Abstract
miR-155 is a prominent microRNA (miRNA) that regulates genes involved in immunity and cancer-related pathways. miR-155 is overexpressed in lung cancer, which correlates with poor patient prognosis. It is unclear how miR-155 becomes increased in lung cancers and how this increase contributes to reduced patient survival. Here, we show that hypoxic conditions induce miR-155 expression in lung cancer cells and trigger a corresponding decrease in a validated target, FOXO3A. Furthermore, we find that increased levels of miR-155 radioprotects lung cancer cells, while inhibition of miR-155 radiosensitizes these cells. Moreover, we reveal a therapeutically important link between miR-155 expression, hypoxia, and irradiation by demonstrating that anti-miR-155 molecules also sensitize hypoxic lung cancer cells to irradiation. Our study helps explain how miR-155 becomes elevated in lung cancers, which contain extensive hypoxic microenvironments, and demonstrates that inhibition of miR-155 may have important therapeutic potential as a means to radiosensitize hypoxic lung cancer cells.
Introduction
Tumor microenvironments have a prominent role in shaping malignant progression, and in some cases may be as important as genetic and epigenetic factors in influencing cancer development.Citation1 Tumor microenvironments, which are often characterized by low pH, nutrient deprivation, and hypoxia, confer tumor cells with a marked ability to survive in adverse conditions.Citation2,Citation3 Hypoxia in particular causes tumors to acquire aggressive characteristics such as increased metastatic potential,Citation4 decreased drug sensitivity,Citation5 diminished p53-dependent apoptosis,Citation6 and increased genetic instability and mutation rates.Citation7 Notably, these hypoxia-induced characteristics have been shown to independently and significantly correlate with reduced survival in patients with cervical and head and neck cancers.Citation8,Citation9
Additionally, hypoxia greatly affects tumor responsiveness to radiation therapy. In normoxic conditions, radiation treatment generates oxygen radicals that mediate DNA damage and tumor cell eradication. However, in hypoxic tumor microenvironments, where intratumoral oxygen is reduced or absent, tumor cells are 2.5–3 times more resistant to radiation therapy, largely due to the lack of oxygen radical-mediated DNA damage.Citation2,Citation10 Thus, poor outcome in cancer patients is often a direct result of hypoxia-induced radioresistance in tumor cells.Citation2,Citation8,Citation9
Interestingly, in the last several years, numerous studies have identified important links between hypoxia, cancer, and miRNAs. MiRNAs are negative modulators of gene expression that have been implicated in almost every mammalian biological process investigated. A number of miRNAs have been shown to be regulated by hypoxia and many of them confer cancer cells with increased ability to adapt to hypoxic conditions and survive. The most prolific miRNA vis-à-vis hypoxia is miR-210; miR-210 is upregulated in many cancers and is induced in hypoxic conditions.Citation11 This miRNA has an overall effect of increasing tumor cell survival by regulating genes involved in DNA repair, angiogenesis, cell cycle progression, and mitochondrial metabolism.Citation11-Citation13 Additionally, high miR-210 is associated with poor prognosis in pancreatic and breast cancer patients,Citation13,Citation14 further implicating it as a crucial link between hypoxia and cancer outcome. Other miRNAs regulate or are regulated by hypoxia including miR-373,Citation15 miR-424Citation16 and miR-17–92.Citation17
Another miRNA, miR-155, has emerged as a “master-regulator” of numerous biological processes, most notably those involved in immune function and cancer development. Interestingly, miR-155 is overexpressed in lung cancersCitation18,Citation19 and its increased expression is associated with poor prognosis in lung cancer patients.Citation19,Citation20 However, the mechanism for miR-155 upregulation in lung cancer is currently unknown, and it is unclear how increased miR-155 levels contribute to poor prognosis in patients. Here we show that miR-155 is induced by hypoxia in lung cancer cells. Furthermore, miR-155 radioprotects lung cancer cells, which may help explain why patients with increased levels have worse prognosis. Importantly, we find that inhibition of miR-155 radiosensitizes hypoxic lung cancer cells, demonstrating a potentially novel therapeutic approach for lung cancer patients undergoing radiotherapy.
