Abstract
Caspases play a key role in the apoptotic pathway by virtue of their ability to cleave key protein substrates within the dying cell. Caspases are produced as inactive zymogens, and need to become proteolytically processed in order to become active. A key executioner caspase, caspase-3, has previously been found to exist in both the cytosol and the mitochondria. At the mitochondria, caspase-3 is associated with both the inner and outer mitochondrial membranes, where it interacts with heat shock proteins Hsp60 and Hsp10. Like caspase-3, a small portion of the p53 tumor suppressor protein is localized to mitochondria, particularly after genotoxic stress. p53 interacts with various members of the Bcl2 family at the mitochondria, and this interaction is key to its ability to induce apoptosis. In this study, we sought to determine the identity of other mitochondrial p53-interacting proteins. Using immunoprecipitation from purified mitochondria followed by mass spectrometry we identified caspase-3 as a mitochondrial p53-interacting protein. Interestingly, we find that tumor-derived mutant forms of p53 retain the ability to interact with mitochondrial caspase-3. Further, we find evidence that these mutant forms of p53 may interfere with the ability of procaspase-3 to become proteolytically activated by caspase-9. The combined data suggest that tumor-derived mutants of p53 may be selected for in tumor cells due to their ability to bind and inhibit the activation of caspase-3.
</br> See commentary: Caspase-3 joins the p53 interactome
Introduction
Caspases are a family of cysteine proteases that play important roles in apoptosis and inflammation. These proteases are normally produced as inactive zymogens, referred to as pro-caspases; these need to have their prodomain cleaved by other caspases in order to become active. Active caspases are responsible for the membrane blebbing, cell shrinkage, nuclear fragmentation and chromatin condensation that occur during apoptosis. The apoptotic caspases can be classified as either initiator caspases, which contain long prodomains and are typically activated by oligomerization or effector caspases, which can be activated by upstream initiator caspases. Initiator caspases, which include caspases-2, -8, -9 and -10, activate the caspase cascade through the removal of the inactive prodomains of the effector caspases. Once activated, effector caspases, including caspases-3, -6 and -7, cleave several dozen key substrates within the cell in order to carry out the apoptotic process.Citation1–Citation3
Caspases normally are located in the cytoplasm; however they have also been found in other organelles within the cell. In particular, pro-caspase-2, -9 and -3 have each been observed at the mitochondria.Citation4 The distribution of pro-caspase-3 at the mitochondria is found in a variety of rat tissues including the thymus, brain, liver, heart, spleen and kidney, indicating that this a common occurrence and not specific to a certain cell type.Citation5 Mitochondrial pro-caspase-3 is found associated with both the inner and outer mitochondrial membranes, in a complex with heat shock proteins Hsp60 and Hsp10.Citation6,Citation7 Upon induction of apoptosis, caspase-3 becomes activated, dissociates from the heat shock complex and traffics to the cytosol. Additionally, a small fraction of both active caspase-9 and caspase-3 remain at the mitochondria, presumably to proteolytically cleave mitochondrial substrates.Citation4,Citation7
The tumor suppressor p53 is one of the main inducers of the apoptotic pathway. When a cell becomes exposed to genotoxic or metabolic stress, p53 protein is stabilized, allowing it to transcriptionally activate or repress various downstream target genes involved in apoptosis, such as Bax, PUMA and Noxa.Citation8 A small portion of p53 also traffics to the mitochondria where it interacts with members of the Bcl2 family, including the anti-apoptotic members Bcl2 and Bcl-xL, and the pro-apoptotic members Bax and Bak. Interaction with anti-apoptotic Bcl2 family members inhibits their function, while interaction of p53 with pro-apoptotic Bax and Bak induces their oligomerization. This oligomerization forms a pore in the outer mitochondrial membrane, allowing the release of cytochrome c into the cytoplasm and leading to the induction of apoptosis.Citation9–Citation11 Whether p53 also interacts with other mitochondrial proteins influential in apoptosis has not been clear. In an effort to identify other activities of p53 at the mitochondria, we performed immunoprecipitation of this protein from purified mitochondria, and we used mass spectrometry of tryptic peptides in order to identify p53-interacting proteins. This study identified pro-caspase-3 as a mitochondrial p53-interacting protein.
