vector (pEF#3; A and B), wtNrf2 (A) or dnNrf2 (B) were treated with vehicle (control) or the agents indicated in (C), for 24 h. Whole cell lysates were analyzed by western blotting. Calreticulin (CRN) was used as a protein loading control. Representative blots of three similar experiments are shown. The identification numbers of wtNrf2 and dnNrf2 clones are indicated next to the corresponding blots. Note that the exposure of the RXRα blot from dnNrf2 #13 cells (B) to X-ray film was longer than in the rest of the RXRα blots (A and B) in order to allow visualization of lower RXRα expression in this dnNrf2 clone compared to that in the pEF #3 clone. (C) VDRE-Luc reporter activity was determined in untransfected U937 cells (U937) and in the indicated stable clones following transient transfection with VDREx6-Luc and Renilla luciferase and treatment with vehicle (control) or indicated test agent for 24 h. The relative VDRE-Luc activity (means ± SE) was calculated from the data of three individual experiments performed in triplicate. The treatment groups in (A and B) are designated “1” to “5” as indicated in (C). §p < 0.05; ¶p < 0.01 and ‡p < 0.001 versus corresponding values of the pEF#3 cells.
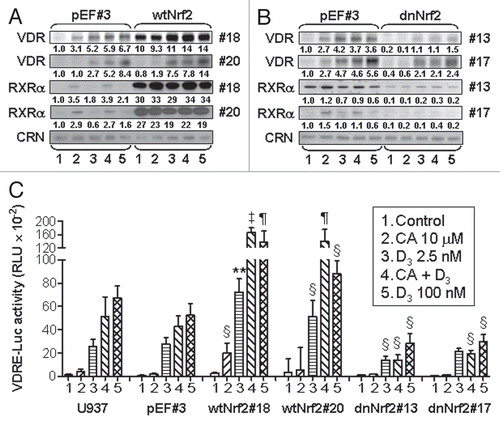
Figure 9 Suggested modes of the involvement of Nrf2/ARE in the regulation of AML cell differentiation. Plant antioxidants, e.g., CA activate Nrf2 by releasing it from the inhibitory partner, Keap-1. As a result, Nrf2 binds to ARE sequences and heterodimerizes with small Maf proteins leading to the induction of phase II enzymes and other redox regulators, e.g., γGCS, responsible for glutathione synthesis, and thioredoxin. 1,25D can further activate the Nrf2/ARE pathway, possibly by post-transcriptionally activating Nrf2 via phosphorylation by signaling protein kinases (PKC, MAPKs and/or PI-3-K). In addition, 1,25D can probably further enhance γGCS expression by Nrf2/ARE-independent mechanism. The resulting generation of reducing conditions can increase the activity of AP-1 by maintaining the cysteine residues (C in KCR) in their reduced (SH) form, which is required for DNA binding. Ref-1 is a redox regulator.Citation54 KCR is a tripeptide in the DNA binding region of both Jun and Fos. Our data indicate that Nrf2 can also positively regulate the expression AP-1 family proteins (cJun, cFos and ATF-2), further upregulating the AP-1 functional activity. Activated AP-1 can increase the expression of VDR and, perhaps, RXRα, resulting in the enhancement of the transcriptional activity of VDR/RXRα complex. A possibility exists that Nrf2 may also facilitate VDR and RXRα expression in an AP-1-inependent manner. Eventually, the enhancement of VDR function by 1,25D/CA is likely to increase the sensitivity of AML cells to the differentiation-inducing effects of lower concentrations of 1,25D.
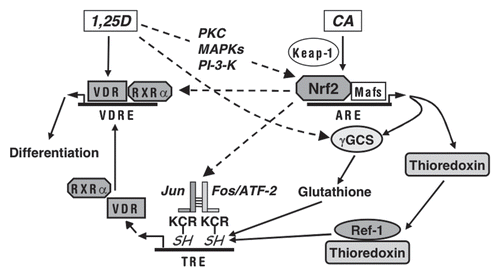
Table 1 Primer sequences used for quantitative RT-PCR
Acknowledgements
We thank Dr. Milan Uskokovic, BioXell, Nutley, NJ, for the gift of 1,25α-dihydroxyvitamin D3. This study was partially supported by a NIH grants from the National Cancer Institute RO1-CA-117942-3 to G.P.S. and M.D., RO1-CA-044722-20 to G.P.S. and the Israel Science Foundation grant 778/07 to M.D. and Y.S.