Results
Hypoxia induces miR-155 in lung cancer cells
miR-155 levels are increased in a number of cancer types, including non-small cell lung cancer (NSCLC).Citation18,Citation19 Given that miR-155 levels are increased in lung cancers, which often contain hypoxic tumor microenvironments,Citation21 we tested whether hypoxia induces miR-155. Non-small cell lung cancer lines (A549 and H460) were placed in an incubated cell culture hypoxia chamber for a time-course of hypoxia (< 0.01% O2) exposure. At various time-points, cells were removed from hypoxia and immediately prepared for molecular analysis. We found that miR-155 levels increased ~2-fold at 8 h of hypoxia exposure and ~8-fold by 48 h of hypoxia exposure in A549 cells (). Interestingly, this corresponded with diminished levels of a validated miR-155 target,Citation22 Forkhead box O3 (FOXO3A), at 24 h hypoxia exposure and beyond (). Similarly, H460 cells demonstrated a ~6-fold induction of miR-155 levels after 48 h of hypoxia exposure and a corresponding decrease in FOXO3a levels ().
Figure 1. Hypoxia induces miR-155 in lung cancer cells. (A) miR-155 fold-change in A549 cells after the demarcated amounts of time in hypoxia as measured by qPCR; protein gel blot showing the corresponding changes in FOXO3A protein levels in A549 cells upon exposure to hypoxia. (B) miR-155 fold-change in H460 cells after exposure to 48 h of hypoxia as measured by qPCR; protein gel bot showing the corresponding decrease in FOXO3A protein expression in H460 cells upon exposure to hypoxia. (C) miR-155 fold-change in A549 cells at the given number of hours after transfer from hypoxic to normoxic conditions as measured by qPCR; protein gel blot showing the corresponding changes of FOXO3A protein levels in A549 cells after transfer from hypoxic to normoxic conditions.
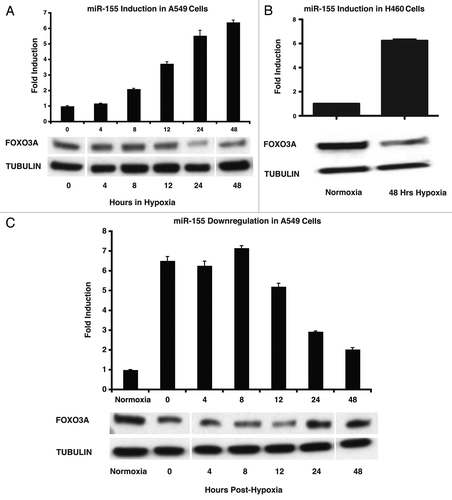
We next determined a time course for miR-155 downregulation after removal from hypoxic conditions. miR-155 levels decreased to ~3-fold by 24 h and approximately 2-fold by 48 h after transfer to normoxic conditions in A549 cells (). This corresponded with a subsequent increase in FOXO3A protein levels by 24 h (). FOXO3A is a member of the forkhead family of transcription factors; when unphosphorylated, FOXO3A localizes to the nucleus where it functions as a tumor suppressor by inducing genes involved in apoptosis.Citation23 Recently, a study showed that FOXO3A is negatively regulated by miR-155 in breast cancer cells, resulting in their reduced chemo-sensitivity.Citation22 Our results combined with this study support the notion that hypoxia-induced miR-155 may confer lung cancer cells with the ability to resist other forms of treatment such as irradiation, in part by modulating FOXO3A.
HIF-1α is necessary for hypoxic miR-155 induction
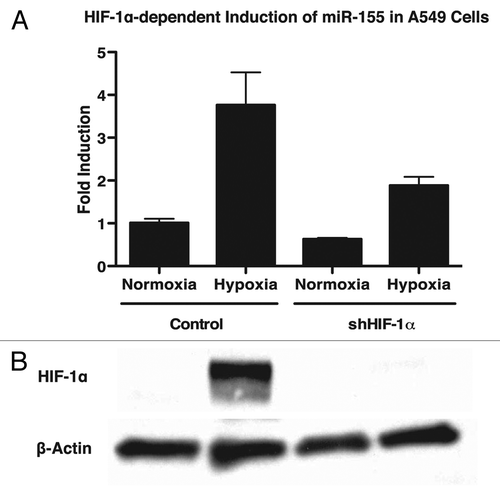
One of the main effectors of hypoxia is the transcription factor HIF-1α, which is responsible for regulating a wide variety of cellular pathways including survival and angiogenesis. Importantly, HIF-1α has been shown to be necessary for the hypoxia-driven induction of a number of miRNAs including miR-210 and miR-373.Citation15 To determine whether HIF-1α is required for the upregulation of miR-155 under hypoxic conditions, we utilized a stable cell line expressing shRNA to HIF-1α. A549 lung cancer cells stably expressing either shHIF-1α or a vector control were exposed to normoxia or hypoxia for 48 h. In the presence of shHIF-1α, we found that the upregulation of miR-155 is attenuated in hypoxia (). These results suggest a role for HIF-1α in the induction of miR-155 under hypoxic conditions. Protein gel blot analysis confirmed induction of HIF-1α in hypoxic conditions and the absence of HIF-1α in hypoxic cells containing shHIF-1α (). We found similar results using MCF7 breast cancer cells (Fig. S1A and B).