Results
Caspase-3 binds to wild-type p53 at the mitochondria.
Having recently identified and characterized the interaction between p53 and Bak at the mitochondria,Citation11,Citation12 we sought to determine the identity of other proteins that interact with p53 at the mitochondria. Toward this goal, mitochondria from Saos2 cells containing a temperature-sensitive inducible form of wild-type p53 were purified on sucrose gradients and used to immunoprecipitate p53. At the same time, mitochondria were isolated from the Wm115 melanoma cell line (wild-type p53) treated with camptothecin, and used to immunoprecipitate p53. Coomassie staining of these immunoprecipitates identified several bands present in the p53-expressing mitochondria, including one at approximately 32 kilodaltons that tryptic peptide mass spectrometry identified as caspase-3 ( and B).
To verify the interaction between caspase-3 and p53, Wm115 cells were treated with Camptothecin for 6 h. Whole cell lysates from these cells were immunoprecipitated with an antibody for caspase-3 or IgG and the blot was probed for associated p53; the latter was readily detectable in Caspase-3 immunoprecipitates (). Notably, this immuno-complex was also detectable in purified mitochondria from Wm115 cells (). We next verified this interaction in U2OS cells (wt p53) following treatment with Adriamycin for 24 h, and determined that this interaction was as readily detectable as the Bak-p53 complex (). A p53-caspase-3 complex was likewise detectable in mitochondria purified from cells with tetracycline-inducible p53 (). From the initial SDS-PAGE gel (), the size of co-precipitating caspase-3 was most consistent with pro-caspase-3; to confirm this interaction, we used antisera specific for pro-caspase-3 for the immunoprecipitation, and noted that both the proline 72 polymorphic variant of p53, as well as the arginine 72 variant, are able to interact with pro-caspase-3 in cells containing temperature-sensitive variants of both proteins (). Interestingly, in these temperature-sensitive cells p53 predominantly assumes a conformation characteristic of tumor-derived mutants of this protein at the uninduced (zero) timepoint. We noted that p53 retained ability to interact with pro-caspase-3 at this temperature, suggesting that mutant forms of p53 might retain the ability to interact with caspase-3.
We next made efforts to determine the percentage of wild-type p53 that interacts with caspase-3 in cells. Toward this end, H1299 cells containing tetracycline-inducible versions of both codon 72 variants of p53 were used for immunoprecipitation analyses. Immunoprecipitation of caspase-3 or p53, followed by western blotting for associated p53, indicated that approximately 1–3% of total wild-type p53 could be found in immunoprecipitates with caspase-3. These data also indicated that there was no difference in the interaction between caspase-3 and either the P72 or R72 variant ().
Caspase-3 localizes to the outer mitochondrial membrane.
Previous studies have reported that caspase-3 localizes to the mitochondria and at this organelle is associated with both the inner and outer mitochondrial membranes.Citation6 To verify the localization of mitochondrial caspase-3, mitochondria were purified from H1299 cells and incubated with increasing amounts of trypsin. The purity of these mitochondria, and lack of cytosolic contamination, was evident from the fact that they were positive for the mitochondrial proteins cytochrome c, Grp75 and Hsp60, but not for the abundant nuclear/cytosolic protein PCNA (). Trypsin digestion experiments of purified mitochondria indicated that the amount of caspase-3 at the mitochondria decreased with increasing amounts of trypsin. Bcl-xl, which is bound to the outer mitochondrial membrane, also decreased in abundance, while the inner mitochondrial proteins cytochrome c and Grp75 were protected from digestion by the mitochondrial membrane (). These data are most consistent with the localization of caspase-3 on the outer mitochondrial membrane, at least in this cell type.
Mutant p53 binds caspase-3.