References
- Shipley JL, Butera JN. Acute myelogenous leukemia. Exp Hematol 2009; 37:649 - 658
- Fathi AT, Grant S, Karp JE. Exploiting cellular pathways to develop new treatment strategies for AML. Cancer Treat Rev 2010; 36:142 - 150
- Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, et al. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988; 72:567 - 572
- Petrie K, Zelent A, Waxman S. Differentiation therapy of acute myeloid leukemia: past, present and future. Curr Opin Hematol 2009; 16:84 - 91
- Gocek E, Studzinski GP. Vitamin D and differentiation in cancer. Crit Rev Clin Lab Sci 2009; 46:190 - 209
- Brown AJ, Slatopolsky E. Vitamin D analogs: therapeutic applications and mechanisms for selectivity. Mol Aspects Med 2008; 29:433 - 452
- Danilenko M, Studzinski GP. Enhancement by other compounds of the anti-cancer activity of vitamin D3 and its analogs. Exp Cell Res 2004; 298:339 - 358
- Sokoloski JA, Shyam K, Sartorelli AC. Induction of the differentiation of HL-60 promyelocytic leukemia cells by curcumin in combination with low levels of vitamin D3. Oncol Res 1997; 9:31 - 39
- Liu Y, Chang RL, Cui XX, Newmark HL, Conney AH. Synergistic effects of curcumin on all-trans retinoic acid- and 1α,25-dihydroxyvitamin D3-induced differentiation in human promyelocytic leukemia HL-60 cells. Oncol Res 1997; 9:19 - 29
- Danilenko M, Wang X, Studzinski GP. Carnosic acid and promotion of monocytic differentiation of HL60-G cells initiated by other agents. J Natl Cancer Inst 2001; 93:1224 - 1233
- Kang SN, Lee MH, Kim KM, Cho D, Kim TS. Induction of human promyelocytic leukemia HL-60 cell differentiation into monocytes by silibinin: involvement of protein kinase C. Biochem Pharmacol 2001; 61:1487 - 1495
- Danilenko M, Wang Q, Wang X, Levy J, Sharoni Y, Studzinski GP. Carnosic acid potentiates the antioxidant and prodifferentiation effects of 1α,25-dihydroxyvitamin D3 in leukemia cells but does not promote elevation of basal levels of intracellular calcium. Cancer Res 2003; 63:1325 - 1332
- Sharabani H, Izumchenko E, Wang Q, Kreinin R, Steiner M, Barvish Z, et al. Cooperative antitumor effects of vitamin D3 derivatives and rosemary preparations in a mouse model of myeloid leukemia. Int J Cancer 2006; 118:3012 - 3021
- Shabtay A, Sharabani H, Barvish Z, Kafka M, Amichay D, Levy J, et al. Synergistic antileukemic activity of carnosic acid-rich rosemary extract and the 19-nor Gemini vitamin D analogue in a mouse model of systemic acute myeloid leukemia. Oncology 2008; 75:203 - 214
- Zhang J, Harrison JS, Uskokovic M, Danilenko M, Studzinski GP. Silibinin can induce differentiation as well as enhance vitamin D3-induced differentiation of human AML cells ex vivo and regulates the levels of differentiation-related transcription factors. Hematol Oncol 2010; 28:124 - 132
- Sokoloski JA, Narayanan R, Sartorelli AC. Enhancement by antisense oligonucleotides to NFκB of the differentiation of HL-60 promyelocytic leukemia cells induced by vitamin D3. Cancer Lett 1998; 125:157 - 164
- Wang Q, Salman H, Danilenko M, Studzinski GP. Cooperation between antioxidants and 1,25-dihydroxyvitamin D3 in induction of leukemia HL60 cell differentiation through the JNK/AP-1/Egr-1 pathway. J Cell Physiol 2005; 204:964 - 974
- Callens C, Coulon S, Naudin J, Radford-Weiss I, Boissel N, Raffoux E, et al. Targeting iron homeostasis induces cellular differentiation and synergizes with differentiating agents in acute myeloid leukemia. J Exp Med 2010; 207:731 - 750
- Ben-Dor A, Steiner M, Gheber L, Danilenko M, Dubi N, Linnewiel K, et al. Carotenoids activate the antioxidant response element transcription system. Mol Cancer Ther 2005; 4:177 - 186