Figure 2. Knockdown of HIF-1α attenuates induction of miR-155 in response to hypoxia in A549 cells. (A) miR-155 fold-change as measured by qPCR in A549 lung cancer cells expressing a vector control or an shRNA to HIF-1α. Cells were exposed to normoxia or hypoxia for 48 h and miR-155 levels were monitored. After hypoxia, miR-155 is induced ~4-fold in control cells, while only a ~2-fold induction of miR-155 is observed in cells expressing shRNA to HIF-1α. (B) Protein gel blot confirms HIF-1α knockdown.
miR-155 modulates irradiation response in lung cancer cells
Several studies have shown that increased miR-155 levels correlate with poor prognosis in lung cancer patients. In a previous study (submitted), we found that miR-155 induction in mouse lungs is not sufficient to initiate tumorigenesis (data not shown). Furthermore, we tested whether miR-155 alters the ability of A549 lung cancer cells to migrate and/or invade using the Boyden chamber assay, and in our hands miR-155 did not have an effect (data not shown).
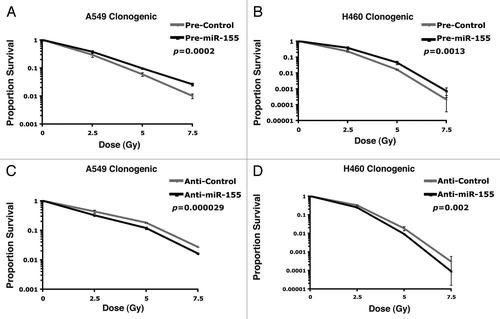
We then asked whether miR-155 influences the ability of lung cancer cells to respond to radiotherapy, which could potentially explain why miR-155 contributes to poor prognosis in lung cancer patients. We found that miR-155 is expressed and induced by hypoxia in the two NSCLC cell lines A549 and H460 (). We then transfected A549 and H460 cancer cells with synthetic pre-miR-155 molecules or pre-control molecules containing a scrambled sequence (Ambion). Sixteen hours post-transfection, cells were treated with varying doses of irradiation and plated for a clonogenic assay, which measures all forms of irradiation-induced cell death. We found that increased levels of miR-155 had a radio-protective effect on A549 cells (; p = 0.0002) and H460 cells (; p = 0.0013) compared with control molecules. We also tested whether inhibition of miR-155 affects radioresistance of these lung cancer cells. Anti-miR-155 or anti-control molecules were transfected into A549 or H460 cells. As before, cells were treated with varying doses of irradiation at 16 h post-transfection and plated for a clonogenic assay. In concordance with our previous results, we found that inhibition of miR-155 significantly sensitized both A549 cells (; p = 0.000029) and H460 cells (; p = 0.002) to radiation treatment. These results suggest that miR-155 alone can significantly impact the ability of lung cancer cells to respond to irradiation.
Figure 3. miR-155 modulates irradiation response in lung cancer cells. (A) Survival curve comparing pre-miR-155 or pre-control treated A549 cells upon exposure to the given doses of irradiation. (B) Survival curve comparing pre-miR-155 or pre-control treated H460 cells upon exposure to the given doses of irradiation. (C) Survival curve comparing anti-miR-155 or anti-control treated A549 cells upon exposure to the given doses of irradiation. (D) Survival curve comparing anti-miR-155 or anti-control treated H460 cells upon exposure to the given doses of irradiation.