We next sought to follow up on the possibility that mutant forms of p53 might likewise interact with caspase-3 at mitochondria. Toward this goal we utilized p53-null Saos2 cells that were stably transfected with CMV vector, temperature sensitive p53 or two different hotspot mutants of p53, R175H and R273H. R175H is a common structural hotspot mutant of p53 that affects the stabilization of the p53 tetramer.Citation13,Citation14 In previous studies, R175H was found to be unable to interact with either Bcl2 or Bcl-xl,Citation9,Citation15 but retains the ability to interact with Bak.Citation12 As shown in , immunoprecipitation with caspase-3 antisera revealed that both the R175H and R273H mutants interact with caspase-3, to levels comparable to wild-type p53.
Mutant p53 interferes with caspase-3 processing.
To determine if the binding of mutant p53 affects the activation of caspase-3, mitochondria were isolated from Saos2 cells that express CMV or the R175H and R273H mutants of p53. To these mitochondria we added increasing amounts of active caspase-9, and the processing of caspase-3 was assessed by western analysis using antisera specific for cleaved caspase-3. As shown in , significantly more cleaved caspase-3 was generated by incubation with caspase-9 in mitochondria isolated from cells transfected with CMV vector alone, compared to mitochondria purified from cells stably-transfected with R175H or R273H. These data were consistent in multiple experiments, and support the contention that interaction with mutant p53 impairs the ability of pro-caspase-3 to be activated by upstream caspases.
Discussion
From this study, we identified pro-caspase-3 as a mitochondrial p53-interacting protein. The ability of p53 to associate with the anti- and pro-apoptotic members of the Bcl-2 family at the mitochondria has been well characterized.Citation9–Citation11 This is the first report of p53 binding to a member of the caspase family. Previously, caspase-3 was found to localize to the mitochondria and to interact with heat shock proteins 60 and 10.Citation7 We confirmed this localization, and used proteolytic digestion studies to support the contention that the majority of mitochondrial caspase-3 appears to interact with the outer mitochondrial membrane. The impact of the interaction of wild-type p53 with caspase-3 is unclear at this time. One intriguing possibility is that, similar to the ability of p53 to induce the oligomerization of BAK, it might also induce the oligomerization of caspase-3, which might then facilitate the activation of the latter. However, through protein cross-linking studies, we were unable to find evidence that mitochondrial p53 influences the oligomerization of pro-caspase-3. A second possibility is that wt p53 binds to caspase-3 and enhances its processing and activation by caspase-9; we are currently making efforts to test this possibility. Others have found that p53 can be cleaved by caspase-3, and that the p53 proteolytic cleavage products can induce the loss of mitochondrial transmembrane potential.Citation16 Therefore, it remains possible that the interaction with pro-caspase-3 facilitates the pro-apoptotic function of wild-type p53 at mitochondria, upon caspase-3 activation.
In this study, we found that two mutant forms of p53 retained the ability to interact with caspase-3 at mitochondria, and in fact that this interaction inhibits the activation of caspase-3 by upstream caspase. A future direction of interest will be to determine whether mutant forms of p53 located in the cytosol can likewise interfere with the processing and activation of cytosolic caspase-3. Mutant p53 has long been known to possess a “gain of function”; in other words, tumor cells containing mutant p53 possess increased transformed properties compared to the identical tumor containing no p53.Citation17 Part of the ability of tumor-derived mutant forms of p53 to enhance the transformed properties of a cell relies on the ability of these proteins to bind and inactivate p73,Citation18 and Mre11.Citation19 Our data indicate that caspase-3 may be added to this list, and that mutant p53 may decrease the apoptotic sensitivity of tumor cells by its ability to bind and inactivate this executioner caspase.
Materials and Methods
Cell culture, drug treatments.