- Satoh T, Kosaka K, Itoh K, Kobayashi A, Yamamoto M, Shimojo Y, et al. Carnosic acid, a catechol-type electrophilic compound, protects neurons both in vitro and in vivo through activation of the Keap1/Nrf2 pathway via S-alkylation of targeted cysteines on Keap1. J Neurochem 2008; 104:1116 - 1131
- Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J 2010; 12:87 - 97
- Kwak MK, Kensler TW. Targeting Nrf2 signaling for cancer chemoprevention. Toxicol Appl Pharmacol 2010; 244:66 - 76
- Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal 2010; 13:1713 - 1748
- Zhao F, Wu T, Lau A, Jiang T, Huang Z, Wang XJ, et al. Nrf2 promotes neuronal cell differentiation. Free Radic Biol Med 2009; 47:867 - 879
- Kosaka K, Mimura J, Itoh K, Satoh T, Shimojo Y, Kitajima C, et al. Role of Nrf2 and p62/ZIP in the neurite outgrowth by carnosic acid in PC12 h cells. J Biochem 2010; 147:73 - 81
- Pi J, Leung L, Xue P, Wang W, Hou Y, Liu D, et al. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J Biol Chem 2010; 285:9292 - 9300
- Hinoi E, Fujimori S, Wang L, Hojo H, Uno K, Yoneda Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J Biol Chem 2006; 281:18015 - 18024
- Hinoi E, Takarada T, Fujimori S, Wang L, Iemata M, Uno K, et al. Nuclear factor E2 p45-related factor 2 negatively regulates chondrogenesis. Bone 2007; 40:337 - 344
- Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth and development. Proc Natl Acad Sci U S A 1996; 93:13943 - 13948
- Kuroha T, Takahashi S, Komeno T, Itoh K, Nagasawa T, Yamamoto M. Ablation of Nrf2 function does not increase the erythroid or megakaryocytic cell lineage dysfunction caused by p45 NF-E2 gene disruption. J Biochem 1998; 123:376 - 379
- Terui K, Takahashi Y, Kitazawa J, Toki T, Yokoyama M, Ito E. Expression of transcription factors during megakaryocytic differentiation of CD34+ cells from human cord blood induced by thrombopoietin. Tohoku J Exp Med 2000; 192:259 - 273
- Maruyama A, Tsukamoto S, Nishikawa K, Yoshida A, Harada N, Motojima K, et al. Nrf2 regulates the alternative first exons of CD36 in macrophages through specific antioxidant response elements. Arch Biochem Biophys 2008; 477:139 - 145
- Mulcahy RT, Wartman MA, Bailey HH, Gipp JJ. Constitutive and β-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J Biol Chem 1997; 272:7445 - 7454
- Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol 2003; 23:7198 - 7209
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap‘n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem 1999; 274:26071 - 26078
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol 1976; 33:451 - 458
- Griffith OW. Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem 1980; 106:207 - 212
- Wang X, Studzinski GP. The requirement for and changing composition of the activating protein-1 transcription factor during differentiation of human leukemia HL60 cells induced by 1,25-dihydroxyvitamin D3. Cancer Res 2006; 66:4402 - 4409
- Yan H, Wang L, Li X, Yu C, Zhang K, Jiang Y, et al. High-performance liquid chromatography method for determination of carnosic acid in rat plasma and its application to pharmacokinetic study. Biomed Chromatogr 2009; 23:776 - 781
- Svedberg H, Chylicki K, Baldetorp B, Rauscher FJ 3rd, Gullberg U. Constitutive expression of the Wilms' tumor gene (WT1) in the leukemic cell line U937 blocks parts of the differentiation program. Oncogene 1998; 16:925 - 932
- Ward JO, McConnell MJ, Carlile GW, Pandolfi PP, Licht JD, Freedman LP. The acute promyelocytic leukemia-associated protein, promyelocytic leukemia zinc finger, regulates 1,25-dihydroxyvitamin D(3)-induced monocytic differentiation of U937 cells through a physical interaction with vitamin D(3) receptor. Blood 2001; 98:3290 - 3300
- Simboli-Campbell M, Narvaez CJ, Tenniswood M, Welsh J. 1,25-Dihydroxyvitamin D3 induces morphological and biochemical markers of apoptosis in MCF-7 breast cancer cells. J Steroid Biochem Mol Biol 1996; 58:367 - 376