Inhibition of miR-155 radiosensitizes hypoxic lung cancer cells
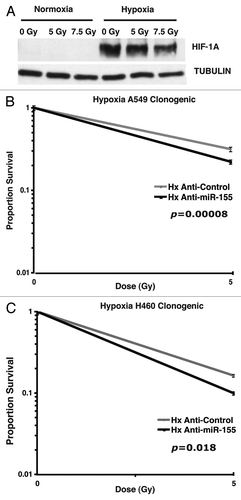
Given that tumor microenvironments are characterized by hypoxia and that hypoxia confers radioresistant properties to cancer cells, we tested the therapeutic effect of miR-155 inhibition combined with irradiation in hypoxic lung cancer cells. A549 and H460 cells were transfected with synthetic anti-miR-155 molecules and anti-control molecules. After 4 h, cells were transferred to mobile irradiation-permissive hypoxia chambers and incubated for 16 h. The cells were then treated with varying doses of irradiation while being maintained in hypoxic conditions. We found that incubation of cells in the mobile irradiation-permissive hypoxia chambers was sufficient to induce hypoxia-inducible factor 1 α (HIF-1α) (), confirming the effective establishment of hypoxia. After irradiation, cells were plated for a clonogenic assay based on colony formation in normoxic conditions as described previously. We found that inhibition of miR-155 significantly radiosensitized hypoxic A549 cells (; p = 0.00008) and hypoxic H460 cells (; p = 0.018). Since hypoxic tumor microenvironments substantially influence radiotherapy in lung cancer patients, these results support miR-155 inhibition as a therapeutically promising approach when combined with radiotherapy.
Figure 4. Inhibition of miR-155 radiosensitizes hypoxic lung cancer cells. (A) Protein gel blot showing HIF-1α induction in cells incubated in mobile irradiation-permissive hypoxia chambers. (B) Survival curve showing anti-miR-155 treatment compared with anti-control treatment in hypoxic A549 cells at the given doses of irradiation. (C) Survival curve showing anti-miR-155 treatment compared with anti-control treatment in hypoxic H460 cells at the given doses of irradiation.
Discussion
As tumor cells rapidly proliferate, they outgrow their blood supply and thus forfeit their normal access to oxygen; this creates an hypoxic tumor microenvironment. It is well established that hypoxia substantially affects tumor cells ability to respond to irradiation. Given that radiotherapy is one of the primary modalities of cancer treatment and that more than 50% of lung cancer patients eventually receive irradiation,Citation24 this contributes to the poor overall survival of lung cancer patients, and lung cancer remains the leading cause of cancer deaths worldwide.
miR-155 has emerged as an important miRNA involved in lung cancer as its increased expression is significantly correlated with poor prognosis.Citation19,Citation20 Here we present one means by which miR-155 is upregulated in lung cancer cells, namely hypoxia. More specifically, we identify a role for HIF-1α in the induction of miR-155. Our data are further supported by the observation that miR-155 is induced in renal cancer cells lacking functional Von Hippel-Lindau (VHL), and that this induction depends in part on HIF-1α expression.Citation25 Intriguingly, the connection between miR-155 and hypoxia is further supported by the observation that both hypoxia and increased miR-155 are associated with increased mutation rates in cancer cellsCitation7,Citation26 and deregulation of DNA repair pathways such as mismatch repair (MMR).Citation27,Citation28 These results suggest that hypoxia-induced genomic instability may in part be mediated via miR-155 induction. Furthermore, the link between increased miR-155 and reduced survival in lung cancer patients may be partially explained by miR-155-mediated radioresistance, which reduces the effectiveness of radiotherapy. Although miR-155 regulates numerous genes in several pathways, its regulation of cellular senescence and apoptosis via targeting of genes such as FOXO3A and tumor protein 53-induced nuclear protein 1 (TP53INP1) in particular is likely to account for its radio-protective effects in lung cancer cells.Citation22,Citation29,Citation30 As miR-155 continues to emerge as a master gene regulator that promotes tumor cell survival, therapies focused on its inhibition will likely prove to be advantageous for cancer patients.
Materials and Methods
Cells
A549 cells were cultured in F12 media supplemented with 10% fetal bovine serum (FBS); H460 cells were cultured in RPMI media + 10% FBS. Cells were maintained in an incubator at 37 degrees with 5% CO2 according to standard procedures. A549, H460, and MCF7 cells were obtained from ATCC, which authenticates cells using short tandem repeat (STR) profiling, morphological analysis, karyotyping, and isoenzyme analysis.