Saos2 temperature sensitive inducible p53 cell lines were grown in DMEM, 10% fetal bovine serum, 1% Penicillin/Streptomycin and 0.4 mg/L G418 at 39°C (mutant conformation p53). To induce wild-type conformation p53, these cells were transferred to a 32°C incubator. H1299 tetracycline inducible p53 cells were grown in DMEM supplemented with 10% Tet-System Approved fetal bovine serum (Clontech) and 1% Penicillin/Streptomycin. Saos2 R175H and R273H mutant cells were grown in DMEM, 10% FBS, 1% Penicillin/Streptomycin and 0.4 g/L G418. WM115, H1299 and U2OS cells were grown in DMEM, 10% FBS, 1% Penicillin/Streptomycin. Adriamycin (Sigma) was used at a concentration of 0.5 µg/ml. Camptothecin (Sigma) was used at a concentration of 5 uM. To induce wild-type p53 in the tetracycline-regulated H1299 cell line, cells were treated with 0.75-µg/mL doxycycline (Sigma) for 3–6 h.
Western analysis, immunoprecipitation-western.
Western analysis was performed exactly as described in reference Citation12. The membranes were blocked and probed with antibodies for p53 Ab6 (Calbiochem) 1:1,000, p53 FL393 Goat (Santa Cruz) 1:200, Actin AC15 (Sigma) 1:10,000, Cytochrome C (BD Pharmingen) 1:250, PCNA PC10 (Santa Cruz) 1:1,000, GRP75 C-19 (Santa Cruz) 1:400, HSP60 K-19 (Santa Cruz) 1:400, Bak NT (Upstate) 1:1,000, Caspase-3 H-277 (Santa Cruz) 1:250, Pro-caspase-3 E-8 (Santa Cruz) 1:250, Cleaved Caspase-3 (Cell Signaling) 1:500. Horseradish peroxidase conjugated secondaries (Jackson Laboratories) were used at 1:10,000. Blots were exposed to ECL (Amersham). For immunoprecipitations, 500 µg of whole cell lysate or 50 µg of mitochondrial lysate was incubated with 1 µg of immunoprecipitating antibody overnight. Protein G agarose beads were added for 1 h followed by washes in NP-40 buffer, exactly as described in reference 20. Horseradish peroxidase conjugated light chain specific secondaries (Jackson Laboratories) were used for mutant p53 immunoprecipitations.
Mitochondria purification, enzymatic treatment, mass spectrometry.
Cells were harvested, spun down and the pellet was resuspended in Mannitol Buffer (10 mM Hepes pH 7.4, 1 mM EDTA, 0.2 mM EGTA pH 7.4, 0.1% BSA, 300 mM Mannitol, supplemented with protease inhibitors). Cells were dounce homogenized 12 times using a motorized homogenizer. Mitochondria were then isolated by differential centrifugation exactly as described in reference Citation12. For trypsin digestion experiments, 20 µg of purified mitochondria were incubated with the indicated units of trypsin (Invitrogen) at 4° for 20 minutes prior to SDS-PAGE. For treatment with caspase-9, 10 µg of mitochondria were incubated with increasing units of active caspase-9 (Abcam) at 37°C for 30 min. western analysis was performed using antisera for p53 (Ab6, Calbiochem) and cleaved caspase-3 (Cell Signaling). For identification of mitochondrial p53-interacting proteins, 500 µg of purified mitochondria was immunoprecipitated with 1 µg of p53 antibody (fl393G, Santa Cruz Biotechnology) and immunoprecipitates were resolved on SDS-PAGE and stained with Coomassie. Bands were isolated, treated and digested with trypsin as described in reference Citation20, prior to analysis by nanoflow RP-HPLC micro electrospray ionization mass spectrometry (Applied Biomics, Inc.).
Figures and Tables
Figure 1 Identification of p53-interacting proteins from gradient-purified mitochondria. (A) 500 µg of mitochondrial lysate was purified from Saos2 cells containing temperature sensitive p53 (mutant conformation at 39°C and wild-type conformation at 32°C), or WM115 cells treated with camptothecin (CPT) and immunoprecipitated with polyclonal p53 antibody, run on SDS-PAGE and stained with Coomassie Blue. Arrows point to bands identified by mass spectrometry to be p53 and caspase-3; the band just above p53 is immunoglobulin heavy chain. (B) Peptide sequence of human caspase-3, with the tryptic peptides identified in (A) outlined in bold.