- Blutt SE, McDonnell TJ, Polek TC, Weigel NL. Calcitriol-induced apoptosis in LNCaP cells is blocked by overexpression of Bcl-2. Endocrinology 2000; 141:10 - 17
- Ma Y, Yu WD, Kong RX, Trump DL, Johnson CS. Role of nongenomic activation of phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase/extracellular signal-regulated kinase kinase/extracellular signal-regulated kinase 1/2 pathways in 1,25D3-mediated apoptosis in squamous cell carcinoma cells. Cancer Res 2006; 66:8131 - 8138
- Wang X, Studzinski GP. Antiapoptotic action of 1,25-dihydroxyvitamin D3 is associated with increased mitochondrial MCL-1 and RAF-1 proteins and reduced release of cytochrome c. Exp Cell Res 1997; 235:210 - 217
- Wang X, Patel R, Studzinski GP. hKSR-2, a vitamin D-regulated gene, inhibits apoptosis in arabinocytosine-treated HL60 leukemia cells. Mol Cancer Ther 2008; 7:2798 - 2806
- Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal 2010; 13:1699 - 1712
- Liebermann DA, Gregory B, Hoffman B. AP-1 (Fos/Jun) transcription factors in hematopoietic differentiation and apoptosis. Int J Oncol 1998; 12:685 - 700
- Pan Q, Granger J, O'Connell TD, Somerman MJ, Simpson RU. Promotion of HL-60 cell differentiation by 1,25-dihydroxyvitamin D3 regulation of protein kinase C levels and activity. Biochem Pharmacol 1997; 54:909 - 915
- Huang HC, Nguyen T, Pickett CB. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor2. Proc Natl Acad Sci U S A 2000; 97:12475 - 12480
- Martin D, Rojo AI, Salinas M, Diaz R, Gallardo G, Alam J, et al. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J Biol Chem 2004; 279:8919 - 8929
- Xu C, Yuan X, Pan Z, Shen G, Kim JH, Yu S, et al. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther 2006; 5:1918 - 1926
- Esposito F, Agosti V, Morrone G, Morra F, Cuomo C, Russo T, et al. Inhibition of the differentiation of human myeloid cell lines by redox changes induced through glutathione depletion. Biochem J 1994; 301:649 - 653
- Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci U S A 1997; 94:3633 - 3638
- Gius D, Botero A, Shah S, Curry HA. Intracellular oxidation/reduction status in the regulation of transcription factors NFκB and AP-1. Toxicol Lett 1999; 106:93 - 106
- Yang H, Magilnick N, Lee C, Kalmaz D, Ou X, Chan JY, et al. Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NFkappaB and AP-1. Mol Cell Biol 2005; 25:5933 - 5946
- Cai DH, Wang D, Keefer J, Yeamans C, Hensley K, Friedman AD. C/EBP alpha:AP-1 leucine zipper heterodimers bind novel DNA elements, activate the PU.1 promoter and direct monocyte lineage commitment more potently than C/EBP alpha homodimers or AP-1. Oncogene 2008; 27:2772 - 2779
- Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet 2006; 38:1269 - 1277
- Yang MY, Liu TC, Chang JG, Lin PM, Lin SF. JunB gene expression is inactivated by methylation in chronic myeloid leukemia. Blood 2003; 101:3205 - 3211
- Qi X, Pramanik R, Wang J, Schultz RM, Maitra RK, Han J, et al. The p38 and JNK pathways cooperate to trans-activate vitamin D receptor via c-Jun/AP-1 and sensitize human breast cancer cells to vitamin D3-induced growth inhibition. J Biol Chem 2002; 277:25884 - 25892
- Steiner M, Priel I, Giat J, Levy J, Sharoni Y, Danilenko M. Carnosic acid inhibits proliferation and augments differentiation of human leukemic cells induced by 1,25-dihydroxyvitamin D3 and retinoic acid. Nutr Cancer 2001; 41:135 - 144
- Garland CF, Gorham ED, Mohr SB, Garland FC. Vitamin D for cancer prevention: global perspective. Ann Epidemiol 2010; 19:468 - 483
- Moiseeva EP, Manson MM. Dietary chemopreventive phytochemicals: too little or too much?. Cancer Prev Res (Phila Pa) 2009; 2:611 - 616