Clonogenic assays
Clonogenic assays were performed according to standard protocols.Citation31 Briefly, cells were reverse transfected with 50 nM of anti- or pre-RNAs using siPORT NeoFX (Ambion). After 16 h cells were treated with increasing doses of irradiation using an X-RAD 320 Biological Irradiator, plated at different dilutions, and incubated for 2 weeks. Two dilutions were used for every treatment group and each dilution was plated in quadruplicate. Cells were stained with crystal violet (0.5% w/v) in 80% methanol, clonogens were blindly counted, and results were compiled and normalized to the no irradiation treatment groups. Statistical analysis was performed using stratified t-tests based on a two-tailed evaluation of data.
Hypoxia
Cells were incubated at 37 degrees in hypoxic chambers infiltrated with 95% nitrogen, 5% carbon dioxide, < 0.01% oxygen for the indicated amounts of time.
qPCR
RNA was extracted using the mirVana miRNA Isolation kit (Applied Biosystems). cDNA was synthesized from total RNA using the Taqman miRNA High-Capactiy cDNA Reverse Transcription kit (Applied Biosystems) with primers specific to miR-155 or RNU6B, an endogenous control. Quantitative PCR was performed according to the manufacturer’s protocol using the Taqman microRNA PCR system (Applied Biosystems) as previously described.Citation15 Briefly, cDNA was combined with Taqman Universal PCR Master Mix and probes specific for miR-155 or RNU6B (Applied Biosystems). PCR was performed in 96-well optical plates. miRNA Ct values were normalized to RNU6B Ct values and relative expression was calculated using the -ΔΔCt method.
Protein gel blot
Cells were removed from normoxia or hypoxia conditions and immediately placed on ice. After rinsing with phosphate buffered solution (PBS) cells were scraped, collected, and used for protein extraction with AZ lysis buffer. 50 μg total protein was loaded and size fractionated via SDS/PAGE and transferred to a PVDF membrane. Antibodies were as follows: mouse monoclonal anti-HIF-1α (BD Transduction Laboratories; Cat: 610958); mouse monoclonal anti-α-tubulin (B-5–1-2; Sigma-Aldrich); rabbit polyclonal anti-FOXO3a [Abcam; ab55010; mouse monoclonal anti-β-actin (Santa Cruz; sc-47778)].
Inhibition of HIF-1α by shRNA
Short hairpin RNA (shRNA) to HIF-1α (shHIF-1α) was expressed in A549 or MCF7 cells as previously described.Citation15 Briefly, lentiviral infection was used to introduce a control vector or an shHIF-1α in A549 or MCF7 cells. Cells expressing the vectors were identified by GFP and collected for further experiments. A549 or MCF7 cells expressing shHIF-1α or a control were exposed to 48 h of normoxia or hypoxia. RNA and protein were isolated for qPCR and protein gel blot, respectively.
Additional material
Download Zip (1.5 MB)Acknowledgments
The authors thank S. Rockwell, Y. Liu, and J. Mendes for lending us the mobile hypoxia chambers and helping with set-up; S. Nallur and D. Steinmetz for assistance with clonogenic assays; D. Hegan and Y. Lu for their advice and assistance. Z. Yun and Q. Lin for their valuable discussions and sharing of reagents. I.A.B. was the recipient of a Gilliam Fellowship (Howard Hughes Medical Institute) and a Harvey Fellowship (Mustard Seed Foundation). This work was supported by NIH grants (CA131301 to J.B.W. and F.J.S) (R01ES005775, R01CA148996 and P01CA129186 to P.M.G).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Ruan K, Song G, Ouyang G. Role of hypoxia in the hallmarks of human cancer. J Cell Biochem 2009; 107:1053 - 62; http://dx.doi.org/10.1002/jcb.22214; PMID: 19479945
- Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer 2004; 4:437 - 47; http://dx.doi.org/10.1038/nrc1367; PMID: 15170446
- Pouysségur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 2006; 441:437 - 43; http://dx.doi.org/10.1038/nature04871; PMID: 16724055
- Subarsky P, Hill RP. The hypoxic tumour microenvironment and metastatic progression. Clin Exp Metastasis 2003; 20:237 - 50; http://dx.doi.org/10.1023/A:1022939318102; PMID: 12741682
- Durand RE. The influence of microenvironmental factors during cancer therapy. In Vivo 1994; 8:691 - 702; PMID: 7727714