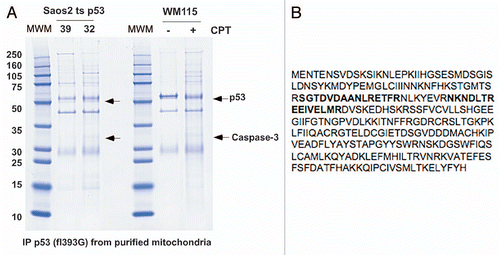
Figure 2 Caspase-3 binds endogenous p53. (A) Whole cell lysates from WM115 cells treated with and without 5 µM Camptothecin (CPT) for 6 h were immunoprecipitated (IP) with antibodies for caspase-3 (Casp3) and IgG. Western blot was performed for both p53 and caspase-3. (B) Mitochondria were purified from WM115 cells treated with and without 5 µM Camptothecin (CPT) for 6 h and immunoprecipitated (IP) with antibodies for IgG and caspase-3 (Casp 3). The blot was probed with an antibody for p53. (C) Whole cell lysates from U2OS cells that were treated with 0.5 µg/ml Adriamycin (Adr) for 24 h and immunoprecipitated (IP) with antibodies for caspase-3 (Casp 3) or Bak. Right: western analysis of the U2OS lysates used in the immunoprecipitation for p53, Caspase-3 and actin. (D) Mitochondria from H1299 Tetracycline (Tet) inducible p53 cells were isolated before and after 0.75 µg/ml Doxycycline (Doxy) treatment for 3 h. The mitochondria were immunoprecipitated (IP) with antibodies for caspase-3 (casp3), Bak and IgG. Right: western analysis was done on cytosolic (Cyto) and mitochondrial (Mito) fractions from lysates used in the immunoprecipitation. Blots were probed for GRP75 (control for mitochondria), p53 and PCNA (control for nuclear/cytosolic contamination). (E) Whole cells lysates from Saos2 cells containing temperature-sensitive forms of p53 encoding proline at amino acid 72 (P72) or arginine 72 (R72) were immunoprecipitated with antisera specific for pro-caspase-3 (E3, Santa Cruz Biotechnology) following temperature shift and wild-type p53 induction for the hours indicated. The bottom panel depicts the steady state level of p53 in whole cell lysate. (F) Immunoprecipitation of caspase-3 and p53 from H1299 cells containing tetracycline-inducible versions of the p53 polymorphic variants P72 and R72, followed by western analysis for associated p53. The right part depicts western analysis of whole cell lysates from these samples.
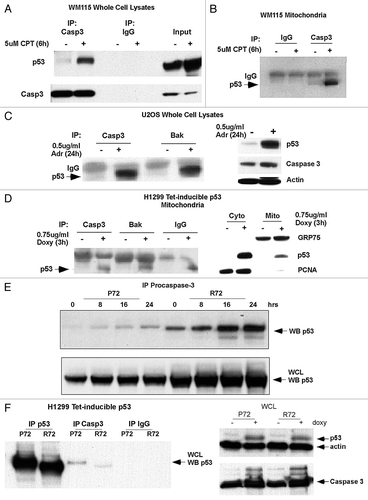
Figure 3 Caspase-3 appears to be predominantly associated with the outer mitochondrial membrane. (A) Cytosolic (Cyto) and mitochondrial (Mito) extracts from H1299 cells were analyzed by western blot for caspase-3 (Casp 3), cytochrome c (Cyto C), GRP75 (control for mitochondria), HSP 60 (control for mitochondria) and PCNA (control for nuclear/cytosolic contamination of mitochondria). (B) Mitochondria from H1299 cells were treated with increasing concentrations of trypsin for 20 min. Western analysis was performed for caspase-3 (Casp 3), Bcl-XL, cytochrome c (Cyto C) and GRP75.
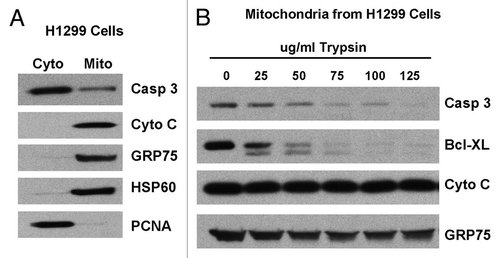
Figure 4 Mutant p53 binds to caspase-3 and inhibits the proteolytic processing of caspase-3 by active caspase-9. (A) Left: immunoprecipitates of whole cell lysate (WCL) from Saos2 cells containing temperature sensitive inducible p53 (at 32°C, wild-type conformation), CMV parental vector, the R175H mutant or the R273H mutant form of p53 with caspase-3 antibody or IgG. Western analysis was performed for p53. Right: western analysis of whole cell lysates from the cell lines indicated for the steady state level of p53, caspase-3 and actin. (B) Purified mitochondria from Saos2 cells stably expressing CMV parental vector or the R175H and R273H mutants of p53 were incubated with increasing units (U) of active caspase-9 for 30 min. Western analysis was performed using antisera specific for p53 and cleaved caspase-3.
Acknowledgements
This work was supported by funding from the National Institutes of Health. The authors declare they have no financial interests in this work.
References
- Nicholson DW. Caspase structure, proteolytic substrates and function during apoptotic death. Cell Death Differ 1999; 6:1028 - 1042
- Stennicke HR, Salvesen GS. Properties of the caspases. Biochimica et Biophysica Acta 1998; 1387:17 - 31
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol 2008; 9:231 - 241
- Zhivotovsky B, Samali A, Gahm A, Orrenius S. Caspases: their intracellular localization and translocation during apoptosis. Cell Death Differ 1999; 6:644 - 651
- Samali A, Zhivotovsky B, Jones DP, Orrenius S. Detection of pro-caspase-3 in cytosol and mitochondria of various tissues. FEBS 1998; 431:167 - 169
- Mancini M, Nicholson DW, Roy S, Thornberry NA, Peterson EP, Casciola-Rosen LA, et al. The caspase-3 precursor has a cytosolic and mitochondrial distribution: implications for apoptotic signaling. J Cell Biol 1998; 140:1485 - 1495
- Samali A, Cai J, Zhivotovsky B, Jones DP, Orrenius S. Presence of a pre-apoptotic complex of pro-caspse-3, Hsp60 and Hsp10 in the mitochondrial fraction of Jurkat cells. EMBO J 1999; 18:2040 - 2048
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell 1997; 88:323 - 331
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 2003; 11:577 - 590
- Chipuk J, Kuwana T, Bouchier-Hayes L, Droin N, Newmeyer D, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 2004; 303:1010 - 1014
- Leu J, Dumont P, Hafey M, Murphy M, George D. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol 2004; 6:443 - 450
- Pietsch EEP, Canutescu A, Wang G, Dunbrack R, Murphy M. Oligomerization of BAK by p53 utilizes conserved residues of the p53 DNA binding domain. J Biol Chem 2008; 283:21294 - 21304
- Cho Y, Gorina S, Jeffrey P, Pavletich N. Crystal structure of a p53 tumor suppressor-DNA complex: uncerstanding tumorigenic mutations. Science 1994; 265:346 - 355
- Hollstein M, Sidransky D, Vogelstein B, Harris C. p53 mutations in human cancers. Science 1991; 253:49 - 53
- Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, et al. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem 2006; 28:8600 - 8606
- Sayan BS, Sayan AE, Knight RA, Melino G, Cohen GM. p53 is cleaved by caspases generating fragments localizing to mitochondria. J Biol Chem 2006; 281:13566 - 13573
- Zambetti GP, Levine AJ. A comparison of the biological activities of wild-type and mutant p53. FASEB J 1993; 7:855 - 865
- Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 2000; 25:47 - 54
- Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 2007; 9:573 - 580
- Pimkina J, Humbey O, Zilfou JT, Jarnik M, Murphy ME. ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem 2009; 284:2803 - 2810