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 1996; 379:88 - 91; http://dx.doi.org/10.1038/379088a0; PMID: 8538748
- Yuan J, Glazer PM. Mutagenesis induced by the tumor microenvironment. Mutat Res 1998; 400:439 - 46; http://dx.doi.org/10.1016/S0027-5107(98)00042-6; PMID: 9685702
- Fyles AW, Milosevic M, Wong R, Kavanagh MC, Pintilie M, Sun A, et al. Oxygenation predicts radiation response and survival in patients with cervix cancer. Radiother Oncol 1998; 48:149 - 56; http://dx.doi.org/10.1016/S0167-8140(98)00044-9; PMID: 9783886
- Nordsmark M, Overgaard M, Overgaard J. Pretreatment oxygenation predicts radiation response in advanced squamous cell carcinoma of the head and neck. Radiother Oncol 1996; 41:31 - 9; PMID: 8961365
- Gray LH, Conger AD, Ebert M, Hornsey S, Scott OC. The concentration of oxygen dissolved in tissues at the time of irradiation as a factor in radiotherapy. Br J Radiol 1953; 26:638 - 48; http://dx.doi.org/10.1259/0007-1285-26-312-638; PMID: 13106296
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Mol Cell Biol 2007; 27:1859 - 67; http://dx.doi.org/10.1128/MCB.01395-06; PMID: 17194750
- Huang X, Le QT, Giaccia AJ. MiR-210–micromanager of the hypoxia pathway. Trends Mol Med 2010; 16:230 - 7; http://dx.doi.org/10.1016/j.molmed.2010.03.004; PMID: 20434954
- Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 2008; 14:1340 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-07-1755; PMID: 18316553
- Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer 2010; 126:73 - 80; http://dx.doi.org/10.1002/ijc.24687; PMID: 19551852
- Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 2009; 69:1221 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-08-2516; PMID: 19141645
- Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest 2010; 120:4141 - 54; http://dx.doi.org/10.1172/JCI42980; PMID: 20972335
- Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, et al. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 2009; 28:2719 - 32; http://dx.doi.org/10.1038/emboj.2009.214; PMID: 19696742
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103:2257 - 61; http://dx.doi.org/10.1073/pnas.0510565103; PMID: 16461460
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006; 9:189 - 98; http://dx.doi.org/10.1016/j.ccr.2006.01.025; PMID: 16530703
- Donnem T, Eklo K, Berg T, Sorbye SW, Lonvik K, Al-Saad S, et al. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J Transl Med 2011; 9:6; http://dx.doi.org/10.1186/1479-5876-9-6; PMID: 21219656
- Graves EE, Maity A, Le QT. The tumor microenvironment in non-small-cell lung cancer. Semin Radiat Oncol 2010; 20:156 - 63; http://dx.doi.org/10.1016/j.semradonc.2010.01.003; PMID: 20685578
- Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 2010; 285:17869 - 79; http://dx.doi.org/10.1074/jbc.M110.101055; PMID: 20371610
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96:857 - 68; http://dx.doi.org/10.1016/S0092-8674(00)80595-4; PMID: 10102273
- Mohiuddin MM, Choi NC. The role of radiation therapy in non-small cell lung cancer. Semin Respir Crit Care Med 2005; 26:278 - 88; http://dx.doi.org/10.1055/s-2005-871986; PMID: 16052429
- Neal CS, Michael MZ, Rawlings LH, Van der Hoek MB, Gleadle JM. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med 2010; 8:64; http://dx.doi.org/10.1186/1741-7015-8-64; PMID: 20964835
- Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci USA 2011; 108:4908 - 13; http://dx.doi.org/10.1073/pnas.1101795108; PMID: 21383199
- Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol 2003; 23:3265 - 73; http://dx.doi.org/10.1128/MCB.23.9.3265-3273.2003; PMID: 12697826
- Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA 2010; 107:6982 - 7; http://dx.doi.org/10.1073/pnas.1002472107; PMID: 20351277
- Wang Y, Scheiber MN, Neumann C, Calin GA, Zhou D. MicroRNA Regulation of Ionizing Radiation-Induced Premature Senescence. Int J Radiat Oncol Biol Phys 2010;
- Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA 2007; 104:16170 - 5; http://dx.doi.org/10.1073/pnas.0703942104; PMID: 17911264
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc 2006; 1:2315 - 9; http://dx.doi.org/10.1038/nprot.2006.339; PMID: 17406473